Abstract
Trehalose fulfils a wide variety of functions in cells, acting as a stress protectant, storage carbohydrate and compatible solute. Recent evidence, however, indicates that trehalose metabolism may exert important regulatory roles in the development of multicellular eukaryotes. Here, we show that in the plant pathogenic fungus Magnaporthe grisea trehalose-6-phosphate (T6P) synthase (Tps1) is responsible for regulating the pentose phosphate pathway, intracellular levels of NADPH and fungal virulence. Tps1 integrates glucose-6-phosphate (G6P) metabolism with nitrogen source utilisation, and thereby regulates the activity of nitrate reductase. Activity of Tps1 requires an associated regulator protein Tps3, which is also necessary for pathogenicity. Tps1 controls expression of the nitrogen metabolite repressor gene, NMR1, and is required for expression of virulence-associated genes. Functional analysis of Tps1 indicates that its regulatory functions are associated with binding of G6P, but independent of Tps1 catalytic activity. Taken together, these results demonstrate that Tps1 is a central regulator for integration of carbon and nitrogen metabolism, and plays a pivotal role in the establishment of plant disease.
Keywords: glucose-6-phosphate, nitrate utilisation, phytopathogen, Pyricularia oryzae, rice blast
Introduction
Trehalose (α-D-glucopyranosyl-α-D-glucopyranoside) is a non-reducing sugar that is among the most widespread disaccharides in living cells, occurring in bacteria, plants, insects, some invertebrates and fungi. Trehalose has been implicated in the cellular response to numerous environmental stresses, such as heat-shock, starvation, hyperosmotic shock and desiccation, but it is becoming increasingly clear that trehalose and its immediate precursor, trehalose-6-phosphate (T6P), also regulate growth and development. The absence of T6P synthase (Tps1) in Arabidopsis thaliana, for example, causes embryo lethality (Eastmond et al, 2002; Gómez et al, 2006), while T6P phosphatase is required for inflorescence development in maize (Satoh-Nagasawa et al, 2006). The sites of action of T6P and trehalose in the regulation of growth and development in multicellular eukaryotes, however, are largely unresolved.
Recently, we discovered that Tps1 is essential for establishment of rice blast disease by the filamentous fungus Magnaporthe grisea (Foster et al, 2003). Rice blast is a devastating problem in cultivated rice (for a review see Talbot, 2003) and the fungus infects host plants using specialised infection structures called appressoria, which develop on the rice leaf surface (Veneault-Fourrey et al, 2006). T6P synthase was found to be essential for development of functional appressoria and M. grisea Δtps1 mutants are non-pathogenic (Foster et al, 2003). In addition, Δtps1 mutants sporulated very poorly, suggesting that Tps1 may play a role in two distinct cellular differentiation events in M. grisea—conidiogenesis and appressorium formation (Foster et al, 2003).
Loss of Tps1 in yeasts has previously been shown to have severe effects on sugar metabolism. Deletion of the homologous TPS1 gene in the yeasts Saccharomyces cerevisiae and Kluyveromyces lactis, for instance, prevents their growth in the presence of glucose. This is associated with overactive hexokinase activity, an increased flux of glucose into glycolysis, irreversible and catastrophic depletion of ATP and free inorganic phosphate (Pi), and accumulation of hexose phosphate intermediates (Luyten et al, 1993; Winderickx et al, 1996; Ernandes et al, 1998; Arguelles, 2000; Gancedo and Flores, 2004). In these yeasts, T6P inhibits hexokinase activity and therefore acts as a means of regulating the entry of glucose into glycolysis. Consequently, glucose-containing media are toxic for tps1Δ mutants. In S. cerevisiae tps1Δ mutants, deletion of HXK2, which encodes hexokinase II, remediates the growth defect on glucose, presumably by slowing entry of glucose into glycolysis (Hohmann et al, 1993). Furthermore Tps1, which is part of a multi-enzyme complex with three additional proteins, Tps2, Tps3 and Tsl1 (Reinders et al, 1997; Voit, 2003; Elbein et al, 2003), may act directly on hexokinase, modulating its activity (Bonini et al, 2003). We previously noted that in addition to the developmental phenotypes associated with loss of Tps1 in M. grisea, Δtps1 mutants were also unable to grow on glucose, but unlike the yeast species their growth could be restored by addition of free amino acids to the medium, suggesting that cross-talk between sugar signalling and nitrogen metabolism occurs in the fungus (Foster et al, 2003).
In this study, we set out to determine the role of trehalose biosynthesis in plant infection by M. grisea. We decided to explore, in particular, the relationship between the role of Tps1 in sugar metabolism, and its requirement for fungal pathogenicity. Here, we present evidence to show that the apparent inability of M. grisea Δtps1 to grow on glucose as a sole carbon source is not due to glycolytic misregulation, as predicted by studies in yeast, but is instead due to a role for Tps1 in the control of NADPH levels, via regulation of the oxidative pentose phosphate pathway. Furthermore, Tps1 regulates nitrogen utilisation by controlling NMR1 gene expression and nitrate reductase activity. We also establish that the role of Tps1 in fungal pathogenicity results from regulatory functions of the protein and its control of virulence gene expression, rather than its catalytic function. Taken together, our results demonstrate that the Tps1 protein is a central regulator of infection-related processes in a plant pathogenic fungus.
Results
M. grisea TPS1 restores glycolytic regulation to a S. cerevisiae tps1Δ mutant
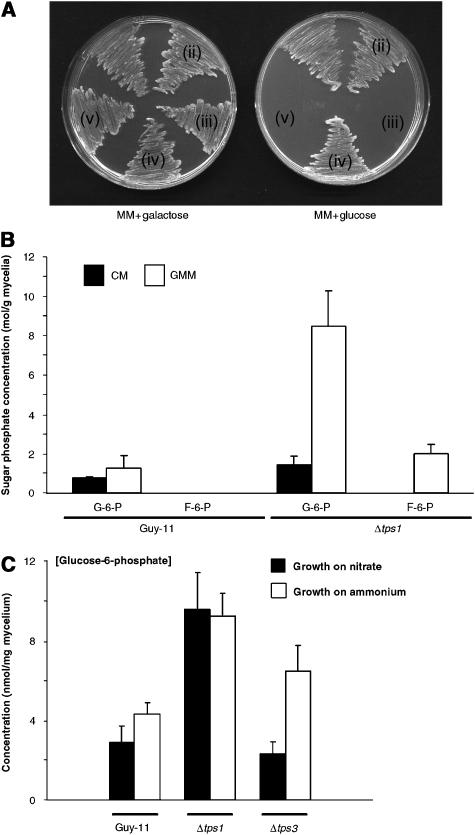
In order to investigate the biological function of Tps1 in M. grisea, we first investigated the functional relationship between M. grisea Tps1 and S. cerevisiae Tps1, where a role in the regulation of glycolysis has been clearly established (Hohmann et al, 1993). The M. grisea TPS1 gene was expressed in S. cerevisiae tps1Δ∷ura3 mutant MB062 under control of the galactose-inducible GAL1 promoter. Transformants were grown on growth medium containing galactose and then replica plated onto medium with glucose as sole carbon source (Figure 1A). All transformed strains, either with or without a functional TPS1 gene, were able to grow on galactose, but only the wild-type W303 strain and the tps1Δ∷ura3:GAL1(p):MgTPS1:URA3 strain expressing M. grisea Tps1 could grow on glucose, when transferred by replica plating. We conclude that expression of M. grisea TPS1 is sufficient to restore glycolytic regulation to a yeast tps1Δ mutant.
Figure 1.
Expression of M. grisea TPS1 in S. cerevisiae and analysis of sugar phosphate metabolism. (A) Complementation of S. cerevisiae tps1Δ strain MB062 with MgTPS1. Yeast strains were plated onto minimal media (MM) with galactose as sole carbon, and compared to growth on glucose yeast medium (GMM). (i, ii) tps1Δ∷ura3:GAL1(p):MgTPS1:URA3 (yeast tps1Δ deletion strain transformed with MgTPS1); (iii) tps1Δ∷ura3:GAL1(p): URA3 (yeast tps1Δ deletion strain transformed with empty vector); (iv) W303 (isogenic strain of S. cerevisiae with functional TPS1 gene); (v) tps1Δ∷ura3 (tps1Δ deletion strain MB062). (B) Sugar phosphate accumulation in mycelium of M. grisea Guy-11 and Δtps1. Mycelium was grown in CM for 48 h before transferring to CM (closed bars) or GMM (open bars) for 24 h. G6P (G-6-P) and fructose-6-phosphate (F-6-P) levels were determined by HPAE-PAD using a PA100 column (Dionex). Error bars represent s.d. of three replications of the experiment.
Characterisation of the relationship between Tps1 and the regulation of glycolysis in M. grisea
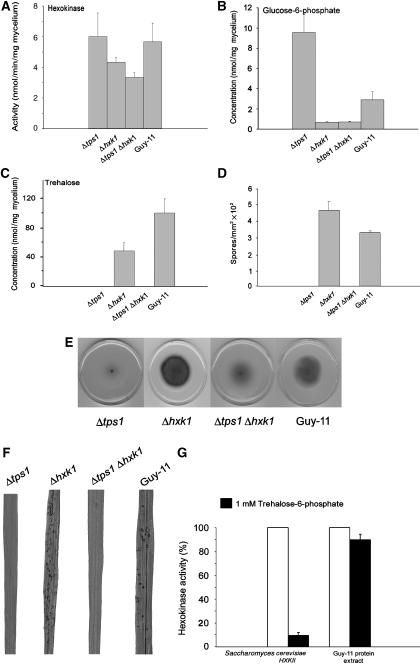
S. cerevisiae tps1Δ mutants accumulate early glycolytic sugar phosphate intermediates when grown with glucose as carbon source, which results from stalled glycolysis (Gancedo and Flores, 2004). We therefore carried out high-performance anion-exchange chromatography with pulsed amperometric detection (HPAE-PAD) to measure sugar phosphate levels in mycelium of a wild-type M. grisea strain Guy-11 and the isogenic Δtps1 mutant (Foster et al, 2003), as shown in Figure 1B. Significantly higher amounts of G6P (P⩽0.01) and fructose-6-phosphate (P⩽0.01) were observed when M. grisea Δtps1 mutants were incubated in glucose minimal medium (GMM), compared to the wild-type strain Guy-11, consistent with misregulation of glycolysis. However, unlike in yeast, fructose-1,6-bisphosphate levels were not elevated in Δtps1 strains (Supplementary Figure 1A, available online). To investigate whether control of glycolysis is altered in Δtps1 mutants in the same way as in yeast (Hohmann et al, 1993), we used bioinformatics to identify the sole hexokinase gene, HXK1, of M. grisea (Dean et al, 2005) and disrupt its function by targeted gene replacement, both in Guy11 and in a Δtps1 mutant background. To study the effect of deleting HXK1 in M. grisea, we grew Guy-11, Δtps1, Δhxk1 and Δtps1 Δhxk1 double mutants in CM for 48 h, followed by growth in GMM, with nitrate as nitrogen source, for 16 h. Hexokinase activity was assayed from all four strains and was reduced in Δhxk1 and Δhxk1 Δtps1 mutants (Figure 2A). Consistent with this, the levels of G6P were reduced four-fold in the Δhxk1 and Δhxk1 Δtps1 mutants compared to Guy-11, and reduced almost 14-fold compared to Δtps1 strains (Figure 2B). The residual enzyme activity observed in our assay (Figure 2A) in the mutants is likely due to G6P production by glucokinase, although this does not appear to play a major role in G6P synthesis in M. grisea (Figure 2B). Trehalose levels were also reduced in Δhxk1 mutants, which may result from reduction in the Tps1 substrate G6P (Figure 2C). The ability of M. grisea Δtps1 Δhxk1 mutants to grow in the presence of glucose was partially restored compared to Δtps1 mutants (Figure 2E), but there was no restoration of sporulation (Figure 2D), or their ability to cause rice blast disease (Figure 2F). Furthermore in S. cerevisiae T6P is a potent inhibitor of HXK2 activity (Gancedo and Flores, 2004), whereas hexokinase activity in a Guy-11 extract is not significantly affected by the presence of T6P (Figure 2G). Interestingly, S. cerevisiae tps1Δ diploids do show a sporulation defect that is not repressible by deletion of Hxk2 (Neves et al, 1995), suggesting that some effects of this mutation are independent of its role in glycolytic regulation. When considered together, our results suggested that, unlike in yeast, M. grisea Δtps1 growth phenotypes cannot be remediated by deletion of HXK1 and that Tps1 may serve distinct functions in sugar metabolism, asexual development and the control of fungal virulence.
Figure 2.
Characterisation of M. grisea Δhxk1 mutants. (A) Hexokinase activity was measured and compared in Δtps1, Δhxk1, Δtps1 Δhxk1 and Guy-11 strains. Strains were grown in complete medium (CM) for 48 h followed by growth in glucose minimal medium (GMM) for 16 h. (B) G6P levels in the same strains and the same conditions. Bars represent standard error of three independent analyses. (C) Trehalose levels in the same strains and conditions. Bars represent standard error of three independent analyses. (D) Effect of Δhxk1 mutation on sporulation. Δtps1, Δhxk1, Δtps1 Δhxk1 and Guy-11 strains were grown on GMM plates (n=4) for 10 days, followed by harvesting of the spores for counting. Bars represent standard error. (E) Effect of Δhxk1 mutation on vegetative growth. Δtps1, Δhxk1, Δtps1 Δhxk1 and Guy-11 strains were grown on GMM plates for 10 days. (F) Virulence of Δtps1, Δhxk1, Δtps1 Δhxk1 and Guy-11 strains on rice seedlings. Rice seedlings were inoculated at a concentration of 5 × 104 spores/ml. (G) Hexokinase inhibition by 1 mM T6P. Bars represent standard error of three independent analyses.
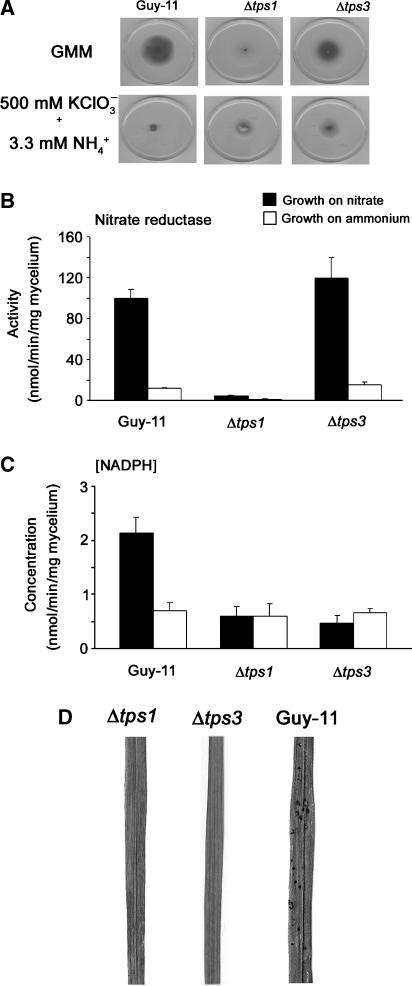
M. grisea tps1 mutants are unable to utilise nitrate as a nitrogen source
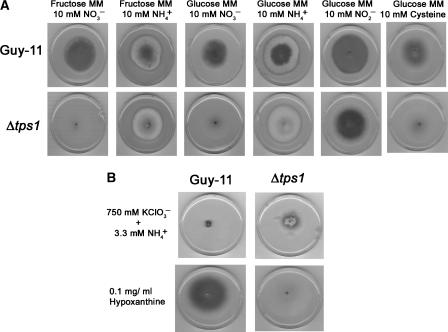
Previously, it was demonstrated that addition of a rich source of amino acids would restore the ability of Δtps1 mutants to grow in the presence of glucose (Foster et al, 2003). We were intrigued by this observation and therefore grew Δtps1 mutants on both GMM supplemented with single amino acids or alternative nitrogen sources (Supplementary Table 1). Δtps1 mutants could grow well on GMM with single amino acids (except cysteine, see below) and also grew normally on GMM with ammonium (NH4+) or nitrite (NO2−), but not nitrate (NO3−), as sole nitrogen source irrespective of carbon source (Figure 3A). Thus, the original suggestion that Δtps1 mutants cannot grow on glucose (Foster et al, 2003) is actually a conditional response mediated by the nature of the available nitrogen source. Because the metabolism of NO2− differs from that of NO3− by a single enzymatic step, nitrate reductase, we reasoned that M. grisea Δtps1 mutants could be nitrate non-utilising due to the absence, or suppression, of nitrate reductase. To test this idea, we assayed nitrate reductase activity in Δtps1 mutants grown in NO3− and ammonium NH4+-containing medium following a switch from CM. Guy-11 showed increased nitrate reductase activity after growth in NO3− compared to growth on NH4+, while under both growth conditions, Δtps1 mutants showed almost undetectable levels of nitrate reductase activity (Figure 4A). Consistent with this, Δtps1 mutants were resistant to toxic concentrations of potassium chlorate (750 mM) when grown under de-repressing conditions for nitrogen source utilisation (Cove, 1976), confirming the absence of functional nitrate reductase activity in Δtps1 (Figure 3B). Therefore, we conclude that the inability of M. grisea Δtps1 mutants to grow on GMM is not due to glycolytic misregulation, but is instead due to NO3− non-utilisation. Supplementation with individual amino acids, ammonium or nitrite thus restores growth by providing an alternative nitrogen source to the fungus.
Figure 3.
Nitrogen source utilisation and chlorate resistance of Δtps1 mutants of M. grisea. (A) Guy-11 and Δtps1 mutants were grown on MM containing 1% fructose or glucose as sole carbon sources. Sole nitrogen sources were provided as indicated, at 10 mM concentrations. M. grisea cultures were grown for 10 days at 26°C. (B) M. grisea strains were grown on GMM supplemented with 750 mM potassium chlorate and 3.3 mM NH4+ for 10 days at 26°C. Potassium chlorate is toxic to strains carrying active nitrate reductase enzymes under nitrogen de-repressing conditions (Cove, 1976). Strains were also grown on GMM with 0.1 mg/ml hypoxanthine as sole nitrogen source for 10 days at 26°C.
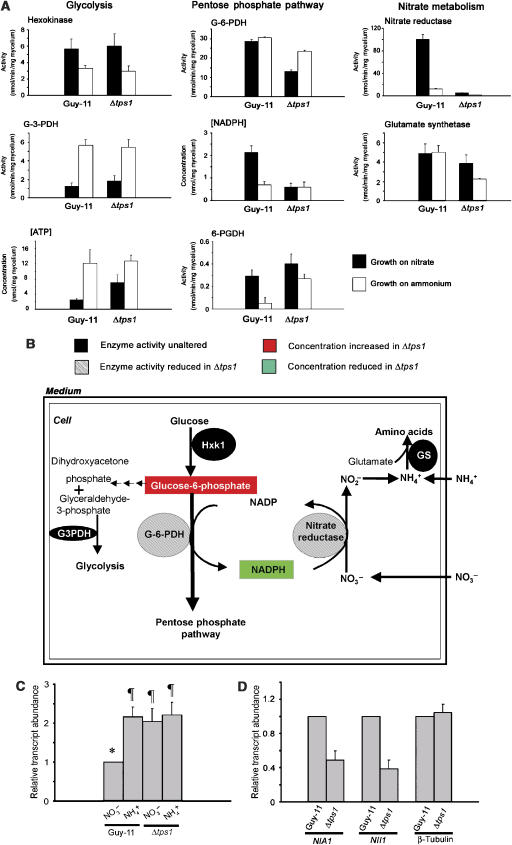
Figure 4.
Carbon and nitrogen metabolism is linked via G6P metabolism and NADPH production. (A) Enzyme activities associated with glycolysis, the pentose phosphate pathway and NO3− metabolism from Guy-11 and Δtps1 strains grown in CM for 48 h followed by incubation in GMM with NO3− (closed bars) or NH4+ (open bars) for 16 h. Bars represent standard error of three independent replicates. G-3-PDH is glyceraldehyde-3-phosphate dehydrogenase. G-6-PDH is G6P dehydrogenase. 6-PGDH is 6-phosphogluconic dehydrogenase. (B) Schematic representation of the effect of Δtps1 mutation on carbon and nitrogen metabolism in M. grisea. Differences in metabolite accumulation and enzyme activity between Guy-11 and Δtps1 strains are shown. (C) Transcript analysis of NMR1. Strains were grown for 48 h in CM followed by incubation in GMM with NO3− or NH4+ as a sole nitrogen source for an additional 16 h. Total RNA was extracted and gene expression analysed by RT–PCR. Analysis was performed in triplicate and normalised using levels of actin gene expression. Transcript abundance was calculated relative to NMR1 expression in Guy-11 on GMM with nitrate. Values with a different letter above are significantly different (Student's t-test, P<0.05). (D) Transcript analysis of NIA1, NII1 and β-tubulin encoding gene. Strains were grown for 48 h in CM followed by incubation in GMM with NO3− as a sole nitrogen source for an additional 16 h. Total RNA was extracted and gene expression analysed by RT–PCR. Analysis was performed in triplicate and normalised using levels of actin gene expression. Transcript abundance was calculated relative to gene expression in Guy-11 on GMM with NO3−.
The role of NADPH in nitrate utilisation
The role of Tps1 in NO3− utilisation was unexpected, and we therefore set out to determine why a trehalose biosynthetic enzyme should be necessary for nitrate reductase activity. In filamentous fungi, NO3− is metabolised to NO2− by the action of nitrate reductase, followed by reduction of NO2− to NH4+ by nitrite reductase (Cove, 1966). Ammonium is assimilated into amino acids through the action of glutamine synthetase and the GOGAT pathway (Kusnan et al, 1987). Nitrate reductase requires NADPH as a cofactor for function, and NADPH is generated from G6P by the action of G6P dehydrogenase (G-6-PDH) in the oxidative pentose phosphate pathway. G6P is, as described previously, also a substrate of Tps1 (Bell et al, 1998). In Aspergillus nidulans, transfer from growth on NH4+ to growth on NO3− leads to increased nitrate reductase and nitrite reductase activities, and also stimulates hexokinase and the enzymes of the pentose phosphate pathway, thereby increasing NADPH production from G6P (Hankinson and Cove, 1974). We therefore assayed the activities of enzymes involved in synthesis of NADPH and catabolism of NO3− in both Guy-11 and Δtps1 mutants (Figure 4A). Incubation in NO3− led to an increase in hexokinase activity in both strains, suggesting that elevated levels of G6P are required by M. grisea under conditions of NO3− utilisation (Dunn-Coleman and Pateman, 1979) and, secondly, that the increase in hexokinase activity does not require Tps1. In addition, other enzyme activities of the glycolytic pathway, such as glyceraldehyde-3-phosphate dehydrogenase (Figure 4A), phosphofructokinase and phosphoglucose isomerase (Supplementary Figure 1B) were not affected by the Δtps1 mutation, while levels of ATP were similar in both Δtps1 and Guy-11 (Figure 4A). These results underline the fact that glycolytic misregulation and ATP depletion does not occur in M. grisea Δtps1 mutants during growth on glucose-containing media.
In the wild-type M. grisea strain Guy-11, a large increase in NADPH levels was observed during growth on NO3− (Figure 4A). In contrast, NADPH levels did not increase in Δtps1 during growth on NO3− and there was also a relative decrease in G-6-PDH activity (Figure 4A) despite the fact that high levels of G6P accumulated in the Δtps1 mutant during growth on both nitrogen sources (Supplementary Figure 1C). We did not detect any inhibitory effect of G6P on G-6-PDH activity in vitro (Supplementary Figure 1D). Glutamine synthetase levels were similar in Guy-11 and Δtps1 strains, suggesting that the GS-GOGAT pathway operates normally in the absence of Tps1. The activity of 6-phosphogluconic dehydrogenase, the next enzyme of the pentose phosphate pathway, is also not reduced in Δtps1. From these data we can conclude that regulation of G6PDH and NADPH production, in response to NO3−, is a Tps1-dependent process in M. grisea (Figure 4B).
It is worth noting that two NADPH-dependent proteins are required for the metabolism of L-cysteine in eukaryotes. Cysteine oxygenase (EC 1.13.11.20) converts L-cysteine to 3-sulfinoalanine (Lombardini and Singer, 1969) and precedes conversion of 3-sulfinoalanine to alanine, while NADPH-dependant coenzyme A disulfide-glutathione reductase (EC 1.8.1.10; Ondarza et al, 1969) is necessary for the reduction of L-cysteine. Interestingly, the only amino acid that Δtps1 mutants are unable to utilise as a sole nitrogen source is cysteine (Figure 3A), consistent with the observed deficiency in NADPH.
Genetic regulation of nitrate utilisation genes via Tps1
The absence of nitrate reductase activity in Δtps1 mutants during growth on NO3− could be due either to an inactive enzyme or to repression of the NIA1 gene, which encodes nitrate reductase. Adding exogenous NADPH to the nitrate reductase activity enzymatic assay did not result in detectable nitrate reductase activity in Δtps1 (Figure 4A), suggesting that there is no nitrate reductase protein present in the cell. We also noted that Δtps1 mutants cannot utilise hypoxanthine as a nitrogen source (Figure 3B). Nitrate reductase and xanthine dehydrogenase (used to metabolise hypoxanthine) both require a molybdenum-containing cofactor, encoded in A. nidulans by the cnx genes (Unkles et al, 1999). Expression of cnx and niaD genes requires a positively acting GATA transcription factor encoded by the areA gene in A. nidulans (Wilson and Arst, 1998) and the NUT1 gene in M. grisea (Froeliger and Carpenter, 1996). The activity of AreA is negatively regulated by the product of nmrA (Andrianopoulos et al, 1998). We performed RT–PCR to analyse the gene expression of NMR1, the equivalent M. grisea gene, in Guy11 and Δtps1 during growth on nitrate and ammonium (Figure 4C). Expression of NMR1 was significantly higher in Δtps1 mutants than in Guy-11 during growth on nitrate, and was comparable to expression levels observed during growth of Guy-11 and Δtps1 on ammonium when it fully represses the action of the AreA GATA factor (Andrianopoulos et al, 1998). Furthermore, high levels of NMR1 transcript in Δtps1 strains corresponded with reduced expression of NIA1 and NII1, encoding nitrate and nitrite reductase respectively, compared to Guy-11 (Figure 4D). In A. nidulans, nitrate and nitrite reductase genes are tightly linked and coordinately regulated from a shared promoter (Cove, 1976). However, M. grisea NIA1 and NII1 genes are not linked and reside on chromosomes III and VI, respectively (Dean et al, 2005). We wondered if, considering Δtps1 strains could grow on NO2− but not NO3− media, there was evidence for differential regulation of nitrate and nitrite reductase activities in M. grisea. Interestingly, unlike in A. nidulans, we were unable to detect nitrate reductase activity in protein extracts of Guy-11 and Δtps1 strains grown in CM followed by a switch to NO2−-containing media (not shown). In addition, NADPH levels of Guy-11 and Δtps1 strains were not elevated during growth on NO2− compared to NH4+ (Supplementary Figure 2). This suggests that nitrate and nitrite reductase activities are not always coordinately regulated, and second that NADPH may not be important for nitrite reductase activity. Studies of Neurospora crassa nitrite reductase (Nason et al, 1954) demonstrated that the enzyme can utilise both NADH and NADPH as cofactor in vitro, and the preferred cofactor for M. grisea NII1 therefore needs to be determined. We conclude that in response to NO3−, Tps1 is required to regulate the activity of G6PDH, which controls levels of G6P, NADPH and nitrate reductase activity, and also to modulate gene expression by control of NMR1.
Genetic regulation of Tps1 activity in M. grisea by Tps3
The catalytic subunit of Tps1, encoded by TPS1, is part of a multi-protein complex in yeast, with the Tps2 T6P phosphohydrolase and two other proteins, Tps3 and Tsl1 (Gancedo and Flores, 2004). To investigate the control of M. grisea Tps1 activity, we analysed the genome sequence of M. grisea (Dean et al, 2005), identified a homologue of TPS3 and generated a Δtps3 mutant. The Δtps3 mutant was impaired in both trehalose (see Figure 6D) and T6P synthesis. HPAE-PAD analysis of mycelial extracts showed concentrations of 99.8±8 pmol per milligram T6P mycelium (n=3) in Guy-11, but no detectable T6P in mycelium of Δtps3 mutants (not shown). The Δtps3 mutant grew better than Δtps1 on NO3−, but was still reduced in sporulation (Figure 5). NADPH was also reduced and showed no increase during growth of Δtps3 mutants on NO3−-containing medium, consistent with the Δtps1 phenotype. Interestingly, however, nitrate reductase activity in Δtps3 mutants was equivalent to that in Guy-11 during growth on NO3− and NH4+ (Figure 5B). Nitrate reductase activity is assayed by addition of exogenous NADPH, so while this result indicates that nitrate reductase is present in Δtps3 mutants, it is unlikely to be active in the cell due to the lack of endogenous cellular NADPH (Figure 5C). Δtps3 mutants are, for example, resistant to potassium chlorate under nitrogen de-repressing conditions (Figure 5A). In addition, Δtps3 does not accumulate G6P when grown on NO3− (Supplementary Figure 1C) or in CM (Supplementary Figure 1E). Taken together, our results indicate that Tps1 and Tps3 are both necessary for T6P synthesis and regulation of the pentose phosphate pathway and NADPH production, while at least Tps1 is also necessary for control of NMR1 expression. Significantly, Δtps3 mutants are also unable to cause rice blast disease (Figure 5D), demonstrating that fungal pathogenicity requires Tps3-dependent Tps1 activity.
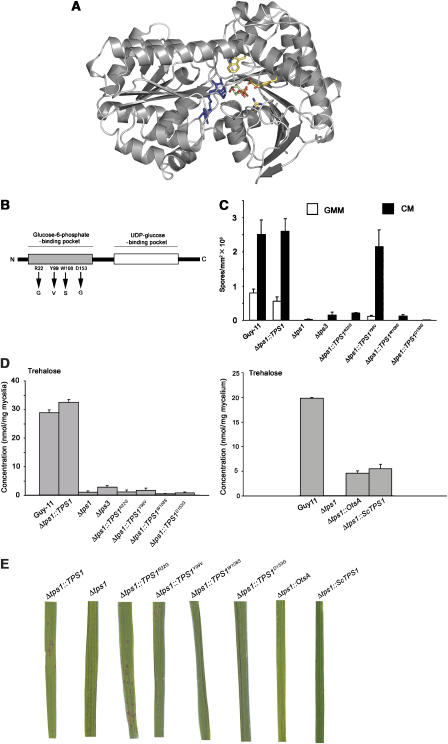
Figure 6.
Site-directed mutagenesis of the G6P-binding pocket of Tps1. (A) Energy-minimised Tps1 homology model structure constructed from OtsA template structures (Gibson et al, 2002) by MODELLER, and showing residues involved in G6P binding. Residues, which were mutated are (clockwise from top) Trp108, Asp153, Tyr99 and Arg22. The locations of G6P (green and red) and UDP-glucose (blue) are based on the template structures to indicate their proximity to the mutated residues. (B) Schematic representation of the relative positions of introduced amino-acid substitutions in the G6P-binding pocket of Tps1. The drawing is not to scale. (C) Spore production was measured for Guy-11, Δtps1∷TPS1, Δtps1, Δtps3, Δtps1∷Tps1R22G, Δtps1∷Tps1Y99V, Δtps1∷Tps1W108S and Δtps1∷Tps1D153G strains. Spores from strains grown for 10 days on CM and GMM were counted from four independent plates. (D) Trehalose concentration in Guy-11, Δtps1∷TPS1, Δtps1, Δtps3, Δtps1∷Tps1R22G, Δtps1∷Tps1Y99V, Δtps1∷Tps1W108S and Δtps1∷Tps1D153G strains (left bar chart) and Guy-11, Δtps1, Δtps1∷otsA and Δtps1∷ScTPS1 (right bar chart). Trehalose was measured in triplicate. Bars represent standard error. (E) The virulence of Δtps1∷TPS1, Δtps1, Δtps1∷Tps1R22G, Δtps1∷Tps1Y99V, Δtps1∷Tps1W108S and Δtps1∷Tps1D153G, Δtps1∷otsA and Δtps1∷ScTPS1 strains was analysed by inoculating spores onto rice seedlings of cultivar CO-39. Spores were inoculated at a concentration of 5 × 104 spores/ml and rice blast disease symptoms observed after 96 h.
Figure 5.
Genetic control of Tps1 activity by Tps3. (A) Guy-11, Δtps1 and Δtps3 strains were grown on GMM and GMM supplemented with 500 mM potassium chlorate and de-repressing concentrations (3.3 mM) of NH4+. (B) Nitrate reductase activity in Guy-11, Δtps1 and Δtps3 strains following 48 h growth in CM and 16 h growth in GMM with NO3− or NH4+. Activities and concentrations were measured in triplicate. Bars represent standard error. (C) NADPH concentration in Guy-11, Δtps1 and Δtps3 strains following 48 h growth in CM and 16 h growth in GMM. Activities and concentrations were measured in triplicate. Bars represent standard error. (D) Guy-11, Δtps1 and Δtps3 strains were inoculated onto rice seedlings to analyse pathogenicity. Spores were inoculated at a rate of 5 × 104 spores/ml.
Functional analysis of M. grisea Tps1
The cellular pool of G6P links carbon and nitrogen metabolism via the pentose phosphate pathway and NADPH generation (see Figure 4B), and so we reasoned that Tps1 might act as a reporter of G6P levels, leading to increased G6PDH activity, increased NADPH levels and de-repression of NO3− utilisation genes in the presence of NO3−. To test this idea, we undertook a functional analysis of the Tps1 protein and characterised the G6P binding pocket of Tps1 by site-directed mutagenesis. A crystal structure for the prokaryotic Tps1 homologue, encoded by otsA in Escherichia coli, has been used to identify amino-acid residues involved in substrate binding (Gibson et al, 2002). We generated a model of M. grisea Tps1 based on the OtsA structure (Figure 6A), to identify residues in the G6P-binding pocket that are conserved between OtsA and Tps1, and introduced nucleotide changes into a full-length genomic clone of TPS1 that would result in four amino-acid substitutions at these positions in Tps1 when the alleles were transformed into a M. grisea Δtps1 mutant (Figure 6B).
The resulting M. grisea Δtps1∷Tps1R22G, Δtps1∷Tps1Y99V, Δtps1∷Tps1W108S and Δtps1∷Tps1D153G strains were all unable to synthesize trehalose (Figure 6D) or T6P (not shown), indicating the importance of each residue for Tps1 activity. Δtps1∷Tps1R22G, Δtps1∷Tps1W108S and Δtps1∷Tps1D153G strains resembled Δtps1 mutants, were NO3− non-utilising (Figure 6C) and did not sporulate on minimal medium—although nitrate reductase activity was detected in extracts from Δtps1∷Tps1R22G strains (Supplementary Figure 1F) suggesting that like Δtps3, this mutant has inactive nitrate reductase present in the cell—whereas strains carrying the Y99V substitution (Δtps1∷Tps1Y99V) produced increased numbers of spores on minimal medium and sporulated almost as well as Guy11 on CM (Figure 6C). G6P levels were not elevated in Δtps1∷Tps1R22G, Δtps1∷Tps1Y99V and Δtps1∷Tps1D153G strains, suggesting that G6P accumulation is related to the absence of Tps1 protein and not to a non-functional Tps1 variant such as Tps1D153G (Supplementary Figure 1E).
Significantly, the expression of TPS1 alleles carrying R22G and Y99V substitutions restored the ability to cause rice blast disease when expressed in a Δtps1 mutant, as shown in Figure 6E. This shows that the catalytic activity of Tps1 is not required for fungal pathogenicity, because Δtps1∷Tps1R22G and Δtps1∷Tps1Y99V strains are unable to synthesize trehalose and T6P, but can still cause disease in rice. In a converse experiment, the S. cerevisiae TPS1 homologue ScTPS1, and the bacterial homologue otsA, were expressed in M. grisea Δtps1 strains under control of the native M. grisea TPS1 promoter. The expression of both genes resulted in elevated trehalose production compared to Δtps1 (Figure 6D). However, neither gene complemented the Δtps1 mutation for growth on GMM (not shown) or for restoration of virulence (Figure 6E). Therefore, trehalose production in the absence of M. grisea Tps1 is not sufficient for virulence.
Analysis of these mutants indicated that NO3− utilisation, sporulation, trehalose production and virulence are four independent processes influenced by the Tps1 protein independently of its catalytic activity. At least two residues in the G6P-binding pocket, W108S and D153, are necessary for all of these processes, suggesting that the regulation of fungal virulence and NO3− utilisation involves efficient G6P binding to Tps1. Indeed, from our model, we see that amino-acid changes resulting in the greatest perturbation of G6P binding correspond to the greatest loss of Tps1-dependent processes. R22G and Y99V are predicted to reduce the number of contacts made to the phosphate moiety of G6P, and R22G would also eliminate a key interaction promoting closure of the catalytic loop over the active site (Supplementary Figure 3A and B; Gibson et al, 2002). However, contacts from other residues remain to position G6P in the active site. W108S and D153G, on the other hand, are predicted to have more profound effects on G6P binding, with both mutations resulting in loss of positioning of the substrate. D153G is predicted to eliminate the only hydroxyl interactions between G6P and the protein (Supplementary Figure 3AII; Gibson et al, 2002), thus permitting a larger range of movement for the bound substrate. Similarly, W108S results in loss of the indole moiety necessary for selection of the α-glucose anomer, and this change could allow the more prevalent β-anomer to bind, hence inhibiting Tps1 function (Supplementary Figure 3BV; Gibson et al, 2002).
Investigating the role of Tps1 in plant infection-related development
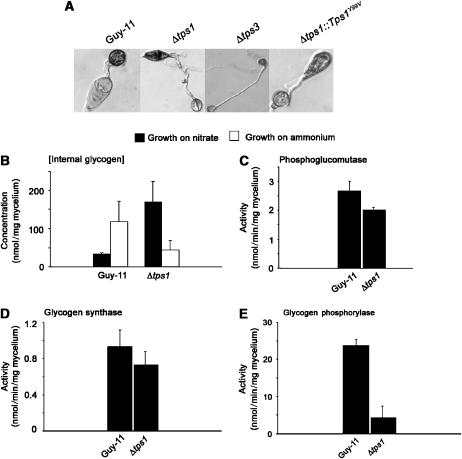
We previously demonstrated that appressoria of Δtps1 mutants are reduced in turgor and cannot infect rice leaves (Foster et al, 2003). Glycogen is present in the conidium of M. grisea and is believed to contribute to cellular turgor as a substrate for glycerol synthesis in the appressorium (de Jong et al, 1997; Thines et al, 2000). During appressorium development, glycogen accumulation occurs before the onset of turgor generation, and is broken down as glycerol is synthesised (Thines et al, 2000). Because glycogen synthesis also requires G6P, we decided to test whether Tps1 is necessary for regulation of glycogen mobilisation in the appressorium. We observed glycogen deposits in conidia of Δtps1 mutants, but could not detect glycogen in appressoria, in contrast to Guy11 (Figure 7A). We therefore quantified glycogen in Guy-11 and Δtps1 mycelium, grown either on NO3− or NH4+-containing medium and found that Δtps1 mutants accumulate more glycogen when grown in the presence of NO3− than NH4+, in contrast to Guy11, which accumulates greater glycogen when grown in NH4+ (Figure 7B). To investigate the process in more detail, we measured activities of two glycogen biosynthetic enzymes: phosphoglucomutase, which converts G6P to G1P for further conversion to UDP-glucose (Figure 7C), and glycogen synthase, which assembles glycogen from UDP-glucose subunits (Figure 7D). The activities of these two enzymes were slightly reduced in Δtps1 compared to Guy-11. Glycogen phosphorylase activity was, however, markedly reduced in Δtps1 mutants compared to Guy11 (Figure 7E). Therefore, Tps1 appears to be required for glycogen metabolism in conidia and mycelium, with reduced glycogen phosphorylase activity in Δtps1 resulting in glycogen accumulation. The Δtps3 mutation also affects glycogen steady-state levels in the conidium (Figure 7A). Interestingly, Δtps1∷Tps1Y99V, which is unable to synthesize trehalose, but is pathogenic, also affects glycogen steady-state levels in the conidium during appressorium development. We can therefore conclude that inappropriate glycogen accumulation in the conidium is associated with loss of Tps1 in M. grisea, but is not sufficient to explain the loss of virulence exhibited by Δtps1 mutants. To determine the wider effect of Tps1 on expression of other virulence-associated functions expressed during rice infection, gene expression of the MPG1 hydrophobin gene and PTH11, a G-protein-coupled receptor-encoding gene, both of which are expressed prior to appressorium development (DeZwaan et al, 1999; Soanes et al, 2002a, 2002b), was analysed. Expression of both MPG1 and PTH11 was reduced in the Δtps1 mutant compared to Guy-11 (Supplementary Figure 4). We conclude that Tps1 is necessary for expression of at least two major virulence-associated genes during appressorium formation by M. grisea.
Figure 7.
Role of Tps1 in glycogen metabolism and infection-related development. (A) At 24 h post-germination, glycogen accumulates in the appressorium of Guy-11 while Δtps1, Δtps3 and Δtps1∷Tps1Y99V strains retain and accumulate glycogen in the conidium. (B) Glycogen concentration in Guy-11 and Δtps1 strains grown in CM for 48 h followed by growth in GMM with nitrate or ammonium as a sole nitrogen source for 16 h. Measurements were performed in triplicate and bars represent standard error. (C) Phosphoglucomutase activities in Guy-11 and Δtps1 strains grown in CM for 48 h followed by growth in GMM with NO3− as a sole nitrogen source for 16 h. Measurements were performed in triplicate and bars represent standard error. (D) Glycogen synthase activities in Guy-11 and Δtps1 strains grown in CM for 48 h followed by growth in GMM with NO3− as a sole nitrogen source for 16 h. Measurements were performed in triplicate and bars represent standard error. (E) Glycogen phosphorylase activities in Guy-11 and Δtps1 strains grown in CM for 48 h followed by growth in GMM with NO3− as a sole nitrogen source for 16 h. Measurements were performed in triplicate and bars represent standard error.
Discussion
In this study, we set out to determine the role of trehalose and T6P synthesis in the establishment of rice blast disease. The absence of Tps1 from M. grisea prevents the fungus from causing disease and also affects spore production, appressorium function and sugar metabolism (Foster et al, 2003). Here, we have shown that the importance of Tps1 to cellular differentiation and fungal virulence results from its wide ranging role as a central regulator of both sugar metabolism and nitrogen source utilisation, acting both post-translationally to regulate enzymatic activities, but also as a regulator of gene expression.
The role of Tps1 in yeast has been extensively studied, and in addition to its biosynthetic function, the Tps1 protein controls entry of glucose into glycolysis by regulating the level of G6P, one of the substrates for Tps1. The M. grisea TPS1 gene has the capacity to fulfil such a function because it is able to complement a yeast tps1Δ strain and regulate glycolysis normally. Unlike M. grisea Δtps1 mutants, however, the growth of yeast tps1Δ mutants cannot be restored on glucose-containing medium regardless of the nitrogen source provided, including ammonium (data not shown). It is worth noting, however, that yeast two-hybrid data (Gavin et al, 2002) suggests that there is an interaction between S. cerevisiae Tps1 and Mks1, a protein involved in regulation of nitrogen utilisation in yeast (Edskes et al, 1999).
It became increasingly clear during the course of this study that Tps1 does not regulate glycolysis in M. grisea in the same manner as occurs in yeast (Hohmann et al, 1993). Deletion of the hexokinase gene HXK1, for example, did not remediate growth or sporulation of Δtps1 on glucose medium and Δtps1 Δhxk1 double mutants were still non-pathogenic. Enzyme assays also showed that the activity of glycolytic enzymes was not affected in Δtps1 mutants, ATP depletion and fructose 1,6-bisphosphate accumulation did not occur in the presence of glucose, and T6P did not inhibit M. grisea hexokinase activity. Moreover, expression of S. cerevisiae TPS1 did not complement a M. grisea Δtps1 mutant. Growth tests meanwhile demonstrated instead that M. grisea Δtps1 mutants were NO3− non-utilising. We postulated that the connection between NO3− utilisation and sugar metabolism in the rice blast fungus was the availability of NADPH, generated from G6P in the oxidative pentose phosphate pathway. Analysis of enzymes involved in generating NADPH showed that in M. grisea, as demonstrated previously in A. nidulans (Hankinson and Cove, 1974; Dunn-Coleman and Pateman, 1979), hexokinase activity and NADPH levels both increase during growth on NO3−, while in Δtps1, G-6-PDH activity and NADPH production are both reduced significantly when the mutant is grown on minimal medium with NO3−. Consequently, nitrate reductase levels were not elevated in Δtps1 mutants during growth on NO3−, explaining why they are unable to grow, and G6P accumulated in cells (Figure 1B). Gene expression analysis, meanwhile, showed that Δtps1 mutants express NMR1, a negative repressor of nitrogen metabolism, at high levels during incubation in NO3−-containing medium (Dunn-Coleman et al, 1981; Andrianopoulos et al, 1998), while expression of the nitrate and nitrite reductase structural genes was reduced in Δtps1 strains. This provides evidence that Tps1 is involved in de-repression, via NMR1, of genes involved in nitrate utilisation, in addition to its role in stimulation of the pentose phosphate pathway and NADPH production. A. nidulans NmrA has been shown to discriminate between oxidised and reduced dinucleotides, including NADP+ and NADPH, and may act as a redox sensor (Lamb et al, 2003). The extracellular nitrate signal could act on Tps1 directly via a nitrogen sensing mechanism, or, more likely, via increased G6P levels, due to stimulated hexokinase activity. Additionally, because glycolytic enzyme activities are unaltered in Δtps1 compared to Guy-11, and are relatively low in both strains compared to G-6-PDH activity in Guy-11, this suggests the major pathway for glucose consumption in M. grisea may be the pentose phosphate pathway. In the absence of TPS1 therefore, G6P accumulates but G6PDH is not active. We conclude that Tps1 integrates carbon and nitrogen metabolism, probably through G6P sensing, resulting in increased NADPH production and induction of gene expression associated with nitrate utilisation.
The role of Tps1 in plant disease
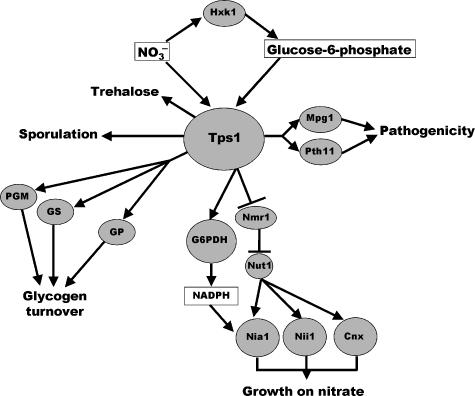
To address the role of Tps1 in pathogenicity and to distinguish this from the other biochemical and regulatory functions of the protein, we decided to carry out a structure–function study. Amino-acid substitutions in Tps1 demonstrated that trehalose synthesis is not required for plant infection by M. grisea, but that the protein itself, and in particular its efficient binding to G6P, is necessary for its regulatory functions and for its role in fungal virulence. The Δtps1∷Tps1R22G and Δtps1∷Tps1Y99V strains, which lack Tps1 catalytic activity, but may allow some G6P binding to Tps1, based on structural modelling of the Tps1 protein with the E. coli otsA structure (Gibson et al, 2002), are still able to cause rice blast disease in response to G6P binding, even though they are unable to synthesize trehalose. Tps1 therefore acts as an integrator of the carbon and nitrogen status of the cell, perhaps acting as a reporter of G6P levels. Consistent with this, Δtps1∷Tps1W108S and Δtps1∷Tps1D153G strains, in which the Tps1 protein is very unlikely to bind to G6P, cannot cause rice blast disease. Additionally, Tps1 can be thought of as being central to a complex network of interactions and cellular processes responding to nitrogen and carbon signals in the cell and resulting in the potential Tps1 interactome depicted in Figure 8. Analysis of Δtps1∷Tps1R22G, Δtps1∷Tps1W108S, Δtps1∷Tps1Y99V, Δtps1∷Tps1D153G and Δtps3 mutants has demonstrated that trehalose biosynthesis, glycogen mobilisation, plant pathogenicity, sporulation, activation of the pentose phosphate pathway and nitrate utilisation gene expression are independent processes, each affected by Tps1.
Figure 8.
Model for the role of Tps1 in Magnaporthe grisea. In response to G6P and nitrate, Tps1 coordinates nitrate utilisation by a genetic response involving NMR1 and a post-translational effect on NADPH levels via stimulation of G6PPDH activity. Tps1 also regulates glycogen metabolism based on its effect on G6P levels and independently controls sporulation-related functions and virulence-associated gene expression. Protein names are explained in the text, except the following: PGM, phosphoglucomutase; GS, glycogen synthase; G6PDH, G6P dehydrogenase; GP, glycogen phosphorylase; Nia1, nitrate reductase and Nii1, nitrite reductase.
An accumulating body of evidence in plants points to trehalose and T6P biosynthesis and metabolism being highly integrated with plant development, a relationship that was somewhat unexpected (Eastmond et al, 2002). Tps1 mutations in Arabidopsis thaliana, for example, lead to embryonic lethality due to loss in coordination in cell wall biosynthesis and cell division, indicating a key role for trehalose metabolism in plant embryonic development (Gómez et al, 2006). T6P levels have also been shown to affect starch synthesis in Arabidopsis by post-translational redox activation of ADP-glucose pyrophosphorylase (Kolbe et al, 2005), while in maize, a recent study demonstrated that T6P phosphatase encoded by TPS2 is necessary for inflorescence architecture (Satoh-Nagasawa et al, 2006). Interestingly, transcriptional profile analysis of Arabidopsis in response to changes in nitrate availability, indicates an effect on the expression of trehalose metabolic genes (Scheible et al, 2004). Our identification of a role for Tps1 in M. grisea in regulating NADPH levels, the oxidative pentose phosphate pathway, and expression of nitrogen source utilisation and virulence genes show how trehalose biosynthesis might act in regulating complex developmental processes in multicellular eukaryotes such as plants and fungi.
Materials and methods
Fungal strains, manipulations and physiological tests
All M. grisea strains used in this study are derived from Guy-11. Standard procedures of M. grisea growth, maintenance, appressoria formation and transformation were performed as described previously (Crawford et al, 1986; Talbot et al, 1993). Gel electrophoresis, restriction enzyme digestions, ligations, DNA and RNA gel blot hybridizations were performed using standard procedures (Sambrook et al, 1989). DNA and RNA probes were radiolabelled using the random primer method (Feinberg and Vogelstein, 1983).
Strains were grown on complete medium (CM)-containing 1% (w/v) glucose, 0.2% (w/v) peptone, 0.1% (w/v) yeast extract and 0.1% (w/v) casamino acids—or GMM—containing 10% glucose and 0.6% sodium nitrate—unless otherwise stated, as described in Foster et al (2003).
Rice plant infections were made as described previously (Talbot et al, 1993). For additional information, see Supplementary data. Nucleic acid, trehalose and sugar phosphate extractions were performed as described in Supplementary data.
Enzyme activity assays
All enzymatic assays were performed at 22°C. All assay components were purchased from Sigma, except NADH, NADPH and NADP (Calbiochem). Enzyme activities were determined spectrophotometrically in triplicate, and are expressed as the concentration of product formed in 1 min by total cell protein from 1 mg of mycelium. For full experimental protocols, see Supplementary data.
Metabolite identification and quantification
Trehalose was detected by hydrolysis to glucose using 3 mU of porcine kidney acidic trehalase (Sigma), followed by determination of glucose concentration using a diagnostic kit (Roche). The concentration of glucose from the trehalase-treated extract was compared to glucose from non-treated control, the difference representing the concentration of glucose liberated from trehalose.
Sugar phosphates were quantified using HPAE-PAD analysis on mycelial extracts using a BioLC system (Dionex). See Supplementary data for details. Mycelial glycogen concentration was determined enzymatically from mycelium, as described in Supplementary data. Glycogen was visualised in appressoria using an iodine stain consisting of 60 mg KI and 10 mg I2, per millilitre, in distilled water. Samples were observed with a Nikon Optiphot microscope connected to a Nikon FX-35 camera. Mycelial concentrations of NADPH and ATP were determined enzymatically, as described in Supplementary data.
Protein modelling
Protein modelling of M. grisea Tps1 and E. coli otsA (Gibson et al, 2002) was performed as described in Supplementary data.
Construction of targeted gene replacement vectors and gene expression studies
A full description of all plasmids used in this study, and their construction, is provided in Supplementary data. MPG1 gene expression was examined by Northern blot analysis. NMR1, PTH11, NIA1, NII1 and β-tubulin gene expression was examined using RT–PCR (Clontech). Following PCR, quantification of the resulting amplicons used the ImageMaster TotalLab software programme and normalisation of expression data was achieved by quantification of actin gene expression. For full details, see Supplementary data.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3A
Supplementary Figure 3B
Supplementary Figure 4
Supplementary data
Acknowledgments
This work was funded by a grant to NJT from the Biotechnology and Biological Sciences Research Council (BBSRC), and by a BBSRC studentship to JMJ. We thank Dr Nick Smirnoff (University of Exeter, UK) for guidance on using Dionex HPAE-PAD, and also thank Professor C Gancedo (Instituto de Investigaciones Biomédicas, Madrid, Spain) for the gift of MB062.
References
- Andrianopoulos A, Kourambas S, Sharp JA, Davis MA, Hynes MJ (1998) Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J Bacteriol 180: 1973–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguelles JC (2000) Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch Microbiol 174: 217–224 [DOI] [PubMed] [Google Scholar]
- Bell W, Sun W, Hohmann S, Wera S, Reinders A, De Virgilio C, Wiemken A, Thevelein JM (1998) Composition and functional analysis of the Saccharomyces cerevisiae trehalose synthase complex. J Biol Chem 273: 33311–33319 [DOI] [PubMed] [Google Scholar]
- Bonini BM, Van Dijck P, Thevelein JM (2003) Uncoupling of the glucose growth defect and the deregulation of glycolysis in Saccharomyces cerevisiae Tps1 mutants expressing trehalose-6-phosphate-insensitive hexokinase from Schizosaccaromyces pombe. Biochim Biophys Acta 1606: 83–93 [DOI] [PubMed] [Google Scholar]
- Cove DJ (1966) The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta 113: 51–56 [DOI] [PubMed] [Google Scholar]
- Cove DJ (1976) Cholorate toxicity in Aspergillus nidulans: the selection and characterisation of chlorate resistant mutants. Heredity 36: 191–203 [DOI] [PubMed] [Google Scholar]
- Crawford MS, Chumley FG, Weaver CG, Valent B (1986) Characterisation of the heterokaryotic and vegetative diploid phases of Magnaporthe grisea. Genetics 114: 1111–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JC, McCormack BJ, Smirnoff N, Talbot NJ (1997) Glycerol generates turgor in rice blast. Nature 389: 471–483 [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, Read ND, Lee YH, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W, Harding M, Kim S, Lebrun MH, Bohnert H, Coughlan S, Butler J, Calvo S, Ma LJ, Nicol R, Purcell S, Nusbaum C, Galagan JE, Birren BW (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986 [DOI] [PubMed] [Google Scholar]
- DeZwaan TM, Carroll AM, Valent B, Sweigard JA (1999) Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11: 2013–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn-Coleman NS, Pateman JA (1979) The regulation of hexokinase and phosphoglucomutase activity in Aspergillus nidulans. Mol Gen Genet 171: 69–73 [DOI] [PubMed] [Google Scholar]
- Dunn-Coleman NS, Tomsett AB, Garrett RH (1981) The regulation of nitrate assimilation in Neurospora crassa: biochemical analysis of the nmr-1 mutants. Mol Gen Genet 182: 234–239 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, van Dijken AJ, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JD, Smeekens SC, Graham IA (2002) Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 29: 225–235 [DOI] [PubMed] [Google Scholar]
- Edskes HK, Hanover JA, Wickner RB (1999) Mks1p is a regulator of nitrogen catabolism upstream of Ure2p in Saccharomyces cerevisiae. Genetics 153: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein AD, Pan YT, Pastuszak I, Carroll D (2003) New insights on trehalose: a multifunctional molecule. Glycobiology 13: 17R–27R [DOI] [PubMed] [Google Scholar]
- Ernandes JR, De Meirsman C, Rolland F, Winderickx J, de Winde J, Brandao RL, Thevelein JM (1998) During the initiation of fermentation overexpression of hexokinase PII in yeast transiently causes a similar deregulation of glycolysis as deletion of Tps1. Yeast 14: 255–269 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Foster AJ, Jenkinson JM, Talbot NJ (2003) Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. EMBO J 22: 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froeliger EH, Carpenter BE (1996) NUT1, a major nitrogen regulatory gene in Magnaporthe grisea, is dispensable for pathogenicity. Mol Gen Genet 251: 647–656 [DOI] [PubMed] [Google Scholar]
- Gancedo C, Flores CL (2004) The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res 4: 351–359 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Gibson RP, Turkenburg JP, Charnock SJ, Lloyd R, Davies GJ (2002) Insights into trehalose synthesis provided by the structure of the retaining glucosyltransferase OtsA. Chem Biol 9: 1337–1346 [DOI] [PubMed] [Google Scholar]
- Gómez LD, Baud S, Gilday A, Li Y, Graham IA (2006) Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J 46: 69–84 [DOI] [PubMed] [Google Scholar]
- Hankinson O, Cove DJ (1974) Regulation of the pentose phosphate pathway in the fungus Aspergillus nidulans. J Biol Chem 249: 2344–2353 [PubMed] [Google Scholar]
- Hohmann S, Neves MJ, de Koning W, Alijo R, Ramos J, Thevelein JM (1993) The growth and signalling defects of the ggs1 (fdp1/byp1) deletion mutant on glucose are suppressed by a deletion of the gene encoding hexokinase PII. Curr Genet 23: 281–289 [DOI] [PubMed] [Google Scholar]
- Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P (2005) Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci USA 102: 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnan MB, Berger MG, Fock HP (1987) The involvement of glutamine synthetase/glutamate synthase in ammonia assimilation by Aspergillus nidulans. J Gen Microbiol 133: 1235–1242 [DOI] [PubMed] [Google Scholar]
- Lamb HK, Leslie K, Dodds AL, Nutley M, Cooper A, Johnson C, Thompson P, Stammers DK, Hawkins AR (2003) The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides. J Biol Chem 278: 32107–32114 [DOI] [PubMed] [Google Scholar]
- Lombardini JB, Singer TP (1969) Cysteine oxygenase. J Biol Chem 244: 1172–1175 [PubMed] [Google Scholar]
- Luyten K, de Koning W, Tesseur I, Ruiz MC, Ramos J, Cobbaert P, Thevelein JM, Hohmann S (1993) Disruption of the Kluyveromyces lactis GGS1 gene causes inability to grow on glucose and fructose and is suppressed by mutations that reduce sugar uptake. Eur J Biochem 217: 701–713 [DOI] [PubMed] [Google Scholar]
- Nason N, Abraham RG, Averbach BC (1954) The enzymatic reduction of nitrite to ammonia by reduced pyridine nucleotides. Biochemica Biophysica Acta 15: 159–161 [DOI] [PubMed] [Google Scholar]
- Neves MJ, Hohmann S, Bell W, Dumortier F, Luyten K, Ramos J, Cobbaert P, de Koning W, Kaneva Z, Thevelein JM (1995) Control of glucose influx into glycolysis and pleiotropic effects studied in different isogenic sets of Saccharomyces cerevisiae mutants in trehalose biosynthesis. Curr Genet 27: 110–122 [DOI] [PubMed] [Google Scholar]
- Ondarza RN, Abney R, Lopez-Colome AM (1969) Characterization of a NADPH-dependent coenzyme A-SS-glutathione reductase from yeast. Biochemica Biophysica Acta 191: 239–248 [DOI] [PubMed] [Google Scholar]
- Reinders A, Burckert N, Hohmann S, Thevelein JM, Boller T, Wiemken A, De Virgilio C (1997) Structural analysis of the subunits of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae and their function during heat shock. Mol Microbiol 24: 687–695 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D (2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 11: 227–230 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soanes DM, Kershaw MJ, Cooley RN, Talbot NJ (2002a) Regulation of the MPG1 hydrophobin gene in the rice blast fungus Magnaporthe grisea. Mol Plant Microbe Interact 15: 1253–1267 [DOI] [PubMed] [Google Scholar]
- Soanes DM, Skinner W, Keon J, Hargreaves J, Talbot NJ (2002b) Genomics of phytopathogenic fungi and the development of bioinformatic resources. Mol Plant Microbe Interact 15: 421–427 [DOI] [PubMed] [Google Scholar]
- Talbot NJ (2003) On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 57: 177–202 [DOI] [PubMed] [Google Scholar]
- Talbot NJ, Ebbole DJ, Hamer JE (1993) Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5: 1575–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines E, Weber RW, Talbot NJ (2000) MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12: 1703–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unkles SE, Heck IS, Appleyard MV, Kinghorn JR (1999) Eukaryotic molybdopterin synthase. Biochemical and molecular studies of Aspergillus nidulans cnxG and cnxH mutants. J Biol Chem 274: 19286–19293 [DOI] [PubMed] [Google Scholar]
- Veneault-Fourrey C, Barooah MK, Egan MJ, Talbot NJ (2006) Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312: 580–583 [DOI] [PubMed] [Google Scholar]
- Voit EO (2003) Biochemical and genomic regulation of the trehalose cycle in yeast: review of observations and canonical model analysis. J Theor Biol 223: 55–78 [DOI] [PubMed] [Google Scholar]
- Wilson RA, Arst HN Jr (1998) Mutational analysis of AREA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the ‘streetwise' GATA family of transcription factors. Microbiol Mol Biol Rev 62: 586–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winderickx J, de Winde JH, Crauwels M, Hino A, Hohmann S, Van Dijck P, Thevelein JM (1996) Regulation of genes encoding subunits of the trehalose synthase complex in Saccharomyces cerevisiae: novel variations of STRE-mediated transcription control? Mol Gen Genet 252: 470–482 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3A
Supplementary Figure 3B
Supplementary Figure 4
Supplementary data