Abstract
The majority of known bacteriophages have long noncontractile tails (Siphoviridae) that serve as a pipeline for genome delivery into the host cytoplasm. The tail extremity distal from the phage head is an adsorption device that recognises the bacterial receptor at the host cell surface. This interaction generates a signal transmitted to the head that leads to DNA release. We have determined structures of the bacteriophage SPP1 tail before and after DNA ejection. The results reveal extensive structural rearrangements in the internal wall of the tail tube. We propose that the adsorption device–receptor interaction triggers a conformational switch that is propagated as a domino-like cascade along the 1600 Å-long helical tail structure to reach the head-to-tail connector. This leads to opening of the connector culminating in DNA exit from the head into the host cell through the tail tube.
Keywords: bacteriophage SPP1, electron microscopy, Siphoviridae, signal transmission, structural analysis
Introduction
Bacterial viruses (bacteriophages or phages) are the most populated biological entity in the Biosphere (Brüssow and Hendrix, 2002). The vast majority of known bacteriophages have an icosahedral capsid and a tail (order Caudovirales). Bacteriophage tails are nanomolecular machines designed to recognise the host cell surface and deliver the viral genome to the bacterial cytoplasm (Molineux, 2006; Vinga et al, 2006b). Tailed bacteriophages are divided into three families according to their tail morphology: Podoviridae (examples are bacteriophages P22, epsilon15 and phi29), Myoviridae (bacteriophage T4) and Siphoviridae (or Styloviridae, representatives of this family are the bacteriophages λ and SPP1). They have short, long contractile or long non-contractile tails, respectively (Ackermann, 2003). The most detailed structural analysis of bacteriophage tails has been performed on T4. The structure has been obtained for both extended and contracted states (Moody 1973; Amos and Klug, 1975; Lepault and Leonard, 1985; Arisaka et al, 1990; Leiman et al, 2004; Rossmann et al, 2004; Kostyuchenko et al, 2005). Recently, structures of the tail machinery of P22, epsilon 15 and phi29 have been elucidated by cryo-electron microscopy (Tang et al, 2005; Chang et al, 2006; Jiang et al, 2006; Xiang et al, 2006). Although more than 60% of presently known phages belong to the family of Siphoviridae, little is known about the three-dimensional (3D) organisation of non-contractile tails of these phages apart from studies of bacteriophage T5 (Böhm et al, 2001; Effantin et al, 2006). Their long and flexible tails are efficient for the recognition of bacterial host cells. The tail bending that results from its flexibility, varies among individual phages (see e.g. Figure 1 of Tavares et al, 1995) complicating structural analysis of this type of tails.
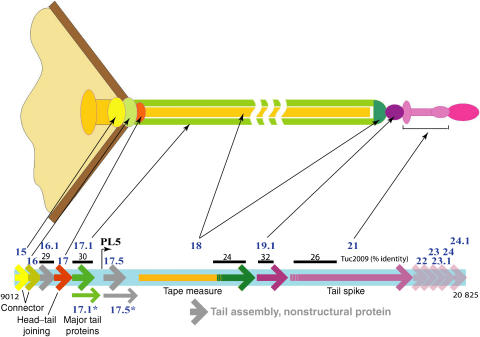
Figure 1.
Bacteriophage SPP1 tail proteins. The location of genes coding for tail proteins is shown in the lower part of the scheme (coordinates 9012–20 825 of the SPP1 nucleotide sequence; access number X97918; Alonso et al, 1997). Genes of proteins (Supplementary Table ST1) with known function or location in the phage structure (indicated by the thin black arrows) are shown as full arrows using the same colour code as in the upper scheme of the phage structure (genes coding for components of the SPP1 tail) or a grey colour (genes coding for nonstructural proteins assisting tail assembly). The gp18 N terminus is shown in gold and its C terminus in dark green to illustrate that these two regions form the tail tube internal tape measure and probably the cap, respectively (see text and Supplementary Table ST1 for details). Proteins with a putative function in tail assembly are represented as semitransparent arrows. Percentage identity between SPP1 proteins (gp16.1 and gp17.1) or segments of those (identified by the dark line above genes coding for gp18, gp19.1 and gp21) and components of the Siphoviridae phage Tuc2009 is also shown.
The long non-contractile tail of bacteriophage SPP1 is formed of an adsorption device (tip) and a tube (Figures 1 and 2A). The tail is attached to the capsid (or head) through the head-to-tail connector, which is composed of gp6, gp15 and gp16 dodecameric rings. The gp16 ring closes the structure to prevent DNA exit from the nucleocapsid (Lurz et al, 2001; Orlova et al, 2003). One end of the double-stranded DNA (dsDNA) coiled within the capsid is attached to the connector, probably bound to gp16 (Tavares et al, 1996). This is the first DNA segment to exit the phage when ejection of the viral genome is triggered (Tavares et al, 1996).
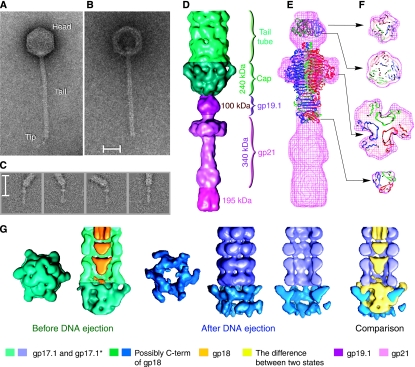
Figure 2.
Structure of bacteriophage SPP1 tail adsorption apparatus composed of the tail tip and cap. (A, B) Phage particles before and after incubation with the viral receptor ectodomain YueB780. Note that emptying of the particles due to DNA release leads to loss of the tail tip and stain accumulation in the centre of the empty capsid. Scale bar corresponds to 300 Å. (C) Image classes showing flexibility at the tail cap–tip interface. (D) Structure of the tail cap (dark green) and tip (variations of rose representing different components identified on the right). (E, F) Side view and cuts through the tip reconstruction with the crystallographic structure of the head binding domain (1LKT) and main domain (1TSP) from bacteriophage P22 tail spike fitted into the EM map. The percent of occupation of the head-binding domain in the EM reconstruction of the tip is of ∼40% for 1LKT. The main domain 1TSP occupies ∼70% of the broad flattened area of the tip and ∼100% in the rod region. The N terminus of 1TSP is anchored in the tip sphere region while the C terminus points towards the rod. (G) Structures of the tail cap before (dark green) and after DNA ejection (dark blue).
The tail tip of SPP1 binds to the Bacillus subtilis receptor YueB, a membrane protein whose extracellular region (ectodomain) crosses the Gram-positive thick cell wall to expose a receptor region at the bacterial surface (São-José et al, 2004, 2006). The SPP1–YueB interaction is the critical event for specific recognition of the host cell surface in vivo that leads to the irreversible commitment of the virus particle to eject its DNA, thus initiating infection (São-José et al, 2004, 2006). This interaction generates a signal that is transmitted along the tail tube to the connector, resulting in the opening of the connector channel allowing DNA to egress from the phage capsid to the bacterial cytoplasm (Tavares et al, 1996; Orlova et al, 2003). This sequence of events was reproduced in vitro by incubation of SPP1 with the dimeric ectodomain YueB780 (São-José et al, 2006) (Figure 2B). To investigate the trigger mechanism of genome expulsion from phage capsids through the tail tube, we determined and compared the structures of the SPP1 non-contractile tail before and after DNA ejection.
Results
Structure of the tail tip
Images of SPP1 bacteriophages that were not treated with receptor showed the presence of a thin appendage, the tip, at the tail end distal from the phage head. The tip is a scavenger device necessary for recognition of the receptor at the bacterial surface and for degradation of the cell wall during phage infection. Analysis of images reveals a hinge connection between the tip and the tail tube, allowing bending angles as high as 50° (Figure 2C). The area of the hinge that closes the tube was named tail cap. The tilting freedom between the tip and the cap likely facilitates recognition and binding to YueB. The flexibility between these two tail components required us to divide the structure into segments corresponding to the tip and to the cap that were treated separately using a single-particle approach.
Images of the tip were extracted from micrographs of SPP1 intact phages. Tips were aligned into a vertical position and the rest of the tail was masked out before orientation determination and 3D reconstruction. The reconstruction with symmetry C3 provided the best match between images and reprojections from the reconstruction (see Materials and methods). The tip is 310 Å long and 95 Å wide at its largest diameter. It can be divided into three, rather compact, main constituents: a sphere-like region, a broad flattened domain and a terminal rod (Figure 2D). The reconstruction shows that it does not have a channel for DNA egress.
The positions of gp19.1 and gp21 in the SPP1 tip (Figure 2D) were derived from the organisation of homologous proteins mapped by immunoelectron microscopy in the tip of the SPP1-related phage Tuc2009 (McGrath et al, 2006; Figure 1 and Supplementary Table ST1). The end of the rod distal from the tail (∼195 kDa) is formed by a yet unidentified protein(s). PSI-BLAST searches showed that the first 400-amino-acid region of the gp21 sequence is homologous to numerous phage host specificity proteins and to putative tail fibre proteins. Prediction of the gp21 beta-strand-rich secondary structure with Jpred and identification of a right-handed beta-helix fold with BetaWrap (Bradley et al, 2001; Supplementary Figure S1) suggested structural similarity between gp21 and the main tail spike domain of bacteriophage P22 (1TSP; Steinbacher et al, 1994). The trimeric P22 tail spike main domain structure fits particularly well into the broadest part and first half of the SPP1 tip rod exhibiting evident three-fold symmetry (Figure 2E and F). Gp21 has twice the mass of the P22 spike main domain. This can be accounted for by some additional density present in the broadest region of the SPP1 tip and by the C-terminal region of a gp21 trimer whose mass accounts for the spike rod region delimited in Figure 2D. The predicted secondary structure of gp19.1 is compatible with the organisation of beta strands found in the head-binding domain of the P22 tail spike (1LKT, Steinbacher et al, 1997). The structure of 1LKT (∼10 kDa × 3), fitted into the density map, occupies only partially the sphere-like region of the tip (Figure 2E and F), whereas three copies of gp19.1 (∼30 kDa × 3) would fully account for the 100 kDa mass of this region.
The tail cap before and after DNA ejection
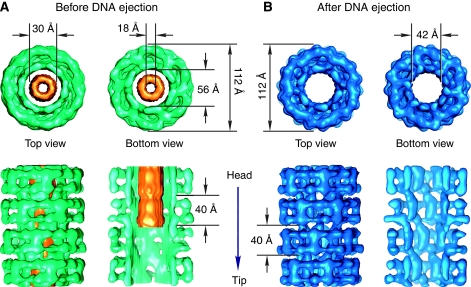
The tail tip is attached to the cap structure that closes the tail tube (Figure 2D). The absence of a channel for DNA traffic in the tip implies that it must dissociate from the cap for DNA passage to the cytoplasm during infection. Binding of YueB780 dimers to SPP1 triggers this necessary step for DNA ejection, leaving the tail tube open (Figure 2B). Reconstructions were performed for two states of the tail: before and after DNA ejection. The cap structure was reconstructed separately from the tip and the main area of the tail. Figure 2G shows reconstructions of the cap together with the first four rings of the tail tube. The tail external diameter (before DNA ejection) tapers from ∼110 to ∼40 Å at the capped extremity and changes symmetry from six-fold to three-fold. This arrangement provides a sturdy interface between the tail tube and the three-fold symmetric tip. Opening of the dome-shaped cap involves loss of the tip and movement of the cap subunits outwards from the tail axis, creating a channel with the same diameter as the inner tail tube. The mass of the cap in the closed and open states (green and blue in Figure 2G, respectively) does not change. The organisation established by immmunoelectron microscopy for phages TP901-1 and Tuc2009 suggests that the cap is formed by a domain of their homologous tape measure protein (gp18 in SPP1) (Figure 1; Vegge et al, 2005; McGrath et al, 2006). If true, gp18 would have to undergo a proteolytic cleavage to separate this domain from its main part that occupies the tail tube interior and is expelled from the tail at the onset of infection (Figure 3), a necessary prelude for DNA transit through the tail. Proteolysis of the tape measure protein was previously described for phages lambda (Tsui and Hendrix, 1983) and Tuc2009 (McGrath et al, 2006). Whereas the tip is apparently responsible for recognition and attack of the target host cell wall and initiation of the signal transmitted through the tail for opening of the head-to-tail connector, the cap structure most likely provides adhesion of the tail to the host membrane.
Figure 3.
Structure of two rings of the SPP1 tail tube in intact phages (green—major tail protein; gold—tape measure protein (A)) and in empty phages that ejected their DNA following incubation with YueB780 (blue—major tail protein (B)). The tape measure protein was removed from the interior of the two bottom rings of the tail tube from DNA-filled phages. This allows the detailed comparison between the major tail protein internal domains organisation before and after DNA ejection.
The helical tail region before DNA ejection
Images of the SPP1 bacteriophage demonstrate that its tail tube can bend. The curvature observed in some regions of the tube and its variation among individual phage particles made it difficult to apply classical approaches used for helical assemblies. Therefore, we extracted small segments of straight regions from the tail tube of a large number of phages. The 3D reconstruction was determined using single-particle analysis. A stack of rings compose the tail tube that provides a flexible link between the receptor recognition apparatus and the capsid containing the viral genome. The tube has a diameter of ∼112 Å and is 1600±35 Å long. This size is typical of the tails of Siphoviridae, which range in length from 1350 to 2600 Å (Katsura and Hendrix, 1984; Pedulla et al, 2003; Vegge et al, 2005; McGrath et al, 2006). The SPP1 tail tube is composed of ∼40 stacked rings, each of 40 Å in height. Each tail ring is rotated by ∼21° relative to the previous one, and comprises six subunits (Figures 3 and 4A). These features were maintained in the tails imaged before and after DNA ejection, respectively (Figure 3). Their structures were determined to ∼14.5 Å resolution (Supplementary Figures S2 and S3) as described in Material and methods.
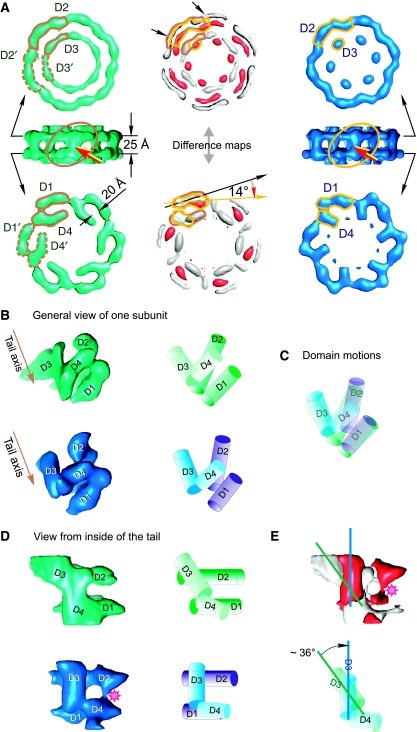
Figure 4.
The tail ring. Conformational states of gp17.1/gp17.1* tail subunits in intact phages (green) and after phage DNA ejection (blue). (A) Comparison of the upper (D2 and D3) and lower domains (D1 and D4) of subunits within the hexameric ring. The domains of one subunit are contoured with brown and gold lines. Changes between the two conformational states are seen on the difference maps shown in white and red (centre). White corresponds to the volume that contains density prior DNA ejection but not after DNA ejection; red corresponds to the volume that contains density only after DNA ejection. Red arrows indicate the link between D1 and D2 domains. (B) General views of the single subunit. The left column shows the subunits from the tip end (tilted view); the right column shows the schematic representation of the four domains. The brown arrow shows the tail axis towards the tip. (C) superposition of schematically represented subunits, revealing the motion of domains. (D) View from the central axis of the tail. Sequence of panels from the left to the right is the same as shown in (B). (E) The upper panel depicts the difference map between the subunits in the left column of (D) using the same colour code as in (A). The pink asterisk indicates the new link between D2 and D4 domains found in tails imaged after DNA ejection. Blue and green lines show the longitudinal axes of the D3 domain at the two conformational states. Superimposed cartoons of D3 and D4 domains indicate the direction of the D3 domain motion. The difference maps are shown at thresholds corresponding to the level of significant variations between the maps (2 σ).
The helical tail of the intact SPP1 phage is composed of two co-axial tubes (Figure 3A). The external tubular structure of the tail delimits a central channel of 56 Å in diameter. The channel is filled by the inner tube of density that has outside and inside diameters of 30 and 18 Å, respectively. This internal tube density is attributed to the tape measure protein gp18 (Figures 1 and 3A). Its featureless continuous appearance in the sixfold averaged reconstruction indicates an organisation, which has neither six-fold symmetry nor any translational periodicity coherent with the tail rings arrangement in the external tube. No connections were found between the structures (Figure 3A). These can be either nonexistent or smeared out by the structural mismatch. Loose contacts between gp18 and the tail channel walls likely facilitate the exit of the tape measure protein that precedes DNA ejection through the tail. The estimated mass of the inner tube is ∼650 kDa, indicating the presence of multiple copies of gp18 subunits.
The outer tube of the tail is formed by stacked rings of six subunits disposed in a helical arrangement with six-fold rotational symmetry (Figure 3 and Supplementary Figure S2). The boundaries of the tail rings were defined by regions of thin connectivity (inter-ring boundaries), whereas a single subunit was defined by areas of low electron density (⩽0.5 σ) between the six identical elements composing each ring (intra-ring subunit boundaries), although it was not always unambiguous. The distribution of density in the adjacent sections was taken in account to delimit the subunit. The volume of one isolated subunit at 1.3 σ threshold is 26 nm3, which corresponds to a protein mass of 21.8 kDa. This mass is in good agreement with the averaged mass of the SPP1 major tail proteins gp17.1 and gp17.1* (∼21.4 kDa=¼(3 × gp17.1 (19.1 kDa)+gp17.1* (28.2 kDa))) that are present at a ratio of ∼3:1 in the structure (Dröge, 1998). gp17.1* is generated by a +1 programmed translational frameshift from the coding frame of gp17.1 (Dröge, 1998; Fraser et al, 2006). We were not able to distinguish these two protein forms in the structures obtained due to averaging of data during processing and the final resolution of the structures. The gp17.1/gp17.1* subunit can be subdivided roughly into four domains of elongated shape that are linked by thin connections. Domains D1 and D2 form the external surface of the ring, whereas domains D3 and D4 line the interior side (Figure 4). The outside domains are arranged in the plane perpendicular to the main axis of the tail. Outer and inner domains are connected through domains D1 and D4 (Figure 4A and B). D2 and D3 do not appear to interact, but establish D2–D2′ and D3–D3′ contacts between subunits within one ring. D1 and D4 provide additional intersubunit connections (Figure 4A). The tail is held together only by connections between the inner domains (D3 and D4) of adjacent rings (Figures 3 and 5).
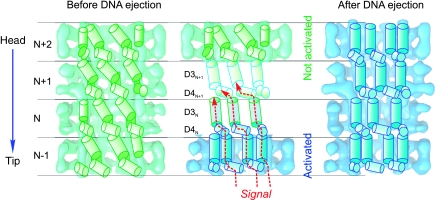
Figure 5.
Model for signal transmission through the internal wall of the tail helical lattice. A cartoon of the helical array formed by the internal domains D3 and D4 is superimposed on cut-open structures of four rings of the tail before (green) and after (blue) DNA ejection. The rings are labelled from N-1 (towards the tip end) through to N+2 (towards the head end). Domains D3 and D4 are shown as cylinders and are at the same level in all figures. The domains in the tail before ejection (left) undergo rearrangements (middle) to form the structure after DNA ejection (right). A central diagram illustrates the mechanism of signal transduction through adjacent rings as a domino-type cascade through the tail towards the head. The tape measure protein density was removed computationally from the tail of intact phages to allow visualisation of the internal tail tube walls.
The helical tail region after DNA ejection
Binding of YueB780 triggers a signal communicated along the tail for DNA release from SPP1 virions. This interaction mimics the situation in vivo at the beginning of viral infection. Empty capsids, characterised by accumulation of stain in their interior, identify the SPP1 particles whose tails have transmitted the signal (São-José et al, 2006) (Figure 2B). Their reconstruction confirmed that ejection of DNA that follows the release of the tape measure protein, leaves the tail channel empty (Figure 3B). The external dimensions and helical features of the tail outer tube were maintained after SPP1 genome exit. In contrast, the internal channel diameter is reduced to ∼42 Å due to major rearrangements in the inner domains of gp17.1/gp17.1*. As random selection of SPP1 tail segments were used for processing, the results show that the helical tail tube adopts one dominant conformation throughout its full length at each of the two states analysed in this work. Our reconstructions demonstrate their changed organisation.
Comparison of the tail tube structure before and after DNA ejection
To best appraise changes between tail structures, we normalised their densities and calculated difference maps. Alignment of subunits of the two reconstructions shows that domains D1 and D4 rotate 14° clockwise towards the tail axis, whereas the D2 domain bends slightly towards the tail axis (Figure 4A). Significantly, more extensive domain motions occur in the inner side of the ring revealing that D3 and D4 have undergone a series of interactions analogous to a domino-like cascade. Domain D3 tilts to a vertical orientation making a side link with the D4 domain of the same subunit (Figure 4B–D). The D3 domain also moves ∼6 Å inwards (inner red blobs located in the upper part of the ring shown in Figure 4A) reducing the diameter of the central channel by 25%. This rearrangement is accompanied by a change of domain D4 from a stretched shape to a more curved one and a new contact with domain D2 (shown by the asterisk in Figure 4D and E).
The structural rearrangements observed in the major tail protein suggests a sequence of interactions that provide a path for directional signal transduction from the tail tip to the head to trigger DNA release (Figure 5). The change in orientation of domain D4 causes a reorientation of domain D3 within the same ring (Figure 5, domains D4N and D3N). Then domain D3 induces a change in orientation in domain D4 in the adjacent ring in the direction of the phage head (Figure 5, domains D3N and D4N+1). This process of interactions continues as a domino-type cascade through the tail towards the head until all domains are in the new conformation (Figure 5, right panel) and the DNA can be ejected. Changes in D3 and D4 do not lead to major variations in their mass, suggesting that the structural rearrangement involves essentially interdomain motions (Figure 4).
Discussion
A key event in infection of bacteria that follows bacteriophage binding to the host cell receptor is communication of a signal across the tail structure to trigger release of DNA from the viral head. Interaction of the SPP1 virus with its active receptor allowed us to mimic this process in vitro. The reconstructions reported here show the initial and the final conformation states of the phage tail providing insight on structural changes that it undergoes for DNA ejection. The configuration of inter-ring bonding along the whole tail changes (Figures 3, 4 and 5). The rotation of internal domains makes the central channel narrower by 25%, but still wide enough to allow for dsDNA to freely slide through the channel (Figure 3B). These domains form a new system of links between the rings through the complete tail tube (Figure 5) providing compelling support for a mechanism in which signal transmission from the tail tip to the connector is achieved by a sequential structural rearrangement of the subunits forming the helical tail tube. The symmetrical nature of tail rings implies that a concerted conformational change of the six subunits acts synchronously on the adjacent ring ensuring a highly reliable ‘six-channel' parallel pathway for signal transmission through the 1600 Å-long structure (Figure 5).
The structural data show that the tail tip does not have a channel for DNA egress and that the signal initiated by interaction of the tip with the bacterial receptor causes release of the tip from the tail cap. This release might trigger the conformational switch, which opens the cap (Figure 2G) and promotes a motion of the major tail protein inner domains. The motion propagates along the stacked tail rings in a domino-cascade manner (Figure 5). The signal reaches the head-to-tail connector, which then becomes open for the DNA outflow from the capsid into the host cell through the tail tube. The conformational change propagating through the internal wall of the tail tube might also serve to promote release of the internal tape measure, which precedes DNA exit. The present data do not contradict the possibility that a change in the contacts between the major tail protein and the tape measure protein throughout the complete tail length cause release of the tape measure, which promotes opening of the connector for DNA exit. The sequence of molecular events described fits well with the process of DNA ejection in vitro. However, during infection of bacteria these steps must be strictly coordinated with the phage attack on the cell wall and its approach to the host membrane. After SPP1 binding to YueB, DNA ejection and dissociation of the tail tip cannot occur until appropriate conditions are reached for efficient traffic of DNA to the bacterial cytoplasm. This requires degradation of the cell wall (probably the tail tip action), attachment to the host cell membrane and creation and use of a hydrophilic pore for DNA to transit efficiently to the cytoplasm (Letellier et al, 2004; Vinga et al, 2006a, 2006b). Otherwise, uncontrolled loss of the tail tip would lead to disruption of the contact with the bacterial cell wall or to abortive ejection of DNA (cf. Vinga et al, 2006a).The temporal programme of events is likely orchestrated to a significant extent by the tail tip whose interaction with the receptor is anticipated to modulate the tip scavenger activity and to initiate signalling.
The molecular strategy of signal transduction leading to viral DNA release found here is most likely shared by the large group of Siphoviridae phages and also by the Myoviridae (Lepault and Leonard, 1985; Rossmann et al, 2004; Kostyuchenko et al, 2005) whose tail sheath surrounds an internal tube resembling the structural organisation of the noncontractile tail. In the well-characterised contractile tail of the Myoviridae phage T4, the internal tube is composed of protein gp19 organised in a six-fold symmetric helical arrangement around the tape measure gp29 (Abuladze et al, 1994; Rossmann et al, 2004; Kostyuchenko et al, 2005). Evolutionarily, the tail organisation found for phages SPP1 and lambda typifies what could have been a primordial long tail structure. Its elaboration with a baseplate originated Siphoviridae such as the SPP1-related phages Tuc2009 and TP901-1 (Vegge et al, 2005; McGrath et al, 2006) and it is tempting to speculate that acquisition of a contractile tail sheath yielded Myoviridae.
The mechanism of signal transduction along the phage helical tail lattice may apply also to other hollow helical structures in response to stimuli. An appealing analogy is the needle of type-III secretion systems. These structures found at the surface of pathogenic bacteria are about 10 nm thick. Like phage tails, they have an internal channel 2–3 nm wide organised around a tape measure protein (Journet et al, 2003). On contact with a target eukaryotic cell, a signal is transmitted through the helical needle structure to activate the type-III secretion machinery that is embedded in the bacterial membrane (Cordes et al, 2003). Protein virulence factors then transit through the hollow needle to enter the mammalian cell cytoplasm, very much like phage DNA traffics through the tail tube to access the host cytoplasm.
Materials and methods
DNA sequencing and bioinformatics
The SPP1 genome region coding for tail proteins (genes 16.1 to 24.1; Figure 1) was sequenced at the DNA sequencing facility of CGM (Gif-sur-Yvette). The protein homology search was carried out using BLAST and PSI-BLAST.
Biochemistry
SPP1 wild-type bacteriophages were purified by isopycnic centrifugation in caesium chloride step gradients, dialysed against 0.5 × TBT buffer (50 mM Tris–Cl (pH 7.5), 50 mM NaCl, 5 mM MgCl2), and further purified by FPLC in a Resource Q column eluted with a 50–500 mM NaCl gradient in phage buffer. Phages were maintained in 0.5 × TBT buffer.
The ectodomain of the SPP1 bacterial receptor YueB780 was purified as described previously (São-José et al, 2006) and run through a Superose 6 size-exclusion column to obtain a homogeneous population of dimers. For DNA ejection reactions, pure SPP1 phages (∼1012 plaque forming units (pfu)) were mixed with YueB780 dimers at a stoichiometric ratio of 1:100 in ejection buffer (50 mM Tris–Cl (pH 7.5), 300 mM NaCl, 5 mM MgCl2) and incubated for 1 h at 37°C. This procedure leads to DNA release from >95% of phage particles (São-José et al, 2006; Vinga et al, 2006a). Phages were diluted to reduce the salt concentration and prepared for EM.
Electron microscopy
Imaging of tails: 3.5 μl of sample was applied onto carbon-coated copper grids, which had been glow-discharged for 30 s. The sample was left for 1–2 min on the grids before blotting and staining with a solution of 2% uranyl acetate. Images were collected with low electron dose on a Tecnai F20 FEG microscope operating at a voltage of 200 kV, a calibrated magnification of 50 000 and a defocus range of 200–1200 Å. These images were also used for analysis of tail ends. Images for analysis of tips were collected with low electron dose on a Tecnai T12 microscope operating at a voltage of 120 kV and a calibrated magnification of 42 000. All micrographs were recorded on Kodak SO-163 films (Supplementary Table ST2).
Image analysis
All micrographs were checked on an optical diffractometer for drift and astigmatism. Suitable micrographs were digitised on a Zeiss SCAI scanner with a step size of 14 μm (2.8 Å/pixel at the specimen level) for the tail. Images for the analysis of the tips were digitised with a final step of 3.33 Å/pixel at the specimen level. The defocus values of all images were determined with the MRC program CTFFIND3 (Mindell and Grigorieff, 2003). Correction for the effect of the contrast transfer function was performed by phase flipping. Image analysis was performed using IMAGIC-5 (van Heel et al, 1996) and SPIDER (Frank et al, 1996) software packages. The resolution of the EM maps was determined by Fourier shell correlation (van Heel and Harauz, 1986) (Supplementary Figure S3).
Image analysis of the tip and cap regions before DNA ejection
In total, 364 images of clear tail ends with tip were extracted from 30 micrographs of intact phages and aligned so that the tips were vertical and the tail then masked out. Multireference alignment was then performed in IMAGIC-5, allowing rotations in the range of ±20°. An initial 3D reconstruction was obtained using a single class with an Euler angle of (0,90,0) assigned to it and applying symmetry C3. After masking, this model was used in an anchor set refinement of all images using three- or six-fold rotational symmetries. Refinement was carried out using anchor set refinement interspersed with rounds of multireference alignment in IMAGIC-5. Because of the small number of images found on the micrographs, we did not perform the classification on final steps of refinement and individual images were used for the reconstruction. There were some images where the tip and tail were coaxial, so these were also included in the 3D reconstruction using Euler angles assigned when the tail was masked out to allow for a later fit with the tail. On average, at evaluation of similarity between images and reprojections, the errors for three-fold symmetry were 20% lower than for six-fold symmetry. Therefore, we have used three-fold symmetry at the final analysis.
For analysis of the tail tube extremity and cap, the tail parts of the images were aligned so that the tail was vertical, the tip part masked out and then the tail aligned rotationally using a small angular range. Classification of images was performed by multivariate statistical analysis (MSA) (van Heel, 1984). In some classes, the tip and tail extremity were straight and these were used to obtain an initial reconstruction for the tail part alone. Angles were assigned only to the tail of these classes. This initial reconstruction was then used for refinement of angles for all classes without the tip part. 3D reconstructions were carried out using C3 and C6 symmetry. Later refinement was performed on single images. Sections of the C3 map were inspected to find where the C6 symmetry characteristic of the helical tail begins.
Image analysis of the cap region after DNA ejection
938 images of tail ends without tip were extracted from a total of 80 micrographs of phages pre-incubated with YueB780 and iteratively aligned until the tail axes were vertical. The data set was processed as described earlier. Classification was performed on individual aligned images. An initial 3D reconstruction was obtained in IMAGIC-5 using selected classes with initial Euler angles of (0,90,0) using symmetry C6. After masking, the averaged model was used in an anchor set refinement of all images using three- or six-fold rotational symmetries. Following Euler angle assignment by projection matching in SPIDER, a final reconstruction was generated with imposed three-fold rotational symmetry.
Image analysis of the tail helical region
As the main population of the tails observed on the micrographs are bent, preventing the use of classical methods that are often used to study objects with helical symmetry (Carragher et al, 1996), a single-particle approach for the analysis of images was adopted. From 15 micrographs ∼7500 segments of tail images, within frames of the size 64 × 64 pixels, from the images of the intact phages were selected and ∼4500 from 12 negatives of the sample incubated with the receptor ectodomain YueB780. Segments were selected along the straight regions of the tail tubes from a large number of phages in each of the two states. The two data sets of images are thus statistically representative of the complete helical tail structure of SPP1 phages before and after DNA ejection. Initial alignment was performed in IMAGIC-5 to bring all images into a vertical orientation. The following iterative procedure of multireference alignment with consequent classification cycles was carried out by combining two software packages: SPIDER for alignment and IMAGIC-5 for MSA and classification of images.
Determination of helical parameters of tail tubes
After bringing all tail segments into the vertical orientation, followed by centring and alignment in the vertical direction, MSA allowed us to obtain several characteristic views that demonstrate clearly that the tails are composed of stacks of rings, with each ring having a height of 4 nm (Supplementary Figure S2A and E). Class averages of the straightest segments with the best contrast were used to calculate Fourier transforms that demonstrated the diffraction pattern characteristic for structures with helical symmetry (Supplementary Figure S2B and F). Indexing of the diffraction pattern gave us estimations of the helical parameters of the tail (Supplementary Figure S2C and G). The layer lines number 5 and 9 have an order of the Bessel function equal to six, which indicates that the helical tail has six-fold symmetry (Moody, 1973). Therefore, each ring also has six-fold rotational symmetry. One ring is ∼40 Å high and is rotated by ∼20° relative to the previous one. Similar procedures have been performed for the phages treated with the receptor ectodomain YueB780. The helical parameters of the YueB780-treated tails were found to have similar values.
Three-dimensional reconstructions of the tail tube
Initial 3D models of the tail in both states were obtained independently using an helical approach. In both cases, the helical parameters of the tail (rise distance h and helical twist Ω from one subunit to the next one) were determined by indexing the diffraction patterns (Moody, 1990) of class averages exhibiting the best vertically aligned tails. Then, a class average with a clear helical diffraction pattern was translationally shifted according to the rise distance of ±h, ±2h and ±3h, so that a set of images was created with the following shifts along Z-axis −3h, −2h, −h, –, h, 2h, 3h. For each copy of the class average, the polar angle (corresponding to the β angle in IMAGIC-5) was set to 90°, and the helical twist with the sequential increment Ω was attributed as the azimuthal angle (corresponding to the γ angle in IMAGIC-5). This set was used as the input later required for the tomographic reconstruction in IMAGIC-5. We used the exact filter back projection approach (Harauz and van Heel, 1986) in all reconstructions described here. Reconstructions were obtained from the four best class averages and then averaged together. This model, after averaging according to the helical parameters h and Ω, was used for further refinement. Angular orientation of the classes obtained was determined using the projection matching procedure in SPIDER. The final reconstruction of the tail not treated with the receptor YueB780 was obtained from 1700 class averages, whereas for the tails after DNA ejection we used 1000 class averages.
The quality of classes was evaluated according to the level of error given by discrepancy between classes and reprojections (Supplementary Figure S2D and H). We have discarded 30–35% of classes, which had an error at comparison with reprojections above the average level of errors (∼45%). The same criterion was used for reconstructions of tail tubes from nontreated phages and from YueB780-treated phages. Typically, such statistics and the number of images used reflect reconstructions of homogeneous populations of macromolecules within the resolution obtained.
Graphics
Illustrations were generated using PyMOL (DeLano, 2002. http://www.pymol.org). Surface representations (unless stated otherwise) are displayed at a threshold level of 1.1 σ corresponding to ∼100% of the expected mass.
Author information: The 3D density maps are being deposited into the EBI-EMD database for the structures of the tail before and after DNA ejection and the tip. The accession codes will be provided. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to EVO (e.orlova@mail.cryst.bbk.ac.uk).
Supplementary Material
Supplementary Tables and Information
Supplementary Figures S1 and S2
Supplementary Figure S3
Acknowledgments
We are indebted to Mário A Santos for sharing with us his pioneer research on the SPP1 receptor that made the present work possible. We thank Helen Saibil for critically reading the manuscript and for continuous encouragement to the SPP1 project. We acknowledge Anja Dröge and Frank Weise for communication of unpublished results, Henry M Krisch for guidance in T4 evolution and Jaime Mota for discussions on type-III secretion systems. This work was supported by grants from the BBRSC (to EVO), the Human Frontier for CP, the CNRS (ATIP to PT), the FCT (to CS-J) and the exchange program Pessoa (to PT and MA Santos).
References
- Abuladze NK, Gingery M, Tsai J, Eiserling FA (1994) Tail length determination in bacteriophage T4. Virology 199: 301–310 [DOI] [PubMed] [Google Scholar]
- Ackermann HW (2003) Bacteriophage observations and evolution. Res Microbiol 154: 245–251 [DOI] [PubMed] [Google Scholar]
- Alonso JC, Lüder G, Stiege AC, Chai S, Weise F, Trautner TA (1997) The complete nucleotide sequence and functional organization of Bacillus subtilis bacteriophage SPP1. Gene 204: 201–212 [DOI] [PubMed] [Google Scholar]
- Amos L, Klug A (1975) Three-dimensional image reconstructions of the contractile tail of T4 bacteriophage. J Mol Biol 99: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisaka F, Takeda S, Funane K, Nishijima N, Ishii S (1990) Structural studies of the contractile tail sheath protein of bacteriophage T4. 2. Structural analyses of the tail sheath protein, Gp18, by limited proteolysis, immunoblotting, and immunoelectron microscopy. Biochemistry 29: 5057–5062 [DOI] [PubMed] [Google Scholar]
- Böhm J, Lambert O, Frangakis AS, Letellier L, Baumeister W, Rigaud JL (2001) FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr Biol 11: 1168–1175 [DOI] [PubMed] [Google Scholar]
- Bradley P, Cowen L, Menke M, King J, Berger B (2001) BETAWRAP: successful prediction of parallel â-helices from primary sequence reveals an association with many microbial pathogens. Proc Natl Acad Sci USA 98: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H, Hendrix RW (2002) Phage genomics: small is beautiful. Cell 108: 13–16 [DOI] [PubMed] [Google Scholar]
- Carragher B, Whittaker M, Milligan RA (1996) Helical processing using PHOELIX. J Struct Biol 116: 107–112 [DOI] [PubMed] [Google Scholar]
- Chang J, Weigele P, King J, Chiu W, Jiang W (2006) Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14: 1073–1082 [DOI] [PubMed] [Google Scholar]
- Cordes FS, Komoriya K, Larquet E, Yang S, Egelman EH, Blocker A, Lea SM (2003) Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem 278: 17103–17107 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific http://www.pymol.org [Google Scholar]
- Dröge A (1998) Capsidmorphogenese des Bakteriophagen SPP1. PhD thesis. Mensch&Bach Verlag, Berlin, Germany [Google Scholar]
- Effantin G, Boulanger P, Neumann E, Letellier L, Conway JF (2006) Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J Mol Biol 361: 993–1002 [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Fraser JS, Yu Z, Maxwell KL, Davidson AR (2006) Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol 359: 496–507 [DOI] [PubMed] [Google Scholar]
- Harauz G, van Heel M (1986) Exact filters for general geometry three dimensional reconstruction. Optik 73: 146–156 [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W (2006) Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet L, Agrain C, Broz P, Cornelis GR (2003) The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302: 1757–1760 [DOI] [PubMed] [Google Scholar]
- Katsura I, Hendrix RW (1984) Length determination in bacteriophage lambda tails. Cell 39: 691–698 [DOI] [PubMed] [Google Scholar]
- Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG (2005) The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol 12: 810–813 [DOI] [PubMed] [Google Scholar]
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG (2004) Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118: 419–429 [DOI] [PubMed] [Google Scholar]
- Lepault J, Leonard K (1985) Three-dimensional structure of unstained, frozen-hydrated extended tails of bacteriophage T4. J Mol Biol 182: 431–441 [DOI] [PubMed] [Google Scholar]
- Letellier L, Boulanger P, Plançon L, Jacquot P, Santamaria M (2004) Main features on tailed phage, host recognition and DNA uptake. Front Biosci 9: 1228–1239 [DOI] [PubMed] [Google Scholar]
- Lurz R, Orlova EV, Gunther D, Dube P, Droge A, Weise F, van Heel M, Tavares P (2001) Structural organisation of the head-to-tail interface of a bacterial virus. J Mol Biol 310: 1027–1037 [DOI] [PubMed] [Google Scholar]
- McGrath S, Neve H, Seegers JF, Eijlander R, Vegge CS, Brondsted L, Heller KJ, Fitzgerald GF, Vogensen FK, van Sinderen D (2006) Anatomy of a lactococcal phage tail. J Bacteriol 188: 3972–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347 [DOI] [PubMed] [Google Scholar]
- Molineux IJ (2006) Fifty-three years since Hershey and Chase; much ado about pressure but which pressure is it? Virology 344: 221–229 [DOI] [PubMed] [Google Scholar]
- Moody MF (1973) Sheath of bacteriophage T4. 3. Contraction mechanism deduced from partially contracted sheaths. J Mol Biol 80: 613–635 [DOI] [PubMed] [Google Scholar]
- Moody MF (1990) Image analysis of electron micrographs. In Biophysical Electron Microscopy: Basic Concepts and Modern Techniques, Hawkes PW, Valdre U (eds), pp 145–287. New York: Academic Press [Google Scholar]
- Orlova EV, Gowen B, Dröge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P (2003) Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J 22: 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR Jr, Hendrix RW, Hatfull GF (2003) Origins of highly mosaic mycobacteriophage genomes. Cell 113: 171–182 [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG (2004) The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol 14: 171–180 [DOI] [PubMed] [Google Scholar]
- São-José C, Baptista C, Santos MA (2004) Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J Bacteriol 186: 8337–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- São-José C, Lhuillier S, Lurz R, Melki R, Lepault J, Santos MA, Tavares P (2006) The ectodomain of the viral receptor YueB forms a fiber that triggers DNA ejection of bacteriophage SPP1 DNA. J Biol Chem 281: 11464–11470 [DOI] [PubMed] [Google Scholar]
- Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R (1997) Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol 267: 865–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P (1994) Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265: 383–386 [DOI] [PubMed] [Google Scholar]
- Tang L, Marion WR, Cingolani G, Prevelige PE, Johnson JE (2005) Three-dimensional structure of the bacteriophage P22 tail machine. EMBO J 24: 2087–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares P, Dröge A, Lurz R, Graeber I, Orlova EV, Dube P, van Heel M (1995) The SPP1 connection. FEMS Microbiol Rev 17: 47–56 [Google Scholar]
- Tavares P, Lurz R, Stiege A, Rückert B, Trautner TA (1996) Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. J Mol Biol 264: 954–967 [DOI] [PubMed] [Google Scholar]
- Tsui LC, Hendrix RW (1983) Proteolytic processing of phage lambda tail protein gpH: timing of the cleavage. Virology 125: 257–264 [DOI] [PubMed] [Google Scholar]
- van Heel M (1984) Multivariate statistical classification of noisy images (randomly oriented biological macromolecules). Ultramicroscopy 13: 165–183 [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G (1986) Resolution criteria for three-dimensional reconstructions. Optik 73: 119–122 [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- Vegge CS, Brøndsted L, Neve H, McGrath S, van Sinderen D, Vogensen FK (2005) Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1. J Bacteriol 187: 4187–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinga I, Dröge A, Stiege AC, Lurz R, Santos MA, Daugelavièius R, Tavares P (2006a) The minor capsid protein gp7 of bacteriophage SPP1 is required for efficient infection of Bacillus subtilis. Mol Microbiol 61: 1609–1621 [DOI] [PubMed] [Google Scholar]
- Vinga I, São-José C, Tavares P, Santos MA (2006b) Bacteriophage entry in the host cell. In Modern Bacteriophage Biology and Biotechnology, Wegrzyn G (ed), pp 165–203. Kerala, India: Research Signpost [Google Scholar]
- Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG (2006) Structural changes of bacteriophage phi29 upon DNA packaging and release. EMBO J 25: 5229–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables and Information
Supplementary Figures S1 and S2
Supplementary Figure S3