Abstract
The membrane-embedded domain of the unusual electron transporter DsbD (DsbDβ) uses two redox-active cysteines to catalyze electron transfer between thioredoxin-fold polypeptides on opposite sides of the bacterial cytoplasmic membrane. How the electrons are transferred across the membrane is unknown. Here, we show that DsbDβ displays an inherent functional and structural symmetry: first, the two cysteines of DsbDβ can be alkylated from both the cytoplasm and the periplasm. Second, when the two cysteines are disulfide-bonded, cysteine scanning shows that the C-terminal halves of the cysteine-containing transmembrane segments 1 and 4 are exposed to the aqueous environment while the N-terminal halves are not. Third, proline residues located pseudo-symmetrically around the two cysteines are required for redox activity and accessibility of the cysteines. Fourth, mixed disulfide complexes, apparent intermediates in the electron transfer process, are detected between DsbDβ and thioredoxin molecules on each side of the membrane. We propose a model where the two redox-active cysteines are located at the center of the membrane, accessible on both sides of the membrane to the thioredoxin proteins.
Keywords: disulfide bond, disulfide reductase, DsbD, thioredoxin, transmembrane electron transporter
Introduction
In both prokaryotes and eukaryotes, the assembly of the correct set of structural disulfide bonds in a protein depends on two distinct processes: the formation of disulfide bonds and their rearrangement (isomerization) when incorrect bonds are formed. In bacteria, two separate electron transfer pathways carry out these processes. In the formation step, the protein DsbA transfers its disulfide bond to a substrate protein by removing electrons from pairs of cysteines within that protein (Bardwell et al, 1991; Kadokura et al, 2004). The now-reduced DsbA is regenerated as an active enzyme by the passage of electrons from DsbA to the membrane protein DsbB via redox-active cysteines in both proteins. DsbB, in turn transfers electrons to oxygen via the intermediaries of quinones and cytochromes (Bader et al, 1999).
However, DsbA is relatively indiscriminate in making disulfide bonds in proteins with multiple cysteine residues (Berkmen et al, 2005). This indiscriminacy, which leads to misfolded proteins with non-native disulfide bonds, explains the need for an isomerization step. DsbC, a protein disulfide isomerase, which also exhibits chaperone activity, is thought to recognize proteins that are misfolded because of incorrectly formed disulfide bonds. DsbC may either reduce these bonds, allowing DsbA to reoxidize the protein, or directly promote isomerization of disulfide bonds (Zapun et al, 1995; Rietsch et al, 1996; Joly and Swartz, 1997; Schwaller et al, 2003; Berkmen et al, 2005). Either process could permit the formation of correct disulfide bonds. When DsbC acts as a reductant, its cysteines become joined in a disulfide bond, necessitating a reductive pathway for maintaining the enzyme in the active form. The electrons for this pathway are transferred from thioredoxin-1 (Trx1) in the cytoplasm to the cytoplasmic membrane protein DsbD and thence to the protein disulfide bond isomerase DsbC (Figure 1) (Rietsch et al, 1996, 1997).
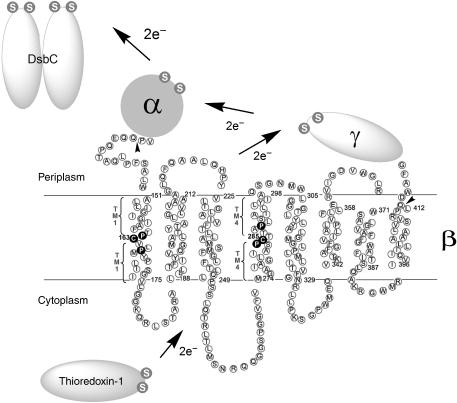
Figure 1.
Electron transfer pathway through transmembrane domain (β) of DsbD and its membrane topology predicted from the primary sequence. The topology of DsbDβ was predicted using HMMTOP (Supplementary data). The essential two cysteines are shown in white in black circles, and are numbered. Studies on the residues in the periplasmic-proximal and cytoplasmic-proximal parts of TM1 and TM4 are shown in Figure 3 and Figure 4, respectively. Studies on the four conserved prolines around the two cysteines, shown in white in black circles, are shown in Figure 5. The essential cysteines in the other domains (α and γ) and interacting proteins (Trx1 and DsbC) are shown in white S (Sulfur of thiol) in gray circles. The tail-less arrows indicate where a signal sequence of DsbD and 3 hemagglutinin (HA) epitopes are fused at the N-terminus of DsbDβ, and a c-Myc epitope is fused at the C-terminus of DsbDβ when DsbDβ was studied as a separate protein.
DsbD is composed of three domains, each containing two redox-active cysteines. All six of these cysteines are required for DsbD activity (Figure 1). The N-terminal periplasmic domain, DsbDα, is the domain that directly reduces DsbC (Katzen and Beckwith, 2000; Goulding et al, 2002; Haebel et al, 2002). DsbDα is then itself reduced by the C-terminal periplasmic domain, DsbDγ, a thioredoxin-like polypeptide (Kim et al, 2003). The resulting oxidized DsbDγ is reduced by the membrane-embedded DsbDβ domain that contains eight transmembrane segments (TMs) (Stewart et al, 1999; Chung et al, 2000; Gordon et al, 2000; Katzen and Beckwith, 2003). Electrons passed from cytoplasmic Trx1 restore DsbDβ to the reduced form, thus allowing it to continue to transfer electrons to DsbDγ.
DsbDβ is a transmembrane two-electron transporter, which appears to require only its two cysteines to mediate electron transfer. In contrast, other examples of transmembrane electron transporters, NADPH oxidase and eukaryotic cytochrome b561 (Saier et al, 2006; http://www.tcdb.org/), are one-electron transporters and contain cofactors, heme or FAD as electron carriers (Bashtovyy et al, 2003; Groemping and Rittinger, 2005).
Much is now understood about the electron transfer steps for both the DsbA/DsbB (Kadokura and Beckwith, 2002; Regeimbal and Bardwell, 2002; Grauschopf et al, 2003; Inaba et al, 2006) and Trx1/DsbD/DsbC pathways (see the above references). However, the mechanism by which DsbDβ is able to transfer electrons across the cytoplasmic membrane via its pair of cysteines (Cys163 and Cys285) has remained particularly elusive. The two cysteines of DsbDβ are predicted to be within TM segments. As an intermediate step in the action of DsbD, the two cysteines form a disulfide bond with each other. Such bonds within membrane proteins have not been seen before (Katzen and Beckwith, 2003). Further, while the cysteines must be close when they are disulfide-bonded, during the electron transfer process, one of the cysteines (Cys163) can form a mixed disulfide with cytoplasmic Trx1, while the other (Cys285) is presumed to interact directly with periplasmic DsbDγ. We have shown that a reverse reaction can occur when DsbDβ is expressed without the periplasmic domains of DsbD in a thioredoxin reductase (trxB) minus background, where the accumulated oxidized Trx1 acts as an oxidant rather than a reductant (Cho and Beckwith, 2006). Cys163 nucleophilically attacks the disulfide bond in Trx1 and the resulting mixed disulfide of DsbDβ(Cys163)-Trx1 is resolved by Cys285 (Katzen and Beckwith, 2000). This reversal of the process generates DsbDβ containing the Cys163–Cys285 disulfide bond (Katzen and Beckwith, 2003).
We previously suggested that the two cysteines of DsbDβ are accessible only to the cytoplasmic side of the membrane (Katzen and Beckwith, 2003). This conclusion was based on the analysis of accessibility of these cysteines to a high-molecular-weight (5 kDa) alkylating agent only from the cytoplasmic side of the membrane.
These previous findings arose the problem of how electrons can be transferred via these cysteines to the periplasmic side of the membrane. Here, we use cysteine-scanning mutagenesis and alkylation methods to resolve this question and others. Our data show that DsbDβ is symmetrical, with the two cysteines accessible to a low-molecular-weight alkylating agent added from either side of the membrane. In addition, we show that the C-terminal halves of the putative TM1 and TM4, which are cytoplasmic-proximal and periplasmic proximal, respectively, are water-exposed when the Cys163 and Cys285 are disulfide-bonded. We report the detection of a previously undetected intermediate, a mixed disulfide complex between DsbDβ and DsbDγ. We propose a model for DsbDβ, where the two cysteines of DsbDβ are water-exposed in the center of the membrane, allowing them to become accessible to both partner thioredoxin-fold proteins (Trx1 from the cytoplasm and DsbDγ from the periplasm). This model permits thiol-disulfide exchange reactions to occur on both sides of the membrane.
Results
Physical location of the two reactive cysteines of DsbDβ
We had previously shown that the two reactive cysteines of DsbDβ are accessible to the high-molecular-weight alkylating agent, methoxypoly(ethylene glycol)-maleimide (malPEG, 5 kDa) from the cytoplasmic side, but not the periplasmic side of the cytoplasmic membrane (Katzen and Beckwith, 2003). Here, we chose to further assess the accessibility of these cysteines by using a lower-molecular-weight (0.5 kDa), membrane-impermeable alkylating agent, 4-acetamido-4′maleimidylstilbene-2,2′-disulfonic acid (AMS) (Nagamori et al, 2002).
In these studies and many others in this paper, we expressed DsbDβ (C-terminally c-Myc tagged) and the two periplasmic domains of DsbD from separate plasmids. We have reported that when the three domains of DsbD are subcloned so that each is expressed from a separate plasmid in the same cell, they reconstitute DsbD activity toward DsbC (Katzen and Beckwith, 2000). Thus, individually expressed DsbDβ is functional and can be used to study the process of electron transfer. To avoid complications in cysteine alkylation studies, we used a version of DsbDβ that has the non-essential Cys282 (Stewart et al, 1999) replaced with alanine and refer to the C282A protein as wild type.
The membrane-impermeable nature of AMS was confirmed by showing that cytoplasmic EF-Tu was only alkylated upon sonication, and not in untreated spheroplasts (Figure 2A and B, compare lanes 1–4 with 5). To test whether the two DsbDβ cysteines are accessible to AMS from the periplasmic side, spheroplasts were treated with AMS. The spheroplast proteins were then denatured and a second alkylating agent, malPEG, was added to determine whether any of the cysteines was still free to be alkylated (Figure 2A). Since malPEG is a 5-kDa alkylating agent, a large shift in band mobility is observed when a cysteine of a protein is alkylated by it. Alkylation of the denatured protein by malPEG in this second step would indicate that the cysteines were not accessible to AMS in spheroplasts in the first step; lack of alkylation would indicate that they were accessible to AMS in spheroplasts. This approach, previously used by Mori et al (2004), is very useful for determining cysteine accessibility, and we refer to it as counter-alkylation.
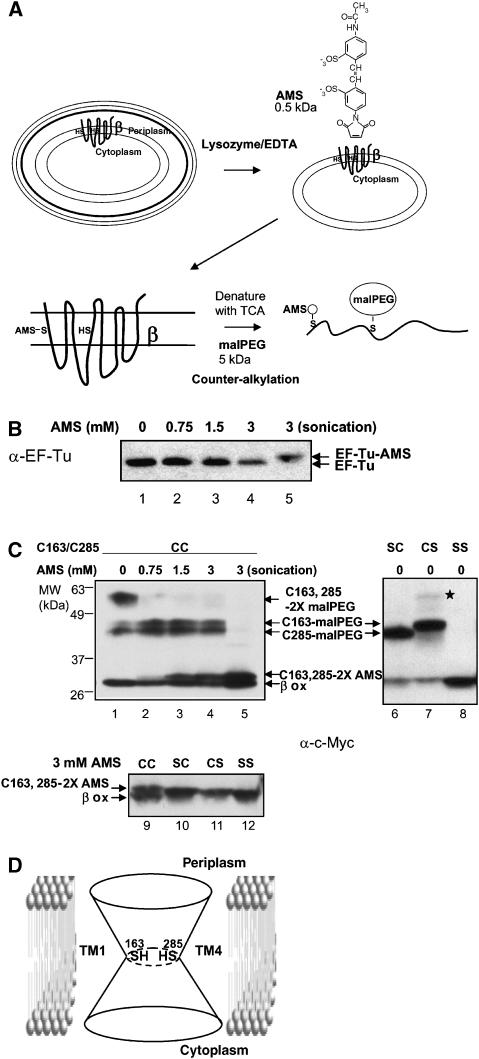
Figure 2.
AMS alkylation and malPEG counter-alkylation of DsbDβ. (A) Experimental scheme. Protein expression was induced by 0.2% arabinose and cells were harvested at mid-log phase, treated with lysozyme and EDTA, and subjected to AMS alkylation. After alkylation, spheroplasts were precipitated with trichloroacetic acid (TCA) (denaturation) and subjected to malPEG alkylation. The cysteines in this figure are chosen arbitrarily for purposes of illustrating AMS and malPEG alkylation. (B) AMS alkylation of EF-Tu. One sample was not treated with AMS (lane 1) and the others were treated with 0.75, 1.5, and 3 mM AMS (lanes 2–4, respectively) without malPEG counter-alkylation. In lane 5, the sample was sonicated before AMS alkylation. Western blotting was performed using an antibody against EF-Tu. (C) AMS alkylation and malPEG counter-alkylation of DsbDβ. Some samples were not treated with AMS (lanes 1 and 6–8), and the others were treated with 0.75 (lane 2), 1.5 (lane 3), and 3 mM AMS (lanes 4 and 5, and 9–12). After AMS treatment, they were counter-alkylated by malPEG. In lane 5, the sample was sonicated before AMS alkylation. The plasmids used were pAP06 (wild-type; CC, lanes 1–5 and 9), pAP06C163S (SC, lanes 6 and 10), pAP06C285S (CS, lanes 7 and 11), and pAP06C163S/C285S (SS, lanes 8 and 12). Western blotting was performed using an antibody against c-Myc. See Results for a discussion of the band marked by a star in lane 7. The strain background used was FED126 (the dsbD mutant). (D) A model of the reduced DsbDβ. The two cysteines are located in the middle of the protein at the juncture of the two cavities, and are exposed to both sides of the membrane, forming an hourglass-like structure.
Without prior incubation with AMS, most of the wild-type DsbDβ, when denatured, was modified with two malPEG molecules, as expected from the presence of two free cysteines (Figure 2C, lane 1). Some of the DsbDβ protein was in the oxidized state and could not be modified by malPEG. Although in vivo DsbDβ is mainly in the reduced state (Cho and Beckwith, 2006), in the experiments described below, of necessity, the DsbDβ was analyzed in non-denaturing conditions rather than after acid denaturation, and thus, the protein became partially oxidized during sample preparation. As expected, mutants of DsbDβ that lacked one or both of the two cysteines (termed CS, SC and SS; S indicates replacement of Cys by Ser) were modified with only one malPEG molecule or not at all (Figure 2C, lanes 6–8). A faint band in the CS mutant (Figure 2C, star indicated in lane 7), which might be thought to represent a doubly malPEG-alkylated species because of its apparent molecular weight, is instead a mixed disulfide between DsbDβ and another protein as it is dithiothreitol (DTT) sensitive, unlike the adducts of a cysteine with an alkylating reagent (data not shown).
When AMS was added to spheroplasts of a strain expressing wild-type DsbDβ, a shifted AMS-alkylated band of DsbDβ was observed, its intensity increasing with increase in concentration of AMS (Figure 2C, lanes 1–4). The following argument indicates that this band is doubly AMS-alkylated. When the wild-type sample prepared after 3 mM AMS alkylation and malPEG counter-alkylation (Figure 2C, lanes 4 and 9) was compared with those of SC, CS, and SS mutants (Figure 2C, lanes 10–12), the upper band from wild type moved more slowly than the bands from mutants, and those from SC and CS mutants moved more slowly than that from the SS mutants. These observations indicate that the upper band from wild type and the bands from SC and CS mutants are doubly AMS-alkylated and singly AMS-alkylated, respectively. The finding that AMS can alkylate both cysteines from the periplasmic side of the membrane contrasts with what was found with the much larger malPEG molecule in analogous experiments.
When alkylation with AMS was followed by denaturation of the AMS-alkylated protein and addition of malPEG, the intensity of a doubly malPEG-alkylated band decreased compared with the sample not treated with AMS. The intensity of singly malPEG-alkylated bands increased at 0.75 mM AMS and remained constant or decreased somewhat above that concentration (Figure 2C, compare lane 1 with 2–4). These results show that one of the cysteines becomes predominantly alkylated by AMS at 0.75 mM, and that at higher concentrations both cysteines begin to be alkylated by AMS (Figure 2C, lanes 2–4). A small amount of singly malPEG-alkylated species was detected even in the absence of AMS (Figure 2C, lane 1), probably due to inefficient alkylation of both cysteines by the bulky malPEG (5 kDa). When the cells were broken to make both sides of the membrane accessible to AMS, all malPEG-alkylated bands, including the singly alkylated band, disappeared and the intensity of a doubly AMS-alkylated band became higher than in the spheroplast sample (Figure 2C, compare lane 5 with 4). These findings are consistent with previous results that malPEG alkylation can occur from the cytoplasmic side. One possible interpretation of these results is that only one of the two cysteines is accessible from the periplasmic side, but that alkylation of one accessible cysteine alters the structure of the protein, so that the second, formerly inaccessible cysteine, can now be alkylated. The following paragraph rules out this explanation.
Interestingly, the two singly malPEG-alkylated bands observed with the CS and SC mutant proteins could be distinguished from each other by their differing mobility on gels (Figure 2C, lanes 6 and 7). In the wild-type protein, these two bands appeared with almost the same intensity (Figure 2C, lanes 1–4), indicating that both cysteines are accessible to AMS from the periplasmic side to approximately the same extent.
These results indicate that the two cysteines are accessible to a low-molecular-weight alkylating agent from both sides of the membrane. An hourglass-like structure for DsbDβ, where both cysteines are at the juncture of the two cavities (Figure 2D), would explain our observations. This structure need not be static, as some motion of the cysteines toward either the periplasmic or cytoplasmic surface of the membrane may be required for them to be accessible to the much larger thioredoxin molecules (see Discussion).
The pseudo-symmetrical arrangement of TM1 and TM4
Sequence alignment indicates that DsbDβ exhibits pseudo-two-fold symmetry, as noted previously (Kimball et al, 2003). Sequence alignment shows that TM1–3 show significant similarity with TM4–6 (Figure 3A). A similar pseudo-symmetry is seen in the DsbDβ homologues, CcdA (Kimball et al, 2003), most of which have six TMs (Katzen et al, 2002). This alignment indicates that the two redox-active cysteines are located in TM1 and TM4, and that there is pseudo-two-fold symmetry between TM1–3 and TM4–6, TMs that are oriented in opposite directions in the membrane. The analysis below reveals important features of this pseudo-symmetry that contribute to an understanding of the mechanism of electron transfer by DsbD.
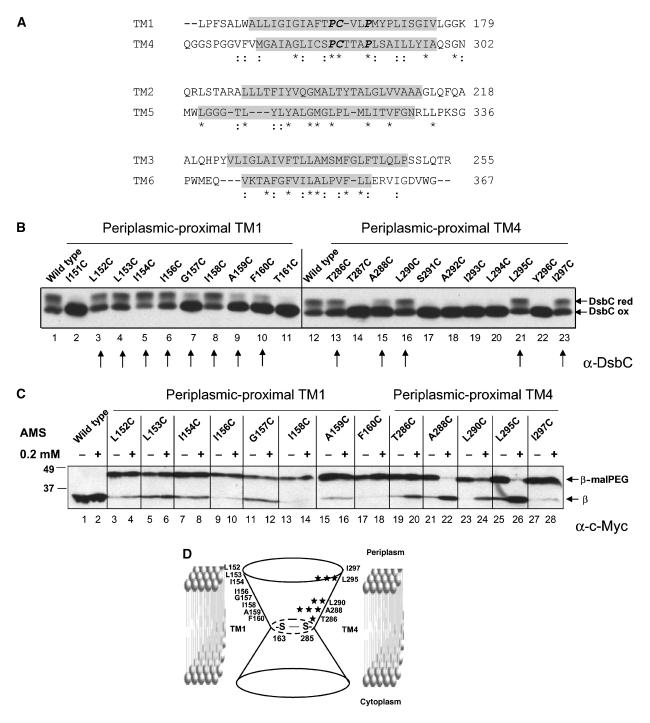
Figure 3.
Sequence alignment of TMs and AMS accessibility to the periplasmic-proximal parts of TM1 and TM4. (A) Amino-acid sequence alignment of the first three TMs with the next three TMs of DsbDβ shows pseudo-two-fold symmetry. The amino-acid sequences containing TM1–3 (residues 144–255) and TM4–6 (residues 264–367) of DsbD were aligned by ClustalW. They share about 18% sequence identity. Similar residues are shown with two dots and identical ones with an asterisk. The four conserved prolines and the two cysteines are shown in bold italic. The predicted TM regions shown in Figure 1 are indicated in gray. (B) The assessment of the in vivo redox states of DsbC in the strains (FED126) expressing the single cysteine-substituted DsbDβs (pAP06 derivatives), DsbDα (pFK051), and DsbDγ (pFK017). Cells were precipitated by TCA and subjected to AMS alkylation. Western blotting was performed using an antibody against DsbC. The arrows indicate the functional cysteine substitutions analyzed in panel C. (C) The assessment of AMS accessibility of single cysteine-substituted DsbDβs (pAP06 derivatives). The samples were analyzed according to the procedure in Figure 2A, except that the strain background was FED513 (the dsbD and trxB double mutant), where oxidized Trx1 is accumulated and Cys163 and Cys285 are disulfide-bonded. In the even-numbered lanes, 0.2 mM AMS was added, while no AMS was added in the odd-numbered lanes, before malPEG counter-alkylation. Western blotting was performed using an antibody against c-Myc. (D) The degree of AMS accessibility to each cysteine-substituted residue in panel C is indicated as the number of stars.
The model for DsbDβ presented in Figure 2D suggests that at least some amino-acid residues in the cysteine-containing TM1 and TM4 should be exposed to the aqueous environment. Because of the ease of analysis using spheroplasts, we first tested the portions of TM1 and TM4 that were predicted to be periplasmic-proximal. We determined which residues are exposed to the aqueous environment by using cysteine-scanning mutagenesis, alkylation, and counter-alkylation, as shown in Figure 2A. The residues to be altered by cysteine-scanning mutagenesis were chosen from the topology prediction in Figure 1.
We analyzed only those residues that, when altered by the cysteine substitutions, resulted in a fully or partially functional DsbDβ. Functionality was assessed by determining how effectively DsbC was reduced when the mutant DsbDβs were expressed along with the separated periplasmic domains of DsbD (α and γ) (Figure 3B, arrows; see Materials and methods for the missing mutants). In considering the in vivo functionality of mutant DsbDβs, we point out that this three-part system itself is not fully functional in the reduction of DsbC, even with wild-type DsbDβ being present (Figure 3B, lanes 1 and 12). In contrast, DsbC is fully reduced in the presence of wild-type, full-length DsbD in vivo (Cho and Beckwith, 2006). Thus, assessing functionality in the three-part system may overestimate the mutational defects of the single-Cys replacements (Figure 3B, lanes 7, 9, 10, and 15), which would likely exhibit greater activity when incorporated into the full-length DsbD.
To examine alkylation of the newly introduced cysteines, it was essential that we prevented alkylation of the two cysteine residues native to DsbDβ. This was accomplished by expressing the mutant DsbDβs in a trxB background, where DsbDβ Cys163 and Cys285 are disulfide-bonded (Katzen and Beckwith, 2003).
Without prior incubation with AMS, no shifts were observed in the oxidized wild-type protein after malPEG alkylation, since the two cysteines are disulfide-bonded. However, from single cysteine-substituted DsbDβs, the newly introduced cysteines were alkylated by malPEG (Figure 3C, odd-numbered lanes). When we repeated these experiments with AMS-treated samples, we observed a striking contrast in results between those obtained with mutants of the periplasmic-proximal portion of TM1 and those of the analogous portion of TM4. None of the single-Cys replacements in the periplasmic-proximal portion of TM1 exhibited accessibility to AMS, while many in TM4 did so (Figure 3C, even-numbered lanes). In TM4, the residues exhibited the following order of accessibility: (A288C and L295C) >L290C>T286C>I297C (Figure 3C, lanes 19–28 and Figure 3D). When these samples were analyzed using broken cells where both sides of the cytoplasmic membrane fragments are accessible, very similar patterns of alkylation with those in Figure 3C were observed (data not shown). These results suggest that when the Cys163 and Cys285 are disulfide-bonded, the periplasmic-proximal portion of TM1 is not water-exposed, surrounded by lipids, by other TMs, and/or loop regions between TMs, while many residues in the periplasmic-proximal portion of TM4 are water-exposed.
Because TM1 and TM4 have high homology in the amino-acid sequences around the two cysteines, but are oriented in opposite directions in the membrane, and the two reactive cysteines have their own redox partners on each side of membrane (Figures 1 and 3A), we considered that the opposite accessibility effects from those described in the previous paragraph might be observed with the cytoplasmic-proximal parts of the protein. Thus, cysteine-scanning mutagenesis and alkylation experiments were performed on the cytoplasmic-proximal portions of these two TMs (Figure 1). As above, we chose those cysteine-scanning mutant DsbDβs that are fully or partially functional in reducing DsbC (Figure 4A, arrows; see Materials and methods for the missing mutants). Before determining the accessibility of the cytoplasmically oriented regions of the TMs, we first wanted to assure that newly introduced cysteines in those regions were not accessible to AMS added to the periplasmic side of the cytoplasmic membrane. We used cells expressing the mutant DsbDβ A288C as a positive control for alkylation from the periplasmic side of the membrane, as it is one of the most accessible residues in the periplasmic-proximal region of TM4 (Figure 3C, lanes 21 and 22). While DsbDβ A288C was well alkylated, none of the single-Cys replacements in the cytoplasmic-proximal regions of the two TMs exhibited accessibility to AMS (Figure 4B, compare lanes 35 and 36 with others). When we then assessed accessibility of these residues in broken cells, the proposed pseudo-symmetrical effects were observed. Most single-Cys replacements in TM4 exhibited very low accessibility to AMS, while many in TM1 exhibited high accessibility to AMS (Figure 4C). The residues in the cytoplasmic-proximal portion of TM1 exhibited the following order of accessibility: (L165C, L170C, I171C, I174C, and V175C) >(M167C and S172C) >V164C (Figure 4C, lanes 3–18; Figure 4D). In analogy with our previous interpretation of such findings, these results suggest that when the Cys163 and Cys285 are disulfide-bonded, the cytoplasmic-proximal portion of TM4 is surrounded by lipids, by other TMs, and/or loop regions between TMs, while many residues in the cytoplasmic-proximal portion of TM1 are water-exposed.
Figure 4.
AMS accessibility to the cytoplasmic-proximal parts of TM1 and TM4. (A, B, D) Same as panels B, C, and D in Figure 3, respectively, except that the cysteine substitutions were generated in the residues of the cytoplasmic-proximal parts of TM1 and TM4. (C) The assessment of AMS accessibility in broken spheroplasts. The same spheroplasts prepared in panel B were broken by sonication. The procedures followed including alkylation are the same as in panel B.
Conserved and pseudo-symmetrically arranged prolines are important for the accessibility to alkylation of cysteines 163 and 285
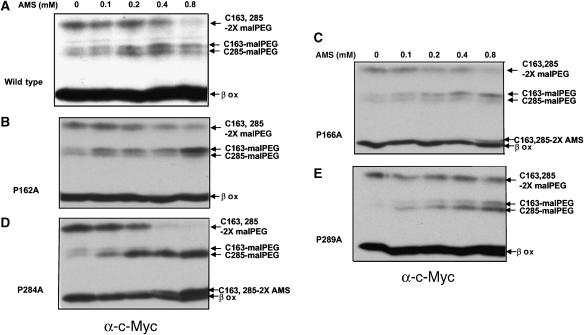
Certain highly conserved proline residues in DsbDβ show pseudo-symmetrical locations around the two cysteines in TM1 and TM4 (Figures 1 and 3A). In previous mutational studies, we found that the proline residues near Cys163 and Cys285 are important for the interaction of DsbDβ with the cytoplasmic Trx1 and the periplasmic DsbDγ (thioredoxin-like), respectively (Cho and Beckwith, 2006). When we assessed the redox states of DsbC from the proline-to-alanine mutants, using the three separately expressed domains of DsbD previously described, P166A, P284A, and P289A mutations caused strong defects, while some reducing activity was observed from P162A mutant (Supplementary Figure 1). We asked whether alterations of these prolines might change the accessibility of the two cysteines to AMS from the periplasmic side, using the AMS alkylation and malPEG counter-alkylation approach, as shown in Figure 2A. Since the two cysteines may be quite close in the three-dimensional structure, we were concerned that the AMS alkylation of one of the cysteines might retard the kinetics of addition of a second AMS. To avoid that possibility, in most tests, we used low AMS concentrations at which no doubly AMS-alkylated band is observed, allowing us to determine whether any of the cysteines has reacted with AMS.
While the degree of AMS alkylation of Cys163 and Cys285 in the wild-type DsbDβ is comparable to that previously shown (Figure 2C, lanes 1–4), the proline mutants result in significant differences in alkylation (Figure 5, compare panel A with B–E). For the DsbDβs that contain the changes P162A and P166A (residues surrounding Cys163), the Cys163 residue becomes less accessible to AMS than the wild type, as shown by the greater degree of alkylation of this residue by malPEG after protein denaturation. In contrast, for P284A and P289A (residues surrounding Cys285), it is the Cys285 that becomes less accessible to AMS than it is in the wild-type protein.
Figure 5.
AMS alkylation of DsbDβ with proline mutations. The samples were analyzed as in Figure 2A and C, except that no AMS, and 0.1, 0.2, 0.4, and 0.8 mM AMS were added to the samples. The plasmids used were pAP06 (A), pAP06P162A (B), pAP06P166A (C), pAP06P284A (D), and pAP06P289A (E).
Detection of the DsbDβ–DsbDγ mixed disulfide complex
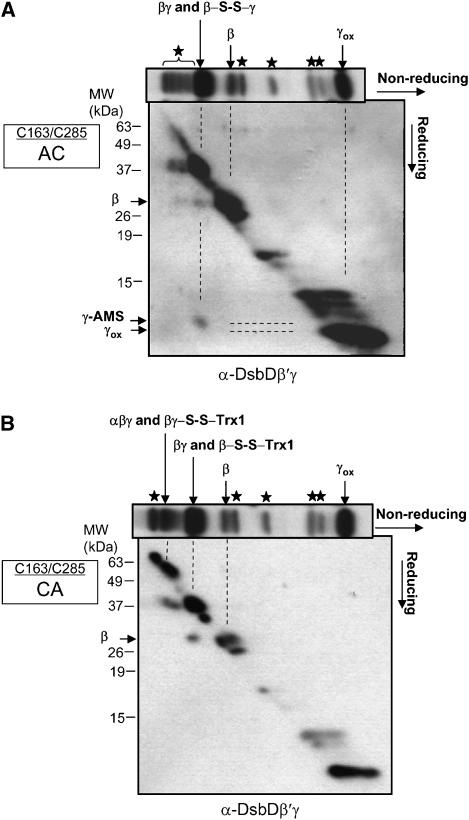
We have previously detected in vivo a Trx1–DsbDβ mixed disulfide complex, a presumed intermediate in the transfer of electrons by DsbD (Katzen and Beckwith, 2000). However, another presumed intermediate, a mixed disulfide complex between DsbDβ and DsbDγ has not been detected. The finding of such a complex would strengthen the importance of the pseudo-symmetrical structure of DsbDβ, in which this domain must interact directly with thioredoxin proteins on both sides of the membrane. To analyze the potential complexes, we used a version of DsbD, DsbDTH, in which we could proteolytically cleave the protein into its sub-domains after making cell extracts. DsbDTH contains thrombin cleavage sites between DsbD domains and a c-Myc epitope before the β domain to allow immunological detection of DsbDβ (Cho and Beckwith, 2006). After thrombin cleavage, a complex where two domains are covalently joined by a disulfide bond should remain intact and only be converted to its component polypeptides after addition of a reducing agent. Detecting such complexes is often facilitated by mutationally eliminating one of the two cysteines involved in the protein's redox reactions, in this way preventing resolution of such mixed disulfide by that cysteine. Thus, to enhance detection of a DsbDβ–DsbDγ complex, we introduced mutations resulting in either a C163A or C285A change into DsbDTH.
During the course of these experiments, we discovered that, because of the way in which the anti-DsbDγ sera were obtained, they contained antibodies both to DsbDγ and to a C-terminal portion of the DsbDβ generated by thrombin cleavage. We refer to this antibody as anti-DsbDβ′γ antibody (see Materials and methods). To overcome the problems generated (see Materials and methods), for the detection of the complex and its reduced products, we performed diagonal two-dimensional gel analyses, where thrombin-treated extracts were run in the first dimension under non-reducing conditions, and the second dimension was run after reduction of the first gel. This approach allows to identify mixed disulfide complexes by reducing these complexes into their components, which then run faster in the second dimension. In our experiments, it was impossible to obtain complete cleavage with thrombin, so that we still saw some amount of intact uncleaved DsbD and uncleaved DsbDβγ on gels. In addition, there were a few contaminating bands that are not mixed disulfide complexes, since they did not alter in mobility after reduction.
In extracts of the C163A (AC) mutant, using anti-DsbDβ′γ antibody (see above), we detected both DsbDβ and DsbDγ being released after reduction from a band in the first dimension that corresponds in its 37-kDa molecular weight to a DsbDβ–DsbDγ complex (Figure 6A), although we could not detect the released DsbDβ using anti-c-Myc antibody, probably due to a small amount of the mixed disulfide complex and lower sensitivity of the antibody (Supplementary Figure 2A). Only DsbDβ was released by reduction of the band in the same position seen in the first dimension from extracts of the C285A (CA) mutant (Figure 6B, and see next paragraph). DsbDγ disulfide-bonded with DsbDβ, should have one free cysteine alkylatable by AMS, resulting in a small shift of the band compared to that of the oxidized form (Figure 6A). These results indicate the existence of a DsbDβ(Cys285)–DsbDγ complex. The detection of this mixed disulfide complex suggests, as predicted, that Cys285 performs the nucleophilic attack on the disulfide bond in DsbDγ.
Figure 6.
Detection of mixed disulfide complexes in C163A and C285A DsbDs. Diagonal two-dimensional analyses were performed on the C163A (AC) (A) and the C285A (CA) sample (B), where the first non-reducing electrophoresis was followed by reduction and electrophoresis in a second dimension. Western blotting was performed using an antibody against DsbDβ′γ (see the text, and Materials and methods). Each non-reducing one-dimensional blot is placed on top of each corresponding two-dimensional blot. Stars indicate contaminating bands that are not mixed disulfide complexes. The plasmids used were pSC51C163A and pSC51C285A, and the strain used was FED126. Cells were precipitated by TCA, and subjected to AMS alkylation and thrombin cleavage.
We also found a mixed disulfide complex of DsbDβ and Trx1 in extracts of the C285A (CA) mutant, using anti-DsbDβ′γ antibody (Figure 6B). The release of the two components of this complex was confirmed by Western blots using anti-c-Myc and anti-Trx1 antibodies (Supplementary Figure 2B and D). This complex presumably represents the Trx1–DsbDβ(Cys163) mixed disulfide seen previously (Katzen and Beckwith, 2000) and it accumulates to levels comparable to those described in that report. No such complex was seen in extracts of the C163A (AC) mutant, as a Western blot with anti-Trx1 antibody showed no release of Trx1 from the complex (Supplementary Figure 2C).
Discussion
DsbD represents a novel class of electron transporters in which no apparent extrinsic factors are required for electron transfer. Only two redox-active cysteines and structural features of the protein are needed to transfer electrons across the membrane. Our results present a potential explanation for this protein's function. We propose that the membrane-embedded domain of DsbD, DsbDβ, interacts in symmetrical manner with structurally near-identical thioredoxin-type proteins on both sides of the membrane. We find that the pseudo-symmetry observed in the amino-acid sequence of DsbDβ extends to the arrangement and water-accessibility of portions of the TM segments of this domain, and to its direct interactions with redox partners.
First, we showed that the two redox-active cysteines of DsbDβ are accessible from both sides of the membrane to the low-molecular-weight (0.5 kDa) alkylating agent AMS. Combined with previous results on accessibility to a higher-molecular-weight alkylating agent, these results indicate (1) that openings in DsbDβ on both sides of the membrane allow accessibility to the two cysteines and (2) that the sizes or specificities of these openings differ.
Second, our cysteine-scanning mutagenesis and counter-alkylation experiments demonstrate the pseudo-symmetrical arrangement of those portions of TM1 and TM4 of DsbDβ that are water-exposed and those portions that are not exposed when the Cys163 and Cys285, are disulfide-bonded. This arrangement is such that the water-exposed portion of TM1 is facing the cytoplasm and that of TM4 is facing the periplasm. These findings are consistent with results showing that cytoplasmic Trx1 reacts with Cys163 (TM1) and periplasmic DsbDγ interacts with Cys285 (TM4). Furthermore, mutations that alter four proline residues, two each of which are pseudo-symmetrically located in TM1 and TM4, make the cysteines of their respective TM less accessible to an alkylating agent. Finally, we have detected a mixed disulfide complex, DsbDβ(Cys285)–DsbDγ, as a likely intermediate in electron transfer. Given that a Trx1–DsbDβ(Cys163) presumed intermediate has been identified previously, the combined findings are consistent with symmetrical interactions of DsbDβ with thioredoxin molecules on both sides of the cytoplasmic membrane.
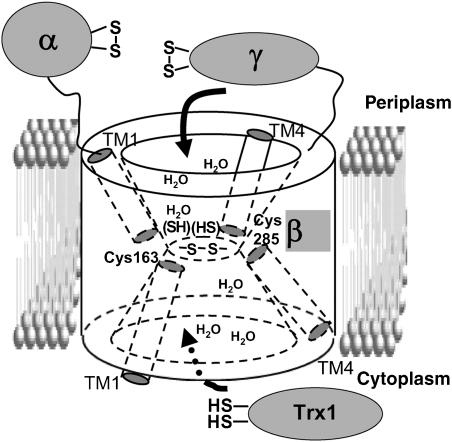
Based on these results, we present a structural model where DsbDβ forms two cavities that are inside the membrane portion of the protein, but are water-exposed to one or the other side of the membrane (Figure 7). The two active site cysteines of DsbDβ are located at the juncture of the two cavities. The cytoplasmic-proximal portion of TM1 and the periplasmic-proximal portion of TM4 form portions of these cavities, as most of the tested residues in them are water-exposed when the Cys163 and Cys285 are disulfide-bonded. In contrast, the periplasmic-proximal portion of TM1 and the cytoplasmic-proximal portion of TM4 are not water-exposed, and therefore, appear to lie outside the cavities. Hydrophilic portions of other TMs or loop regions between TMs may contribute to the water-exposed surface of the cavities.
Figure 7.
Structural model of DsbDβ. The model shows two cavities in DsbDβ that on each side of the membrane, accommodate the appropriate interacting thioredoxin proteins (see Discussion).
An hourglass-like structure containing a constriction in the center of the channel and widening at the membrane surface has been observed in the structures of Aquaporin-1 (Murata et al, 2000), a ClC chloride channel (Dutzler et al, 2002), a protein-conducting channel (Van den Berg et al, 2004), and an ammonia channel (Khademi et al, 2004). The N- and C-terminal halves of the proteins show pseudo-symmetry, but have inverted topologies. In the former two proteins, two pseudo-symmetric but topologically opposite halves of TMs from each half of the protein are situated from the middle of the membrane to the each surface of the membrane and are close each other at their N-termini, and their physical natures are important for the functions of the proteins. The properties of these channels are strikingly similar to the DsbDβ model that we propose. In contrast, major facilitator superfamily transporters such as the lactose permease (Abramson et al, 2003) and the oxalate transporter (Hirai et al, 2003) also show homology between their N- and C-terminal halves, but, in these cases, the two segments are not inverted but assume the same topologies.
Based on these structural models, we propose a mechanism of electron transfer as follows (Figure 7): reduced cytoplasmic Trx1 can interact with the cytoplasmic cavity of DsbDβ, partly by recognition of sequences within the cytoplasmic-proximal portion of TM1. Trx1 then initiates a nucleophilic attack on the disulfide bond in DsbDβ, yielding a mixed disulfide, Cys32(Trx1)–Cys163(DsbDβ), which is resolved by Cys35(Trx1) producing reduced DsbDβ. Analogously, the periplasmic oxidized form of DsbDγ accesses the periplasmic cavity interacting with the periplasmic-proximal portion of TM4 which we propose is water-exposed in the reduced form, the disulfide bond is attacked by Cys285(DsbDβ) nucleophilically, and a mixed disulfide complex is formed. The complex is resolved by Cys163, yielding reduced DsbDγ and a Cys163–Cys285 disulfide bond (DsbDβ).
It is quite possible that the sizes of cavities formed by the 6 TMs (see Results) are not large enough to accommodate fully each partner thioredoxin protein, so that they fail to reach each cysteine. Thus, the two cysteines may not be statically located in the center, but rather can move to approach the periplasmic or cytoplasmic surface of the membrane, allowing each cysteine to be accessible to each partner protein. Although we propose a model that shows openings to both sides of the membrane regardless of redox states, some conformational changes in the proteins during redox reactions could occur, thus affecting the openings to some degree.
We have observed lowered reactivity of Cys163 and Cys285 to an alkylating agent when we introduce amino-acid substitutions for the surrounding prolines. The conserved prolines are important in determining the degree of accessibility of the cysteines, thus, presumably allowing them to be located at the right positions of DsbDβ for interaction with the appropriate thioredoxin proteins (Cho and Beckwith, 2006). Proline residues in the middle of a TM helix can distort geometry of an α-helix, or introduce flexibility into the helix that may result in substantial kinks (Senes et al, 2004). Thus, replacing prolines can have strong effects on the secondary structure of proteins. For example, changes in accessibility of the cysteines near proline-to-alanine mutations could result from altered orientations of them, moving them close to or into the hydrophobic environment (Figure 5B–E). Alternatively, the substitutions may increase the pKa value of the cysteines, making them less reactive. Mutant P284A, which causes a very strong defect in DsbDβ function (Supplementary Figure 1) (Cho and Beckwith, 2006; Hiniker et al, 2006), shows the strongest effect on accessibility (Figure 5D).
The redox-active cysteine pair of DsbDβ, Cys163 and Cys285, behaves quite differently from thioredoxin family members' redox-active cysteines. In thioredoxin-fold proteins, usually the N-terminal cysteine of the Cys-x-x-Cys motif is exposed, and attacks disulfide bonds. The C-terminal cysteine is often buried and resolves the mixed disulfide between thioredoxin-fold proteins and substrates (Kallis and Holmgren, 1980; Nelson and Creighton, 1994). In contrast, each of the two cysteines of DsbDβ can act either as an attacking or a resolving cysteine. When participating in the normal electron transfer reaction of DsbD, Cys285 attacks the disulfide bond in DsbDγ and the resulting mixed disulfide, DsbDβ(Cys285)–DsbDγ, is resolved by Cys163. However, as we describe in the Introduction, under conditions where Trx1 accumulates in the oxidized form, one of the normal DsbD reactions is reversed and oxidized Trx1 acts as an oxidant of DsbDβ (Cho and Beckwith, 2006). In this case, Cys163 nucleophilically attacks the disulfide bond in Trx1 and the resulting mixed disulfide of DsbDβ(Cys163)–Trx1 is resolved by Cys285, thus generating a Cys163–Cys285 disulfide bond in DsbDβ (Katzen and Beckwith, 2000, 2003). We suspect that the bifunctionality of the two cysteines in DsbDβ is related to the water-exposed property of them, and the pseudo-symmetrical organization of the cysteines and the amino acid sequences within which they lie. In particular, each of the cysteines is at the N-terminus of an amino-acid sequence, which is exposed to the aqueous environment, one of the sequences being oriented toward the cytoplasm and one toward the periplasm.
Finally, we point out that this study was performed without purification and separation of DsbD derivatives from their membrane location. These studies should reflect the in vivo arrangement and functioning of these proteins, which might be obscured in vitro by detergent effects or denaturation of the protein during preparation. On the other hand, many of the experiments here are performed with mutant versions of the proteins (e.g., elimination of cysteines or cysteine-scanning), which may alter the normal structure of the proteins. Nevertheless, we believe that the consistency of the results is striking enough to lend credence to our interpretation.
Materials and methods
Strains and media
Strains and plasmids used in this work are listed in Table I. Escherichia coli cells were grown in NZ medium (Rietsch et al, 1996) at 37°C. When needed, 200 μg/ml of ampicillin, 30 μg/ml of chloramphenicol, and 100 μg/ml of spectinomycin were added. Expression of the genes was induced for 1 h by adding 0.2% L-arabinose.
Table 1.
Strains and plasmids used in this work
| Strains or plasmids | Relevant genotype or features | Source or reference |
|---|---|---|
| Strains | ||
| MC1000 | araD139(araABC-leu)7679 galU galK Δ(lac)X74 rpsL thi | Laboratory collection |
| FED126 | MC1000 ΔdsbD | Stewart et al, 1999 |
| FED513 | FED126 trxB::Kan | Katzen and Beckwith, 2000 |
| Plasmids | ||
| pBAD43 | Arabinose regulation, pSC101-based, spectinomycin | Stewart et al, 1999 |
| pBAD18 | Arabinose regulation, pBR based, ampicillin | Guzman et al, 1995 |
| pBAD33 | Arabinose regulation, pACYC184-based, chloramphenicol | Guzman et al, 1995 |
| pFK017 | pBAD43 with APss-DsbDγ | Katzen and Beckwith, 2000 |
| pFK051 | pBAD33 with DsbDα | Katzen and Beckwith, 2000 |
| pAP06 | pBAD18 with DsbDss-3HA-DsbDβ-c-Myc | This work |
| pSC51 | pBAD18 with DsbDTH | Cho and Beckwith, 2006 |
Antibodies
Anti-c-Myc (A-14) rabbit polyclonal antiserum was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Anti-EF-Tu goat polyclonal antiserum was generously provided by Dr David L Miller (Institute for Basic Research in Developmental Disabilities, Staten Island, NY). Curiously, even though anti-DsbDγ antibody was used for the analyses, the 27-kDa band (Figure 6A and B) was confirmed to be DsbDβ, by comparing the Western blots performed using anti-c-Myc antibody to detect DsbDβ from DsbDTH (data not shown). We believe this result is explained by the following: the thrombin cleavage site inserted between DsbDβ and DsbDγ was generated by replacement of the sequence from Leu444 to Ala447 of DsbD (Cho and Beckwith, 2006). However, the anti-DsbDγ antibody was raised in a rabbit using a different version of DsbDγ that started at residue Ala423 (Stewart et al, 1999). Thus, the C-terminal end of DsbDβ that would be generated by thrombin cleavage in DsbDTH was part of the protein used to raise this anti-DsbDγ antibody. We refer to this antibody as anti-DsbDβ′γ antibody.
AMS alkylation and malPEG counter-alkylation of proteins
A 5 ml volume of cells was harvested at mid-log phase, and resuspended in 1 ml of ice-cold buffer containing 50 mM Tris–HCl (pH 8.0), 1 mM CaCl2, 3 mM EDTA, 18% sucrose, and 30 μg/ml lysozyme. Subsequently, various concentrations of AMS (Invitrogen) were added and the samples were incubated for 30 min at 4°C. When indicated, samples were divided into two portions, one to be used for the alkylation experiment using intact spheroplasts, and the other for analysis of broken cell lysates. The latter lysates were prepared by 10-s sonication of the spheroplasts. After AMS treatment, samples were TCA-precipitated, washed with acetone, and resuspended in 50 μl of sodium dodecyl sulfate (SDS)/sample buffer containing 1.7 mM malPEG (Nektar Therapeutics, CA; NOF corporation, Japan). For EF-Tu detection, malPEG was not added. When indicated, 50 mM DTT was added to reduce samples.
Thiol-redox state analyses
To analyze the in vivo redox states of the proteins, free thiols were acid-trapped by TCA and alkylated with AMS, as previously described (Stewart et al, 1999). The single-Cys replacements in the conserved residues Gly155, Pro162, and Pro289 in the periplasmic-proximal parts of TM1 and TM4, caused severe defects in DsbC reduction (data not shown). The single-Ala replacements of these residues in the full-length DsbD in our previous study (Cho and Beckwith, 2006) caused varying defects in DsbC reduction. Given that the changes in some conserved residues cause defects in functionality, for the cytoplasmic-proximal parts of TM1 and TM4, the single-Cys replacements in the conserved residues Pro166, Pro169, and Pro284 were not generated. The single-Ala replacements of these residues in our previous study (Cho and Beckwith, 2006) caused varying defects in DsbC reduction. DsbDTH samples were AMS-alkylated as above and digested with bovine thrombin (Amersham) as previously described (Cho and Beckwith, 2006). When indicated, 50 mM DTT was added to reduce samples. Usually a mixed disulfide complex can be specified by its disappearance after reduction and the consequent appearance of its low-molecular-weight components. However, since in the one-dimensional analysis without reduction (Figure 6A and B), the uncleaved DsbDβγ and the components (DsbDβ and DsbDγ) are detected in significant amounts, we could not demonstrate the presence of the mixed disulfide complex, DsbDβ–DsbDγ, nor could we verify generation of its components after reduction in one-dimensional analysis (data not shown).
Diagonal two-dimensional gel analysis
Samples were run on SDS–PAGE gels under non-reducing conditions and the gels were sliced after running. We reduced the proteins in the sliced gel by incubating it in buffer containing 100 mM DTT and 2% SDS, at 37 °C for 1 h. The consequently reduced first dimension was then placed horizontally on new gels for the second dimensional runs (Sommer and Traut, 1974).
Supplementary Material
Supplementary data
Supplementary Figure 1
Supplementary Figure 2
Acknowledgments
We gratefully acknowledge Federico Katzen for providing many strains and plasmids, and also acknowledge Tom A Rapoport and Hiroshi Kadokura for critical comments on the manuscript. This work was supported by a grant to JB from the National Institutes of Health, GMO41883. JB is an American Cancer Society Professor.
References
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science 301: 610–615 [DOI] [PubMed] [Google Scholar]
- Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC (1999) Oxidative protein folding is driven by the electron transport system. Cell 98: 217–227 [DOI] [PubMed] [Google Scholar]
- Bardwell JC, McGovern K, Beckwith J (1991) Identification of a protein required for disulfide bond formation in vivo. Cell 67: 581–589 [DOI] [PubMed] [Google Scholar]
- Bashtovyy D, Bérczi A, Asard H, Páli T (2003) Structure prediction for the di-heme cytochrome b561 protein family. Protoplasma 221: 31–40 [DOI] [PubMed] [Google Scholar]
- Berkmen M, Boyd D, Beckwith J (2005) The nonconsecutive disulfide bond of Escherichia coli phytase (AppA) renders it dependent on the protein-disulfide isomerase, DsbC. J Biol Chem 280: 11387–11394 [DOI] [PubMed] [Google Scholar]
- Cho S–H, Beckwith J (2006) Mutations of the membrane-bound disulfide reductase DsbD that block electron transfer steps from cytoplasm to periplasm in Escherichia coli. J Bacteriol 188: 5066–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Chen T, Missiakas D (2000) Transfer of electrons across the cytoplasmic membrane by DsbD, a membrane protein involved in thiol-disulphide exchange and protein folding in the bacterial periplasm. Mol Microbiol 35: 1099–1109 [DOI] [PubMed] [Google Scholar]
- Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R (2002) X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415: 287–294 [DOI] [PubMed] [Google Scholar]
- Gordon EH, Page MD, Willis AC, Ferguson SJ (2000) Escherichia coli DipZ: anatomy of a transmembrane protein disulphide reductase in which three pairs of cysteine residues, one in each of three domains, contribute differentially to function. Mol Microbiol 35: 1360–1374 [DOI] [PubMed] [Google Scholar]
- Goulding CW, Sawaya MR, Parseghian A, Lim V, Eisenberg D, Missiakas D (2002) Thiol-disulfide exchange in an immunoglobulin-like fold: structure of the N-terminal domain of DsbD. Biochemistry 41: 6920–6927 [DOI] [PubMed] [Google Scholar]
- Grauschopf U, Fritz A, Glockshuber R (2003) Mechanism of the electron transfer catalyst DsbB from Escherichia coli. EMBO J 22: 3503–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groemping Y, Rittinger K (2005) Activation and assembly of the NADPH oxidase: structural perspective. Biochem J 386: 401–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haebel PW, Goldstone D, Katzen F, Beckwith J, Metcalf P (2002) The disulfide bond isomerase DsbC is activated by an immunoglobulin-fold thiol oxidoreductase: crystal structure of the DsbC–DsbDalpha complex. EMBO J 21: 4774–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiniker A, Vertommen D, Bardwell JC, Collet JF (2006) Evidence for conformational changes within DsbD: possible role for mebrane-embedded proline residues. J Bacteriol 188: 7317–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai T, Heymann JA, Maloney PC, Subramaniam S (2003) Structural model for 12-helix transporters belonging to the major facilitator superfamily. J Bacteriol 185: 1712–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Murakami S, Suzuki M, Nakagawa A, Yamashita E, Okada K, Ito K (2006) Crystal structure of the DsbB–DsbA complex reveals a mechanism of disulfide bond generation. Cell 127: 789–801 [DOI] [PubMed] [Google Scholar]
- Joly JC, Swartz JR (1997) In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry 36: 10067–10072 [DOI] [PubMed] [Google Scholar]
- Kadokura H, Beckwith J (2002) Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J 21: 2354–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Tian H, Zander T, Bardwell JC, Beckwith J (2004) Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 303: 534–537 [DOI] [PubMed] [Google Scholar]
- Kallis GB, Holmgren A (1980) Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J Biol Chem 255: 10261–10265 [PubMed] [Google Scholar]
- Katzen F, Beckwith J (2000) Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell 103: 769–779 [DOI] [PubMed] [Google Scholar]
- Katzen F, Beckwith J (2003) Role and location of the unusual redox-active cysteines in the hydrophobic domain of the transmembrane electron transporter DsbD. Proc Natl Acad Sci USA 100: 10471–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen F, Deshmukh M, Daldal F, Beckwith J (2002) Evolutionary domain fusion expanded the substrate specificity of the transmembrane electron transporter DsbD. EMBO J 21: 3960–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi S, O'Connell J III, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM (2004) Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 305: 1587–1594 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim SJ, Jeong DG, Son JH, Ryu SE (2003) Crystal structure of DsbDgamma reveals the mechanism of redox potential shift and substrate specificity. FEBS Lett 543: 164–169 [DOI] [PubMed] [Google Scholar]
- Kimball RA, Martin L, Saier MH Jr (2003) Reversing transmembrane electron flow: the DsbD and DsbB protein families. J Mol Microbiol Biotechnol 5: 133–149 [DOI] [PubMed] [Google Scholar]
- Mori H, Shimokawa N, Satoh Y, Ito K (2004) Mutational analysis of transmembrane regions 3 and 4 of SecY, a central component of protein translocase. J Bacteriol 186: 3960–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y (2000) Structural determinants of water permeation through aquaporin-1. Nature 407: 599–605 [DOI] [PubMed] [Google Scholar]
- Nagamori S, Nishiyama K–I, Tokuda H (2002) Membrane topology inversion of SecG detected by labeling with a membrane-impermeable sulfhydryl reagent that causes a close association of SecG with SecA. J Biochem 132: 629–634 [DOI] [PubMed] [Google Scholar]
- Nelson JW, Creighton TE (1994) Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry 33: 5974–5983 [DOI] [PubMed] [Google Scholar]
- Regeimbal J, Bardwell JC (2002) DsbB catalyzes disulfide bond formation de novo. J Biol Chem 277: 32706–32713 [DOI] [PubMed] [Google Scholar]
- Rietsch A, Belin D, Martin N, Beckwith J (1996) An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci USA 93: 13048–13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietsch A, Bessette P, Georgiou G, Beckwith J (1997) Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol 179: 6602–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr, Tran CV, Barabote RD (2006) TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res 34 (Database issue): D181–D186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaller M, Wilkinson B, Gilbert HF (2003) Reduction-reoxidation cycles contribute to catalysis of disulfide isomerization by protein-disulfide isomerase. J Biol Chem 278: 7154–7159 [DOI] [PubMed] [Google Scholar]
- Senes A, Engel DE, DeGrado WF (2004) Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol 14: 465–479 [DOI] [PubMed] [Google Scholar]
- Sommer A, Traut RR (1974) Diagonal polyacrylamide-dodecyl sulfate gel electrophoresis for the identification of ribosomal proteins crosslinked with methyl-4-mercaptobutyrimidate. Proc Natl Acad Sci USA 71: 3946–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EJ, Katzen F, Beckwith J (1999) Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. EMBO J 18: 5963–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA (2004) X-ray structure of a protein-conducting channel. Nature 427: 36–44 [DOI] [PubMed] [Google Scholar]
- Zapun A, Missiakas D, Raina S, Creighton TE (1995) Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry 34: 5075–5089 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary Figure 1
Supplementary Figure 2