Abstract
Protection from NO gas, a toxic byproduct of anaerobic respiration in Pseudomonas aeruginosa, is mediated by nitric oxide (NO) reductase (NOR), the norCB gene product. Nevertheless, a norCB mutant that accumulated ∼13.6 μM NO paradoxically survived anaerobic growth. Transcription of genes encoding nitrate and nitrite reductases, the enzymes responsible for NO production, was reduced >50- and 2.5-fold in the norCB mutant. This was due, in part, to a predicted compromise of the [4Fe–4S]2+ cluster in the anaerobic regulator ANR by physiological NO levels, resulting in an inability to bind to its cognate promoter DNA sequences. Remarkably, two O2-dependent dioxygenases, homogentisate-1,2-dioxygenase (HmgA) and 4-hydroxyphenylpyruvate dioxygenase (Hpd), were derepressed in the norCB mutant. Electron paramagnetic resonance studies showed that HmgA and Hpd bound NO avidly, and helped protect the norCB mutant in anaerobic biofilms. These data suggest that protection of a P. aeruginosa norCB mutant against anaerobic NO toxicity occurs by both control of NO supply and reassignment of metabolic enzymes to the task of NO sequestration.
Keywords: anaerobic nitrate regulator (ANR), anaerobic respiration, biofilms, nitric oxide, Pseudomonas aeruginosa
Introduction
The pathogen Pseudomonas aeruginosa (PA) (Holloway, 1969) is capable of aerobic or anaerobic growth. The latter is either via anaerobic respiration, using an inorganic oxynitrogen terminal electron acceptor (Gennis and Stewart, 1996), or slow growth by arginine substrate level phosphorylation (Mercenier et al, 1980). Anaerobic respiration requires nitrate (NO3−), nitrite (NO2−) or nitrous oxide (N2O) as terminal electron acceptors. NO3− reduction occurs via two routes, through an assimilatory pathway, where NO3− is reduced to NH3 and used as a nitrogen source, or by a dissimilatory pathway, where NO3− is reduced to N2 by respiration (Sias et al, 1980). Dissimilatory NO3− reduction occurs only under anaerobic conditions and involves a sequential eight-electron reduction of NO3− to N2, with intermediates that include NO2−, nitric oxide (NO) and N2O (Figure 1A). These reactions are catalyzed by metalloenzymes embedded in the inner membrane and periplasm. Loci involved in the four-step reduction of NO3− to N2 are termed nar (nitrate reductase) (Sias et al, 1980), nir (nitrite reductase) (Silvestrini et al, 1989), nor (nitric oxide reductase) (Arai et al, 1995), and, finally, nos (nitrous oxide reductase) genes (Arai et al, 2003), respectively.
Figure 1.
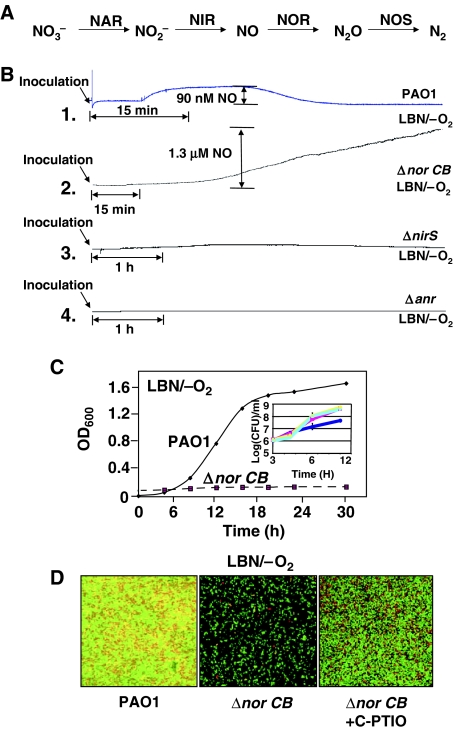
A PA norCB mutant maintains anaerobic viability despite high in vivo NO levels. (A) Anaerobic respiratory (denitrification) pathway. Enzymes involved in each reduction step are termed nitrate reductase (NAR), nitrite reductase (NIR), nitric oxide reductase (NOR) and nitrous oxide reductase (NOS), respectively. (B) NO tracings of wild type, norCB, nirS and anr mutants during anaerobic culture. Aerobic, stationary phase precultures were diluted 30-fold in 3 ml of LBN and placed in the NO electrode chamber. Before inoculation, the NO signal baseline was stabilized for at least 30 min. Note the different time scale between PAO1 and the mutant strains. (C) Anaerobic growth curve of wild-type and norCB mutant grown in LBN. Aerobic starter cultures were diluted 100- and 10-fold for wild-type and norCB mutant bacteria, respectively. Because of this, the initial turbidity was higher in the culture of the norCB mutant to demonstrate that anaerobic growth of the norCB mutant is virtually ceased. A 10-fold more initial bacterial load in norCB mutant, but OD600 remained constant. The inset indicates anaerobic wild-type and norCB mutant bacteria colony forming units in presence versus absence of 10 mM C-PTIO when grown in medium containing 10 mM KNO3 at 3, 6 and 12 h. Because the C-PTIO is stoichiometrically exhausted after 12 h, the final viable cell count was enumerated at this time point. Gray line (PAO1), white line (norcB mutant+C-PTIO), black line (norCB mutant). (D) Confocal images of anaerobic biofilms of wild-type PAO1 and the norCB mutant. Bacteria were grown anaerobically for 24 h in LBN and stained for viability assessment using the BacLite® live/dead stain. Green and red organisms represent those that are either live or dead, respectively.
Transcriptional control of genes involved in anaerobic respiration and arginine substrate level phosphorylation in PA is dependent upon ANR (anaerobic nitrate regulator), an FNR (fumarate/nitrate regulator/CRP)-like transactivator (Galimand et al, 1991; Lu et al, 1999). ANR is also required for transcription of the downstream regulator DNR (Arai et al, 1997). ANR also participates in controlling PA hydrogen cyanide production under reduced oxygen tension (Laville et al, 1998; Pessi and Haas, 2000), conditions that exist in the thick mucus lining the airways of cystic fibrosis (CF) patients (Worlitzsch et al, 2002; Yoon et al, 2002).
In denitrifying bacteria, the denitrification pathway is tightly controlled to minimize the adverse effects of NO, a toxic byproduct. During anaerobiosis, NO levels are maintained between 1 and 65 nM, depending on the bacterial species and the experimental conditions implemented (Goretski et al, 1990). Although NO toxicity mechanism(s) are unclear, accumulating evidence indicates that NO-derived nitrosative species can damage DNA (Woodmansee and Imlay, 2003) and compromise protein function by modifying moieties, including Fe–S clusters (Soum and Drapier, 2003), tyrosine residues (Schopfer et al, 2003), heme (Mayburd and Kassner, 2002) and sulfhydryl groups (Spallarossa et al, 2003). The potency of high intracellular NO was demonstrated by revealing that a mutant lacking the global quorum sensing regulator, RhlR, committed a metabolic suicide by overproduction of anaerobic NO (Yoon et al, 2002). More recently, acidified NO2− contributed to killing the mucoid form of PA, a variant that severely complicates the prognosis for CF patients, through NO evolution and/or through nitrosation of sulfur or metal centers downstream from HNO2 (Yoon et al, 2006).
The molecular basis underlying NO resistance in bacteria is poorly understood. In human disease, some intracellular pathogens replicate within host macrophages that produce high NO levels (Nathan and Shiloh, 2000). The most extensively studied proteins involved in NO detoxification are flavohemoglobin (Hmp or NOD (Hausladen et al, 2001) and flavorubredoxin (FlavoRb) in Escherichia coli. Hmp possesses an oxygen-dependent NO dioxygenase (NOD) activity (Gardner et al, 1998; Poole and Hughes, 2000), while FlavoRb possesses anaerobically induced NO reductase (NOR) activity (Gardner et al, 2002). Recent data indicate that Hmp is activated in response to NO even under anaerobic conditions, and NO-mediated modification of FNR, an E. coli ANR homolog, was suggested as a link between NO stress and anaerobic hmp transcription (Cruz-Ramos et al, 2002). NO exposure also triggers activation of the soxRS regulon, members of which include the superoxide dismutase (SOD) and antibiotic efflux pumps in both E. coli and Salmonella typhimurium (Vasil'eva et al, 2001; Coban and Durupinar, 2003). Because NO can react with superoxide (O2−) to produce highly destructive peroxynitrite (ONOO−), higher levels of SOD are required for protection of NO-treated bacteria (Nunoshiba et al, 1993). Similar to the binding of NO to FNR (Cruz-Ramos et al, 2002), oxidation of the [2Fe–2S]2+ cluster present in SoxR by NO leads to SoxS activation that subsequently results in transcription of genes under its control (Koo et al, 2003). In the CF airway, although low pH favors ONOO− protonation and ONOOH-associated toxicity (Yoon et al, 2002; Ricciardolo et al, 2004), anaerobic conditions may deprive NADPH oxidase of substrate (Worlitzsch et al, 2002).
Although some bacteria are capable of anaerobic growth using even NO as a terminal electron acceptor (Pichinoty et al, 1978), the major function of NOR is to detoxify NO generated anaerobically by NIR. In P. stutzeri, a norCB mutant perished when forced to respire via anaerobic NO3− reduction (Braun and Zumft, 1991). Previously, we showed that a norCB mutant of PA was also impaired in anaerobic NO3− reduction, yet the bacteria mysteriously survived under such conditions (Yoon et al, 2002). We postulated that these organisms adapted to anaerobic growth by either dramatically slowing their rate of growth or developing countermeasures to combat potentially toxic endogenous NO levels.
In this study, we addressed the basis for survival of the norCB mutant of PA and present evidence that it involves an elegant two-pronged defense mechanism. NO-linked control of ANR-mediated transcription is used to both stem the supply and purge the cytoplasm of NO by derepression of two dioxygenase encoding genes, whose gene products have inherent NO-scavenging capacity.
Results
A PA norCB mutant maintains anaerobic viability despite producing high endogenous NO levels
To determine the role of PA NOR in anaerobic growth, we first measured NO levels produced endogenously by strain PAO1 and a norCB mutant. In wild-type bacteria, NO was detected ∼7 min post-inoculation (where oxygen was depleted), reaching a maximum of ∼90 nM, and returning to baseline levels within 20 min (Figure 1B, line 1). In contrast, NO levels generated by the norCB mutant increased linearly to 1.3 μM over a 2-h incubation (Figure 1B, line 2). As expected, NO was not detected in a nirS mutant, that is deficient in NIR activity, and, as such, is incapable of generating anaerobic NO (Figure 1B, line 3). This supports the notion that the NO generated by the norCB mutant is mediated solely by NIR activity. In an anr mutant, where the enzyme involved in NO production (NIR) is not synthesized, NO was also not detected (Figure 1B, line 4). The amount of NO detected was independent of the initial NO3− concentration, suggesting that additional products downstream of nar and nir may be produced but are dependent on inoculum size, confirming that NO production is of biological origin (data not shown). During anaerobiosis, the growth rate of the norCB mutant was dramatically impaired, whereas wild-type bacteria demonstrated a typical growth curve (Figure 1C). The addition of the NO scavenger (2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (C-PTIO) allowed the norCB mutant to grow at a rate equivalent to that of wild-type bacteria (Figure 1C, inset). However, C-PTIO was stoichiometrically exhausted after 12 h of NO scavenging in the mutant strain (data not shown). No differences in aerobic growth rate or cell yield between PAO1 and the norCB mutant were observed (data not shown).
Others and we have demonstrated via a confocal laser scanning analysis of PA biofilms that a live/dead stain is useful in determining viability of biofilm bacteria (Yoon et al, 2002; Webb et al, 2003). Figure 1D shows that norCB mutant biofilm bacteria were mostly alive, although the cell density in these biofilms was significantly lower when compared to wild-type biofilms that harbored a mixture of live and dead cells. The addition of C-PTIO restored near wild-type biofilm growth. These results suggest that there is a dramatic and sustained increase in NO levels in the norCB mutant that persist for more than 2 h without a loss of viability of this strain. We next dissected the molecular basis underlying this phenomenon.
A PA norCB mutant has reduced NAR/NIR activity and nar/nir gene transcription relative to wild-type bacteria
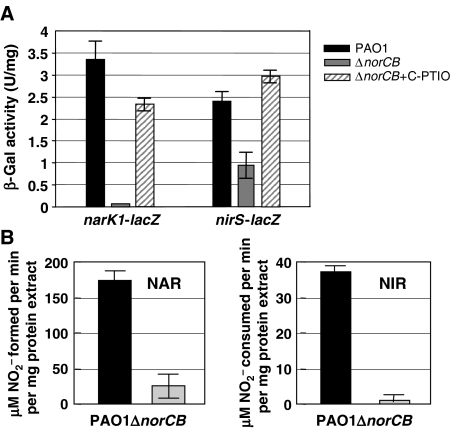
To elucidate the survival mechanism(s) of the norCB mutant under anaerobic conditions, we postulated that one strategy involves a downregulation of nar and nir transcription and concomitant NAR and NIR activities. Collectively, this would reduce the amount of NO that could kill the norCB mutant. First, single-copy narK1-lacZ and nirS-lacZ promoter fusions, representing the first genes of the PA operons involved in NAR and NIR biosynthesis, were constructed and placed in the neutral attB locus on the PA wild-type and norCB mutant genomes. Figure 2A shows that narK1 transcription was barely detectable, while that of nirS was reduced ∼2.5-fold in the norCB mutant when compared to wild-type expression. The reduction in narK1 and nirS transcription was specific, as levels of the constitutively expressed genes katA (encoding the major catalase, KatA (Hassett et al, 1999; Ma et al, 1999)), and crc (encoding the catabolite repressor control protein (MacGregor et al, 1991)) were not significantly altered (data not shown). Biochemical complementation with C-PTIO nearly restored both narK1-lacZ and nirS-lacZ to wild-type levels. Similarly, Figure 2B demonstrates that NAR and NIR activity were reduced ∼85 and 97%, respectively, in the norCB mutant relative to wild-type levels. Not surprisingly, the NO2− levels remaining in the norCB mutant were only ∼16 μM, whereas wild-type supernatants contained 12.3 mM (data not shown).
Figure 2.
The norCB mutant has dramatically reduced NAR/NIR activities and nar/nir gene transcription relative to wild-type bacteria. (A) Cell extracts of wild-type and norCB mutant bacteria were prepared after anaerobic growth in LBN for 24 h. Wild-type PAO1 and norCB mutant bacteria harboring single-copy narK1-lacZ and nirS-lacZ fusions were assayed for β-galactosidase reporter activity in triplicate and mean±s.e.m. is presented. C-PTIO at 10 mM was added for biochemical complementation purposes for the lacZ. (B) Nitrate (NAR) and nitrite (NIR) reductase activity of cell extracts of wild-type and norCB mutant bacteria after anaerobic growth in LBN for 24 h. Each assay was performed in triplicate and the mean±s.e.m. is shown.
Metabolic NO inactivates ANR, altering its metal center, resulting in a loss of DNA binding activity in the norCB mutant
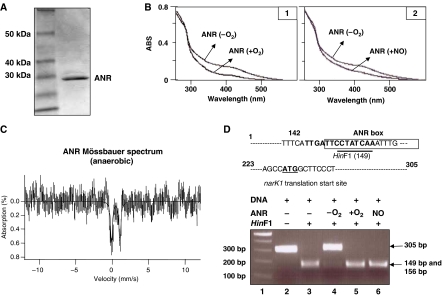
Since there was little or no narK1 and nirS transcription in the anaerobic norCB mutant (Figure 2B), we postulated that ANR-mediated transcription had been compromised. To determine if endogenous NO inactivated ANR in norCB bacteria, anaerobic ANR was first purified (Figure 3A). The physical characteristics of ANR were similar to E. coli FNR (Khoroshilova et al, 1997), with the monomeric Mr being ∼27 kDa (Figure 3A), while dimeric ANR was 54 kDa. ANR is brown, consistent with the presence of a [4Fe–4S]2+ cluster, as found in E. coli FNR (Lazazzera et al, 1996). This was confirmed by the UV/visible spectrum of Figure 3B, which shows the characteristic optical bands of a [4Fe–4S]2+ cluster with a shoulder at 320 nm and a broad peak with an absorption maximum of ∼420 nm. Upon exposure to air, the absorbance decreased in the visible region (Figure 3B1) (Khoroshilova et al, 1997). Because the absorbance at 420 nm did not drop to zero, but rather shifted to a slightly longer wavelength, it is likely that the putative [4Fe–4S]2+ cluster is not destroyed, but is rather converted to a [2Fe–2S]2+ cluster, as observed for FNR. However, we caution our interpretation of these data, for further experimental evidence is necessary to prove this postulate. A similar pattern of protein modification was also mediated by the addition of NO at levels generated endogenously by norCB mutant bacteria (Figure 3B2). To definitively identify the type of iron–sulfur cluster, 57Fe-ANR was prepared for Mössbauer spectroscopy. Fitting of a preliminary spectrum revealed the presence of a quadrupole doublet with an isomer shift δ≈0.43 mm/s and a quadrupole splitting of EQ≈1.2 mm/s, similar to that observed for the [4Fe–4S]2+ cluster of FNR (Figure 3C; Khoroshilova et al, 1997). Addition of O2 or NO to ANR broadened the quadrupole doublet, consistent with a change in the cluster, but the formation of a [2Fe–2S]2+ cluster could not be confirmed due to the poor signal to noise ratio of the spectrum (data not shown).
Figure 3.
Purification of PA ANR and inactivation by NO. (A) SDS–PAGE of purified ANR. Lane 1, molecular weight standard (kDa); lane 2, 1 μg of purified ANR. (B) Absorption spectra of anaerobic ANR (1.2 mg/ml), air-treated and NO-treated ANR. The latter treatment is a 1% gaseous NO (balance) argon exposure for 1 h. The ratio of 1% NO (19 μM NO in solution) to 1.2 mg/ml ANR protein is 1:1 on a molar molar basis. To obtain spectra of anaerobic and NO-treated ANR, samples (200 μl) were placed in sealed cuvettes in an anaerobic chambe and scanned. (C) The 4.2 K Mössbauer spectra of anaerobically prepared ANR. The solid line is a doublet for [4Fe–4S]2+ simulated with EQ=1.2 mm/s and δ=0.43 mm/s. (D) HinF1 restriction protection assay. The narK1 promoter sequence, in which an ANR box overlaps a HinF1 recognition site, was digested with HinF1 in the absence (lane 3) or presence of ANR (lanes 4, 5 and 6). Before digestion, the promoter sequence was incubated with anaerobic (lane 4), air-treated (lane 5) or NO-treated (lane 6) ANR for 30 min. DNA fragments after HinF1 digestion were separated on a 1.5% agarose gel and photographed. Lane 1, DNA ladder (New England Biolabs Inc.); lane 2, DNA only.
We next assessed the effect of NO on the ability of ANR to bind to its cognate recognition promoter sequence using a restriction site protection assay (D'Autreaux et al, 2002). Briefly, a 305-bp DNA fragment (Figure 3D, lane 2) harboring the narK1 promoter was incubated with ANR under anaerobic or aerobic conditions, or with NO-treated ANR under anaerobic conditions prior to HinF1 digestion. Complete HinF1 digestion resulted in two small fragments of 149 and 156-bp (Figure 3D, lane 3). Full-length DNA was expected when the proper binding of ANR to its cognate recognition sequences occurred. Consistent with this expectation, anaerobic ANR successfully protected the narK1 promoter from HinF1 digestion (Figure 3D, lane 4), while air- or NO-treated ANR failed to protect the DNA (Figure 3D, lanes 5 and 6). These results indicate that ANR loses its ability to bind DNA when exposed to physiological concentrations of anaerobic NO.
Two genes encoding oxygen-dependent enzymes are derepressed in the anaerobic norCB mutant
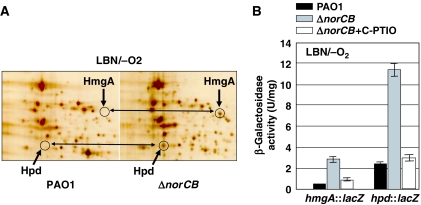
We next tested whether other compensatory mechanisms were used by the norCB mutant that enable it to survive anaerobic growth. We examined whole-cell protein synthesis profiles in anaerobic wild-type versus norCB mutant bacteria similar to Wu et al (2005). A total of 300 spots were reproducibly separated by two-dimensional (2-D) gel electrophoresis, and 16 proteins that showed altered expression were analyzed by Matrix-Associated Laser Desorption Ionozation-Time-of-Flight (MALDI-TOF) mass spectrometry (Supplementary Figure S1). Of these, 14 were identified and are listed in Supplementary Table S1. Several proteins were expected to be absent in the norCB mutant. These included the NirQ regulator (spot 11), NirS (spot 16) and OprE (spot 5) (Supplementary Figure S1). NirQ is required for expression of nir and nor genes and is controlled by ANR (Arai et al, 1994), and OprE expression is induced anaerobically (Yamano et al, 1998). The paucity of NirS further supported the significantly reduced NIR activity in the norCB mutant (Figure 2B). Interestingly, synthesis of two O2-dependent enzymes, homogentisate-1,2-dioxygenase (HmgA) and 4-hydroxyphenylpyruvate dioxygenase (Hpd) was highly induced in anaerobic norCB bacteria, and not detected in wild-type extracts (Figure 4A). Induced synthesis of these proteins in the norCB mutant was also verified transcriptionally by examining hmgA and hpd reporter activity. Figure 4B shows that hmgA and hpd transcription in anaerobic norCB bacteria was ∼4.2- and ∼3.8-fold higher than in anaerobic extracts of wild-type bacteria, respectively. The addition of C-PTIO to the norCB mutant significantly reducted both hmgA-lacZ and hpd-lacZ activity (clear bars).
Figure 4.
Upregulation of two oxygen-dependent enzymes in the anaerobic norCB mutant. (A) A portion of the norCB 2-D gel that contains spots for HmgA and Hpd proteins was compared with the same area of a 2-D gel from wild-type bacteria. A 60 μg weight of whole-cell extracts was separated in the 2-D gel system. The norCB mutant suspension was diluted 10-fold in the main anaerobic culture to obtain comparable amounts of cells with wild-type PAO1. Bacteria were grown in LBN under anaerobic conditions for 24 h. The identity of HmgA and Hpd was confirmed by MALDI-TOF mass spectrometric analysis. For the full 2-D gel images and the complete list of identified proteins, refer to the Supplementary data. (B) Measurements of transcript levels of hmgA and hpd in wild-type PAO1 (black bar), norCB mutant (gray bar) and norCB mutant bacteria +10 mM C-PTIO (white bars) under anaerobic conditions. Cultures were assayed for β-galactosidase activity in triplicate as described in the Materials and Methods and mean±s.e.m. is shown.
Crc was recently found to be upregulated 1.9-fold during anaerobic growth of PA and to negatively control aerobic expression of HmgA and Hpd in P. putida (Morales et al, 2004). In addition, ANR also controls transcription of dnr, encoding a nitrogen oxide-responsive regulator that, when overexpressed in an anr mutant background, restores anaerobic growth and nar, nir and nor transcription (Arai et al, 1997). Thus, we elected to test the hypothesis that the increased transcription of hmgA and hpd might be Crc- and/or DNR-mediated. To test this postulate, we constructed norCB crc and norCB dnr double mutants and measured hmgA and hpd transcription using lacZ reporter fusions. Our hypothesis was refuted, however, when we discovered that double norCB crc and norCB dnr mutants possessed identical hmgA-lacZ and hpd-lacZ as the norCB mutant (data not shown).
We next constructed isogenic mutants in each of the other 12 genes encoding proteins that were identified in our 2-D gel/MALDI-TOF analysis, and tested them for their ability to grow via anaerobic NO3− respiration (Supplementary Table S1, graphic to right). As expected, a nirQ mutant grew poorly when compared to growth of wild-type bacteria. Without NIR (a nirS mutant), PA cannot derive energy from NO2− respiration, although it is still capable of gaining energy from NO3− respiration. Thus, this strain was able to grow, albeit poorly. However, four mutants with defects in flagellin type-B synthesis, a probable aldehyde dehydrogenase (PA5312), the outer membrane protein OprE (PA0291) and an acetyl-CoA acetyltransferase (PA1999) showed a reduction in O.D.600 of ∼0.5.
HmgA and Hpd scavenge NO, thereby contributing to protection of anaerobic norCB bacteria in biofilms
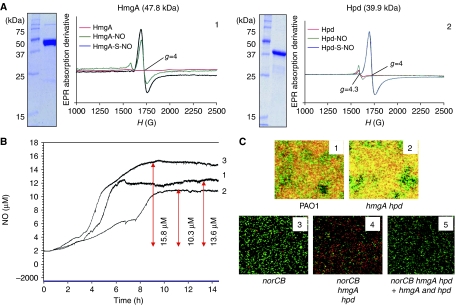
Because HmgA and Hpd are Fe(II) containing enzymes, and NO binds with high affinity to Fe(II) (Arciero et al, 1983; Ford et al, 2002; Ford and Lorkovic, 2002), we postulated that these proteins may contribute to protection of the norCB mutant by their inherent ability to scavenge NO. To test this hypothesis, recombinant HmgA and Hpd were purified (Figure 5A1, 2) and their enzymatic activities were confirmed (data not shown). The NO binding properties of both enzymes were next examined using liquid helium temperature electron paramagnetic resonance (EPR) spectroscopy. Figure 5A1, 2 clearly shows that NO bound avidly to HmgA and Hpd to produce a characteristic spectrum with intense resonances near g=4 (and 2) (Arciero and Lipscomb, 1986). The binding of homogentisate to HmgA caused only a small change in line shape, but the NO affinity is high with or without substrate. In the case of Hpd, 4-hydroxyphenylpyruvate binding caused a large increase in NO affinity, even at near stoichiometric NO levels, as is commonly observed for Fe(II) containing dioxygenases (Arciero et al, 1985). While we do not anticipate high levels of the Hpd substrate to be present under anaerobic conditions, these enzymes typically bind a wide range of aromatic compounds as inhibitors, and these complexes also result in a significant increase in NO affinity (Arciero et al, 1985).
Figure 5.
Binding of NO by HmgA and Hpd helps protect norCB mutant bacteria in anaerobic biofilms. (A) EPR spectra of 13 μM HmgA and Hpd in the absence or presence of NO. To establish that the Fe(II)–NO complex forms specifically at the catalytic center of these enzymes, spectra with the addition of stoichiometric levels of substrate (homogentisate for HmgA (HmgA-S-NO) and 4-hydroxyphenylpyruvate for Hpd (Hpd-S-NO)) are also shown. EPR conditions are as follows: temperature, 2 K; microwave frequency, 9.64 GHz; microwave power, 50 μW; modulation frequency 100 kHz; modulation amplitude, 10 G. Purity of HmgA or Hpd (lane 2) is shown by SDS–PAGE with molecular weight standards in kDa (lane 1). (B) NO tracings of norCB (line 1) and norCB hmgA hpd triple mutants (line 3) and the triple mutant+hmgA and hpd (line 2) during anaerobic culture. Experimental conditions were identical to those in Figure 1B. (C) Confocal images of anaerobic biofilms of wild-type PAO1 (panel 1), hmgA hpd (panel 2), norCB (panel 3), norCB hmgA hpd (panel 4), norCB hmgA hpd+hmgA and hpd in attB site (panel 5).
We next used a genetic approach to test the hypothesis that HmgA and Hpd could help protect the PA norCB mutant growing as anaerobic biofilms by constructing an isogenic norCB hmgA hpd triple mutant. Interestingly, NO levels in the norCB hmgA hpd mutant steadily increased to nearly 16 μM and did not return to baseline, while the norCB mutant produced 13.6 μM NO (Figure 5B). When hmgA and hpd were provided in cis, NO levels were reduced to 10.3 μM. An absence of the two dioxygenases in an hmgA hpd mutant had no adverse effects on anaerobic biofilm structure or viability, where NO was efficiently removed by NOR. However, anaerobic norCB (panel 3) and norCB hmgA hpd (panel 4) mutants formed very poor biofilms resulting from poor anaerobic growth of these strains (compare to wild-type bacteria, panel 1). Note, however, based upon the live:dead ratios, that the amount of dead bacteria was significantly higher in the norCB hmgA hpd mutant (>85%), relative to norCB single mutant (<1%). Finally, genetic in cis complementation of both hpd and hmgA in the attB site restored viability of the triple mutant to the norCB mutant phenotype (Figure 5C).
hmgA and hpd transcription is derepressed in norCB and anr mutants
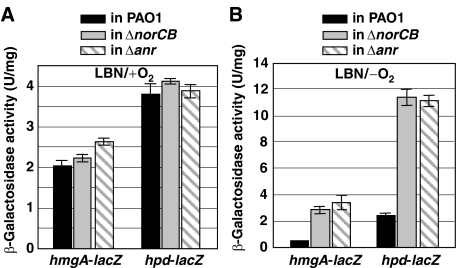
We next tested whether increased anaerobic hmgA and hpd transcription in the norCB mutant resulted from ANR inactivation. To test this postulate, single-copy hmgA-lacZ and hpd-lacZ promoter fusions were introduced into the anr mutant chromosome. In wild-type bacteria, hmgA and hpd transcription was predictably decreased under anaerobic relative to aerobic conditions (Figure 6A and B). Consistently, in separate microarray analyses, we found that hmgA and hpd gene expression was reduced by factors of 14.9- and 7.3-fold in anaerobic versus aerobic wild-type bacteria, respectively (data not shown). Under aerobic conditions, hpd transcription was >2-fold higher than comparable levels of hmgA in either PAO1, norCB and anr mutant strains, and no significant transcriptional differences were observed in these strains (Figure 6A). Under anaerobic conditions, however, hmgA and hpd expression was increased in the anr mutant to the same levels observed in the norCB mutant (Figure 6B), suggesting that anaerobic hmgA and hpd repression is ANR dependent. Taken together, these data suggest that the anaerobic behavior of the norCB mutant is nearly identical to that of the anr mutant vis-à-vis hmgA/hpd gene regulation. Finally, upregulation of hmgA and hpd in the anaerobic norCB mutant is likely because of an NO-mediated inactivation of ANR.
Figure 6.
norCB and anr mutants show similar anaerobic phenotypes with regard to hmgA and hpd regulation. (A, B) Measurements of transcript levels of hmgA and hpd in wild-type PAO1 (black bar), norCB mutant (gray bar) and anr mutant (hatched bar) under both aerobic (A) and anaerobic (B) conditions. Cultures were assayed for β-galactosidase activity in triplicate as described in Materials and Methods.
Discussion
The important CF pathogen PA forms highly problematic and antibiotic/phagocyte refractory biofilms while enmeshed in thick anaerobic airway mucus (Singh et al, 2000; Hassett et al, 2004). Furthermore, when the organism converts to the formidable mucoid form during the chronic stages of the disease, the pulmonary performance of CF patients diminishes dramatically (Govan and Harris, 1986). Although conventional antibiotics keep PA titers at roughly 108 CFU/g sputum, such titers still represent a huge problem, because the antimicrobial power of phagocytes in this complex niche is significantly diminished.
NO is an important antimicrobial component of our innate host defense system and a cell signaling molecule. However, in the context of treatment of recalcitrant PA biofilms enmeshed in the anaerobic airway mucus of CF patients, NO likely has little relevance from an antimicrobial perspective. Recently, however, we have unraveled two important clues to help eradicate this organism that relate to the ability of PA to cope with intracellular or extracellular NO. First, we discovered that paralysis of PA rhl quorum sensing during anaerobic biofilm growth, cause such organisms to commit a metabolic suicide by overproduction of metabolic NO (Yoon et al, 2002). This suggests that novel drugs that can paralyze PA quorum sensing could help eradicate PA from the CF airways. It is for this reason that many laboratories are currently examining quorum sensing inhibitors as potential therapeutic agents for treatment of CF and other infections (Passador et al, 1996; Smith et al, 2003; Suga, 2003; Kim et al, 2004). Second, the results of this study also builds on a major potential therapy for treating what we coined the ‘Achilles' heel' of mucoid PA that specifically involves an inability to cope with nitrogen oxides downstream from extracellular NaNO2 at the slightly acidic pH (6.4–6.5) of the CF airways (Yoon et al, 2006).
This study was initiated because of an intriguing observation that initially appeared to be a glaring biological paradox; a PA norCB mutant, that is incapable of detoxifying endogenously generated NO during anaerobic respiration, did not perish from its inherent toxicity. Our goal was to define the mechanism(s) and the machinery underlying the ability of this microorganism to resist metabolic NO. Our first experiments honed in on the major enzymes involved in PA anaerobic respiration. These include NAR and NIR, the only anaerobic enzymes that collectively mediate metabolic NO production, and the master regulator of nar and nir gene transcription, the anaerobic transcriptional regulator, ANR. Since we found that both nar and nir transcription and NAR and NIR enzymatic activity in the norCB mutant was markedly reduced relative to that of wild-type bacteria, we elected to examine whether the function of ANR was compromised. The E. coli ANR homolog, FNR, is sensitive to O2− and NO-mediated inactivation (Cruz-Ramos et al, 2002). The disruptive poisoning of the E. coli FNR [4Fe–4S]2+ cluster by O2 and NO causes a conversion of the [4Fe–4S]2+ cluster and a change in the oligomeric state of FNR. Thus, exposure of FNR to NO derepressed transcription of hmp, encoding flavohemoglobin (also called NOD (nitric oxide dioxygenase)) that is the major NO detoxifier in E. coli (Gardner et al, 1998; Poole and Hughes, 2000). Reduced DNA binding activity of FNR due to NO-mediated modification of [4Fe–4S]2+ was proposed as a molecular mechanism for NO sensing. In addition to FNR as a NO sensing regulator, CydR protein of Azotobacter vinelandii that is more homologous to ANR (88% identical) was also reported to show decreased binding to its cognate DNA recognition sequence when poisoned by NO (Poole and Hughes, 2000; Wu et al, 2000).
In the case of ANR, our spectrophotometric survey and restriction endonuclease protection assays clearly indicate that NO has deleterious effects on the ANR [4Fe–4S]2+ cluster and its concomitant transcriptional potential. A model of how NO regulates this process is depicted in Figure 7. It is of interest that iron-nitrosyl clusters can both be targets of S-nitrosothiol toxicity and promote S-nitrosothiol metabolism (Vanin et al, 2004), although airway acidification should promote S-nitrosothiol formation in the CF airway (Ricciardolo et al, 2004; Yoon et al, 2006). S-nitrosothiol levels in the CF airway are actually quite low, perhaps—at least in part—because of prokaryotic catabolism (Grasemann et al, 1999). One of the most compelling arguments indicating that the transcriptional capacity of ANR was compromised in the norCB mutant is the near identical behavior of anaerobic anr and norCB mutant bacteria in both planktonic and biofilm cultures.
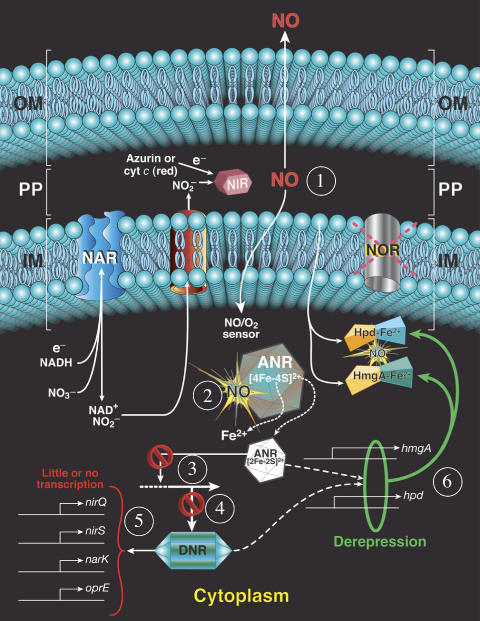
Figure 7.
Summary of the protective mechanisms used by a PA norCB mutant during anaerobic respiration. (1) NO is produced anaerobically in the periplasmic space by NIR and can either exit the cell or enter the cytoplasm; (2) Once in the cytoplasm, NO can inactivate dimeric ANR, causing release of iron from the [4Fe–4S]2+, causing conversion to a putative [2Fe–2S]2+ cluster, thus rendering it incapable of transcriptional activation of genes under its control (3), this is indicated by the ‘no' sign (e.g., 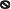 ). In contrast, genes that are anaerobically repressed, including hmgA and hpd, become derepressed, and function as NO scavengers; (4) because ANR is required for transcription of dnr, DNR is not produced; (5) thus, narK1, nirS, nirQ and oprE transcripts are either absent or extremely low (denoted by red X); (6) ANR, and perhaps DNR, denoted as a ‘?', however, loses it activity as a repressor. Thus, genes that would be repressed anaerobically, such as hmgA and hpd, are now derepressed (denoted by green circle). Because of the inherent NO-binding properties of HmgA and Hpd (refer to Figure 5A and B), elevated levels of these enzymes served to help protect the anaerobic norCB mutant against NO-mediated toxicity. OM, outer membrane; PP, periplasmic space; IM, inner membrane; NarK1/2, putative NO2− extrusion pump; CytC (red), reduced cytochrome c; Azurin, periplasmic protein donating electrons to NIR; e−, electron flow; NADH, electron donor for NAR.
). In contrast, genes that are anaerobically repressed, including hmgA and hpd, become derepressed, and function as NO scavengers; (4) because ANR is required for transcription of dnr, DNR is not produced; (5) thus, narK1, nirS, nirQ and oprE transcripts are either absent or extremely low (denoted by red X); (6) ANR, and perhaps DNR, denoted as a ‘?', however, loses it activity as a repressor. Thus, genes that would be repressed anaerobically, such as hmgA and hpd, are now derepressed (denoted by green circle). Because of the inherent NO-binding properties of HmgA and Hpd (refer to Figure 5A and B), elevated levels of these enzymes served to help protect the anaerobic norCB mutant against NO-mediated toxicity. OM, outer membrane; PP, periplasmic space; IM, inner membrane; NarK1/2, putative NO2− extrusion pump; CytC (red), reduced cytochrome c; Azurin, periplasmic protein donating electrons to NIR; e−, electron flow; NADH, electron donor for NAR.
DNR, another transcription factor in the same CRP/FNR family, is also essential for anaerobic growth in PA and is responsive to NO2− and NO, but not NO3− (Arai et al, 1997). Although in trans provision of DNR to the anr mutant restored the ability to denitrify, transcription of dnr is under the control of ANR, suggesting that ANR is the master regulator under anaerobic condition in PA (Arai et al, 1997). Construction of a norCB dnr mutant revealed that hmgA/hpd transcription was identical to that of an anaerobic norCB mutant (data not shown). Interestingly, it was also shown that the transcriptional activity of anr was >10-fold higher than that of dnr in anaerobic bacteria, suggesting that ANR is more abundantly synthesized than DNR under anaerobic conditions and plays a more important role in oxygen sensing and the initiation of the denitrification pathway (Arai et al, 1997). Still, Arai et al (1997) have suggested that the denitrification genes in PA are not directly controlled by ANR, but rather indirectly by DNR. However, the mechanistic basis behind this mode of transcriptional control is unknown. Structural studies have been initiated that hope to determine precisely how the DNR-based regulatory system functions.
Although anaerobic growth was significantly impaired because of repression of genes required for denitrification in the norCB mutant, fundamental biological anaerobic processes of this strain still remained active. This observation forced us to probe potential additional survival mechanisms of the norCB mutant under anaerobic conditions. Using MALDI-TOF analyses, the synthesis of eight of 16 identified proteins was increased in the norCB mutant compared to the wild-type strain. This clearly implies that the bacteria are at least partially capable of responding to high levels of NO and/or downstream metabolites. Two proteins, HmgA and Hpd, are involved in the tyrosine degradation pathway, which occurs under aerobic conditions. Oxidative decarboxylation followed by hydroxylation and ultimately cleavage of the aromatic ring catalyzed by dioxygen splitting are mediated by Hpd and HmgA, respectively, in two consecutive steps of the pathway (Milcamps and de Bruijn, 1999; Fritze et al, 2004). The dioxygenase class of enzymes, of which HmgA and Hpd are members, typically contains iron as a cofactor within their active site (Que and Ho, 1996). Dioxygenases that utilize Fe2+ bind NO with high affinity, and this affinity increases even further when they bind substrate (Arciero et al, 1983). Two well-characterized examples, catechol-2,3-dioxygenase from the related organism P. putida and protocatechuate-4,5-dioxygenase from P. testosteroni, have been shown to bind NO with Kd values well below 1 μM (Arciero et al, 1985). Definitive proof that HmgA and Hpd are capable of scavenging NO emerged from two separate experiments. First, we demonstrated by EPR spectroscopy that NO bound to both purified HmgA and Hpd (Figure 5A). Most importantly, we showed that HmgA and Hpd contributed to protection of the anaerobic norCB mutant in demonstrating that a triple norCB hmgA hpd mutant was killed during the anaerobic biofilm mode of growth because of sustained NO production (Figure 5B and C). The PA genome harbors 22 genes that could potentially produce at least 16 different dioxygenases (www.pseudomonas.com) Although at this juncture, it is not clear whether other dioxygenases or proteins capable of binding NO are also induced in response to NO stress and confer protective roles against NO, the results presented in Figure 5C clearly indicate that there are alternative functions of two aerobic enzymes under anaerobic conditions in the presence of high NO levels. Of note, this increased NO binding of HmgA/Hpd by anaerobic PA could account, at least in part, for the observation that antipseudomonal therapy increases exhaled concentrations of NO in CF patients (Jaffe et al, 2003), in the airways of whom PA growth is largely anaerobic (Worlitzsch et al, 2002; Yoon et al, 2002).
The dramatic upregulation of HmgA and Hpd in the anaerobic norCB mutant was initially perplexing, because it is counterintuitive to synthesize oxygen-dependent enzymes in an anaerobic environment. As expected, in wild-type strain PAO1, transcription of hmgA and hpd was highly repressed under anaerobic conditions (Figure 6A and B). Identical increases in hmgA and hpd transcription was also observed in both norCB and anr mutant strains. These results strongly suggest that repression of hmgA and hpd transcription during anaerobic growth is mediated by intact, dimeric ANR, and derepression of these genes in the norCB mutant is due to an NO-mediated inactivation of ANR. This evidence clearly establishes that the norCB mutant behaves identically to the anr mutant strain under anaerobic conditions.
In summary, this study describes a novel two-tiered, ‘circuit breaker' mechanism involving the paradoxical survival of a mutant lacking the protective enzyme, NOR, during anaerobic respiration by PA (Figure 7). We found that PA possesses an inherent homeostatic mechanism for maintaining NO levels at sublethal concentrations. First, we found that increased NO levels leads to a reduction in the expression of the enzymes that produce NO from NO3− and NO2− (NAR and NIR). Second, we found that the production of enzymes that can sequester NO (HmgA and Hpd) via an ANR-dependent derepression, were significantly increased under these conditions. Furthermore, we found that this unique protective regulatory duality occurs via the direct NO-mediated inactivation of the master anaerobic regulator, ANR, which contains an NO-sensitive [4Fe–4S]2+ cluster.
Materials and methods
Bacterial growth conditions
All bacteria were isogenic derivatives of PA strain PAO1. Bacteria were grown in either L-broth (10 g tryptone, 5 g NaCl, 5 g yeast extract per liter) or LB/1% KNO3 (LBN). Overnight cultures were grown aerobically in LB to stationary phase. Wild-type bacteria were diluted 100-fold and the norCB mutant was diluted 10-fold for anaerobic cultures, unless otherwise indicated. Anaerobic growth was achieved in a Coy anaerobic chamber (Coy labs, Grass Lake, MI).
Construction of isogenic mutants and lacZ transcriptional fusions
Isogenic PA mutants were constructed by two different allelic exchange procedures as described previously (Schweizer and Hoang, 1995). Other PA mutants were obtained from the University of Washington mutant library collection, each of which was confirmed by DNA sequencing. To construct single-copy lacZ transcriptional fusions, DNA fragments containing the promoter region of narK1 (representing the first gene of the nar operon, PA3877) and nirS (representing the first gene of nir operon, PA0519) were flanked by PstI and BspHI restriction sites. Three fragments of (i) an NcoI–SalI fragment of pZ1918 (Schweizer, 1993) containing the lacZ reporter gene with its own ribosomal binding site, (ii) PstI–BspH1 promoter sequences and (iii) PstI–SalI digest of mini-CTX1 (Hoang et al, 2000) were ligated, and putative clones were verified by DNA sequencing. E. coli SM10 harboring each transcriptional lacZ fusion was used as a donor strain in biparental matings with strain PAO1, and isogenic norCB, norCB crc, norCB dnr or anr mutants. Each promoter-lacZ construct was integrated at the nonessential attB locus within the PA chromosome, yielding single-copy transcriptional fusions (Hoang et al, 2000).
NO electrode measurements
NO levels were measured polarimetrically using a NO electrode system (World Precision Instruments Inc., Sarasota, FL). The chamber was prepared with a stably positioned electrode within the cavity of the bacterial suspension, as previously described (Gardner et al, 2003). All NO signals were calibrated from the stoichiometric conversion of acidified NO2− to NO as specified by the vendor. In an experiment to trace NO generated by anaerobically growing PA, strains were inoculated into the closed chamber through a port of ∼0.6 mm. A 100 μl volume of aerobic overnight culture was inoculated into 2.5 ml of LBN at 37°C. The NO signal was recorded and saved with APOLLO 4000™ software (World Precision Instruments Inc., Sarasota, FL). C-PTIO (2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide, Molecular Probes, Eugene, OR) or deoxy-hemoglobin, stoichiometric NO scavengers, were added to confirm that the signal is specific to NO, when needed.
Overexpression and purification of PA ANR, HmgA and Hpd
The hmgA and hpd genes were amplified by PCR and positionally cloned into pET23a (Novagen) and anr into pET19b (Novagen), and transformed into E. coli BL21 (DE3). The resulting pET19b-anr plasmid was transformed into E. coli Tuner™ (DE3, Novagen). For overexpression of HmgA and Hpd, E. coli was grown aerobically in LB, while for ANR overexpression, organisms were grown anaerobically in LBN/0.2% glucose. Expression was induced in mid-logarithmic phase cultures with 0.4 mM IPTG. Recombinant His-tagged ANR, HmgA or Hpd was bound to a nickel-NTA column (Qiagen, Valencia, CA) and eluted with 250 mM imidazole. ANR was purified under anaerobic conditions following the procedure used for purification of E. coli FNR (Sutton and Kiley, 2003). HmgA and Hpd activity assays were performed as previously described (Rodríguez et al, 2000; Kavana and Morna, 2003). ANR was prepared anaerobically and aliquots exposed either to ambient air or 1% NO gas balanced with argon for 1 h under anaerobic conditions.
Reporter assays
β-Galactosidase activity was measured as follows: PA strains were first grown to mid-logarithmic phase and the OD600 of each culture was measured. For norCB and anr mutants, the suspensions were diluted 10-fold from aerobic stationary phase cultures and incubated anaerobically at 37°C. A 100 μl volume of each culture was mixed with 0.9 ml of Z buffer (100 mM sodium phosphate buffer, pH 7.0, 10 mM KCl, 1 mM MgSO4, 25 mM β-mercaptoethanol). A 20 μl volume each of 2% SDS and chloroform were added to 1 ml aliquots of cell suspension, and the mixture was vortexed to facilitate cell lysis. After a 5-min incubation at 28°C, 0.2 ml of a 4 mg/ml solution of o-nitrophenyl-β-D-galactopyranoside was added, and the incubation was continued until development of a yellow color. Reactions were stopped by adding 0.5 ml of 1 M Na2CO3 and the reaction time was recorded. Cell debris was removed and the OD420 was measured immediately. Units of specific activity were expressed as OD420/(OD600 × min × ml of culture).
Microscopic examination of PA biofilms
For examination of biofilm architecture and cell viability, an eight-chambered coverslip system (Lab-Tek Inc., Campbell, CA) was used. LBN (0.4 ml) was inoculated with 4 μl of an aerobically grown overnight LB starter culture of wild-type and various isogenic mutants. All stationary phase strains grown aerobically had approximately the same value of CFU/ml. After 24 h at 37°C, anaerobic biofilms were washed three times with 0.9% saline and stained with 0.4 ml of a LIVE/DEAD BacLight bacterial viability stain for 15 min (Molecular Probes Inc., Eugene, OR). Images were acquired on a Zeiss LSM 510 laser scanning confocal unit attached to an Axiovert microscope using a 63 × 1.4 NA oil-immersion objective (Carl Zeiss MicroImaging Inc., Thornwood, NY). For two-color images, samples were scanned sequentially at 488 and 546 nm. Syto 9 (green fluorescence) was detected through a 505–530 nm bandpass filter and propidium iodine (red fluorescence) was detected through a 560 nm longpass filter and presented in two channels of a 512 × 512 pixel, 8-bit image.
NAR and NIR activity assays
NAR and NIR activities were measured using cell extracts as described previously, with minor modifications (Yoon et al, 2002). Briefly, 0.4 mM methyl viologen reduced by the addition of 2 mM sodium thiosulfite was used as the electron donor in the NAR activity assay, while 5 mM NADH coupled with 20 mM phenazine methosulfate (PMS) was used for NIR assays. The amount of NO2− in the reaction mixture was measured at specified intervals with the Griess reagent (Green et al, 1982). The increase or decrease in NO2− levels was measured using the NAR and NIR assays, respectively.
Optical spectroscopy
A Genesys 5 spectrophotometer (Spectronic, Rochester, NY) was used to record the optical spectrum of purified anaerobic ANR between 250–600 nm. Anaerobiosis was maintained by capping quartz cuvettes containing anaerobically purified and buffered ANR that were previously filled in the anaerobic chamber.
Restriction site protection assays
The 305-bp promoter region of narK1 (PnarK1), in which the ANR box overlaps a HinF1 restriction site, was amplified by PCR and cleaned using a PCR purification kit (Qiagen Inc., Valencia, CA). PnarK1 DNA (100 ng) was incubated with 2 μg of ANR for 30 min in 25 mM Tris–HCl (pH 7.8) and 50 mM NaCl at 37°C. All restriction digestions were performed by adding 1 U of HinF1 (New England Biolabs Inc., Beverly, MA) for 1 h. EDTA (25 mM) was added to stop each reaction. Agarose gel (1.5% agarose) electrophoresis was used to separate the DNA fragments.
2-D gel electrophoresis and MALDI-TOF analysis
2-D gel and in-gel trypsin digestion were performed as described previously (Yoon et al, 2002). MALDI-TOF was used to obtain all peptide mass fingerprinting profiles. Mass-to-charge (m/z) ratios of each fragment generated after trypsin digestion were queried from a public database (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm) and a protein with highest statistical certainty was selected for identification.
EPR and Mössbauer spectroscopy
EPR spectra of the Fe-nitrosyl complex of purified PA HmgA and Hpd was monitored as described previously (Hassett et al, 2000). Mössbauer spectra of 57Fe-labeled ANR were obtained following the procedures described elsewhere (Khoroshilova et al, 1997).
Supplementary Material
Supplementary Table S1
Supplementary Figure S1
Supplementary data
Acknowledgments
This work was supported by NIH grants AI-53079 (DJH), GM-24689 (JDL), Cystic Fibrosis Foundation New Technology grant (DJH), and a Distinguished Dissertation Fellowship from the University of Cincinnati College of Medicine (SSY). Each of the authors of this work declares no conflict of interest.
References
- Arai H, Igarashi Y, Kodama T (1994) Structure and ANR-dependent transcription of the nir genes for denitrification from Pseudomonas aeruginosa. Biosci Biotechnol Biochem 58: 1286–1291 [DOI] [PubMed] [Google Scholar]
- Arai H, Igarashi Y, Kodama T (1995) Expression of the nir and nor genes for denitrification of Pseudomonas aeruginosa requires a novel CRP/FNR-related transcriptional regulator, DNR, in addition to ANR. FEBS Lett 371: 73–76 [DOI] [PubMed] [Google Scholar]
- Arai H, Kodama T, Igarashi Y (1997) Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol Microbiol 25: 1141–1148 [DOI] [PubMed] [Google Scholar]
- Arai H, Mizutani M, Igarashi Y (2003) Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 149: 29–36 [DOI] [PubMed] [Google Scholar]
- Arciero DM, Lipscomb JD (1986) Binding of 17O-labeled substrate and inhibitors to protocatechuate 4,5-dioxygenase-nitrosyl complex. Evidence for direct substrate binding to the active site Fe2+ of extradiol dioxygenases. J Biol Chem 261: 2170–2178 [PubMed] [Google Scholar]
- Arciero DM, Lipscomb JD, Huynh BH, Kent TA, Munck E (1983) EPR and Mossbauer studies of protocatechuate 4,5-dioxygenase. Characterization of a new Fe2+ environment. J Biol Chem 258: 14981–14991 [PubMed] [Google Scholar]
- Arciero DM, Orville AM, Lipscomb JD (1985) [17O]Water and nitric oxide binding by protocatechuate 4,5-dioxygenase and catechol 2,3-dioxygenase. Evidence for binding of exogenous ligands to the active site Fe2+ of extradiol dioxygenases. J Biol Chem 260: 14035–14044 [PubMed] [Google Scholar]
- Braun C, Zumft WG (1991) Marker exchange of the structural genes for nitric oxide reductase blocks the denitrification pathway of Pseudomonas stutzeri at nitric oxide. J Biol Chem 266: 22785–22788 [PubMed] [Google Scholar]
- Coban AY, Durupinar B (2003) The effect of nitric oxide combined with fluoroquinolones against Salmonella enterica serovar Typhimurium in vitro. Mem Inst Oswaldo Cruz 98: 419–423 [DOI] [PubMed] [Google Scholar]
- Cruz-Ramos H, Crack J, Wu G, Hughes MN, Scott C, Thomson AJ, Green J, Poole RK (2002) NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J 21: 3235–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Autreaux B, Touati D, Bersch B, Latour JM, Michaud-Soret I (2002) Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc Natl Acad Sci USA 99: 16619–16624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Hughes MN, Wardman P (2002) Kinetics of the reactions of nitrogen dioxide with glutathione, cysteine, and uric acid at physiological pH. Free Radic Biol Med 32: 1314–1323 [DOI] [PubMed] [Google Scholar]
- Ford PC, Lorkovic IM (2002) Mechanistic aspects of the reactions of nitric oxide with transition-metal complexes. Chem Rev 102: 993–1018 [DOI] [PubMed] [Google Scholar]
- Fritze IM, Linden L, Freigang J, Auerbach G, Huber R, Steinbacher S (2004) The crystal structures of Zea mays and Arabidopsis 4-hydroxyphenylpyruvate dioxygenase. Plant Physiol 134: 1388–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M, Gamper M, Zimmermann A, Hass D (1991) Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol 173: 1598–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner AM, Gessner CR, Gardner PR (2003) Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and sigma54 in the nitric oxide stress response. J Biol Chem 278: 10081–10086 [DOI] [PubMed] [Google Scholar]
- Gardner AM, Helmick RA, Gardner PR (2002) Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J Biol Chem 277: 8172–8177 [DOI] [PubMed] [Google Scholar]
- Gardner PR, Gardner AM, Martin LA, Salzman AL (1998) Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci 95: 10378–10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennis RB, Stewart V (1996) Respiration. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds). Escherichia coli and Salmonella. Cellular and Molecular Biology, 2nd edn, pp 217–261. ASM Press: Washington, DC [Google Scholar]
- Goretski J, Zafiriou OC, Hollocher TC (1990) Steady-state nitric oxide concentrations during denitrification. J Biol Chem 265: 11535–11538 [PubMed] [Google Scholar]
- Govan JRW, Harris GS (1986) Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci 3: 302–308 [PubMed] [Google Scholar]
- Grasemann H, Gaston B, Fang K, Paul K, Ratjen F (1999) Decreased levels of nitrosothiols in the lower airways of patients with cystic fibrosis and normal pulmonary function. J Pediatr 135: 770–772 [DOI] [PubMed] [Google Scholar]
- Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR (1982) Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal Biochem 126: 131–138 [DOI] [PubMed] [Google Scholar]
- Hassett DJ, Lymar SV, Rowe JJ, Schurr MJ, Passador L, Herr AB, Winsor GL, Brinkman FSL, Lau GW, Yoon SS, Hwang SH (2004) Anaerobic metabolism by Pseudomonas aeruginosa in cystic fibrosis airway biofilms: role of nitric oxide, quorum sensing and alginate production. In: MM Nakano and P Zuber, Strict and Facultative Anaerobes: Medical and Environmental Aspects, Horizon Bioscience: Norfolk, England, pp 87–108 [Google Scholar]
- Hassett DJ, Ma J-F, Elkins JG, McDermott TR, Ochsner UA, West SEH, Huang C-T, Fredericks J, Burnett S, Stewart PS, McPheters G, Passador L, Iglewski BH (1999) Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol 34: 1082–1093 [DOI] [PubMed] [Google Scholar]
- Hassett DJ, Ochsner UA, Groce SL, Parvatiyar K, Ma J-F, Lipscomb JD (2000) Hydrogen peroxide sensitivity of catechol-2,3-dioxygenase of Pseudomonas putida: a cautionary note on the use of transcriptional xylE fusions during oxidative stress. Appl Environ Microbiol 66: 4119–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausladen A, Gow A, Stamler JS (2001) Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc Natl Acad Sci USA 98: 10108–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Kutchma AJ, Becher A, Schweizer HP (2000) Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43: 59–72 [DOI] [PubMed] [Google Scholar]
- Holloway BW (1969) Genetics of Pseudomonas. Bacteriol Rev 33: 419–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A, Slade G, Rae J, Laverty A (2003) Exhaled nitric oxide increases following admission for intravenous antibiotics in children with cystic fibrosis. J Cyst Fibros 2: 143–147 [DOI] [PubMed] [Google Scholar]
- Kavana M, Moran GR (2003) Interaction of (4-hydroxyphenyl)pyruvate dioxygenase with the specific inhibitor 2-[2-nitro-4-(trifluoromethyl)benzoyl]-1,3-cycIohexanedione. Biochemistry 42: 10238–10245 [DOI] [PubMed] [Google Scholar]
- Khoroshilova N, Popescu C, Munck E, Beinert H, Kiley PJ (1997) Iron–sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe–4S] to [2Fe–2S] conversion with loss of biological activity. Proc Natl Acad Sci USA 94: 6087–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim JG, Kang Y, Jang JY, Jog GJ, Lim JY, Kim S, Suga H, Nagamatsu T, Hwang I (2004) Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol Microbiol 54: 921–934 [DOI] [PubMed] [Google Scholar]
- Koo MS, Lee JH, Rah SY, Yeo WS, Lee JW, Lee KL, Koh YS, Kang SO, Roe JH (2003) A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J 22: 2614–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laville J, Blumer C, Von Schroetter C, Gaia V, Defago G, Keel C, Haas D (1998) Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J Bacteriol 180: 3187–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera BA, Beinert H, Khoroshilova N, Kennedy MC, Kiley PJ (1996) DNA binding and dimerization of the Fe–S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem 271: 2762–2768 [DOI] [PubMed] [Google Scholar]
- Lu CD, Winteler H, Abdelal A, Haas D (1999) The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J Bacteriol 181: 2459–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J-F, Ochsner UA, Klotz MG, Nanayakkara VK, Howell ML, Johnson Z, Posey J, Vasil ML, Monaco JJ, Hassett DJ (1999) Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J Bacteriol 181: 3730–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor CH, Wolff JA, Arora SK, Phibbs PV Jr (1991) Cloning of a catabolite repression control (crc) gene from Pseudomonas aeruginosa, expression of the gene in Escherichia coli, and identification of the gene product in Pseudomonas aeruginosa. J Bacteriol 173: 7204–7212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayburd AL, Kassner RJ (2002) Mechanism and biological role of nitric oxide binding to cytochrome c. Biochemistry 41: 11582–11591 [DOI] [PubMed] [Google Scholar]
- Mercenier A, Simon J-P, Vander Wauven C, Haas D, Stalon V (1980) Regulation of enzyme synthesis in the arginine deiminase pathway of Pseudomonas aeruginosa. J Bacteriol 144: 159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milcamps A, de Bruijn FJ (1999) Identification of a novel nutrient-deprivation-induced Sinorhizobium meliloti gene (hmgA) involved in the degradation of tyrosine. Microbiology 145 (Part 4): 935–947 [DOI] [PubMed] [Google Scholar]
- Morales G, Linares JF, Beloso A, Albar JP, Martinez JL, Rojo F (2004) The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J Bacteriol 186: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Shiloh MU (2000) Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci USA 97: 8841–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunoshiba T, de Rojas-Walker T, Wishnok JS, Tannenbaum SR, Demple B (1993) Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc Natl Acad Sci USA 90: 9993–9997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passador L, Tucker KD, Guertin KR, Journet MP, Kende AS, Iglewski BH (1996) Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J Bacteriol 178: 5995–6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G, Haas D (2000) Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J Bacteriol 182: 6940–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichinoty F, Garcia JL, Mandel M, Job C, Durand M (1978) Isolation of bacteria that use that use nitric oxide as a respiratory electron acceptor under anaerobiosis. C R Acad Sci Hebd Seances Acad Sci D 286: 1403–1405 [PubMed] [Google Scholar]
- Poole RK, Hughes MN (2000) New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol Microbiol 36: 775–783 [DOI] [PubMed] [Google Scholar]
- Que L Jr, Ho RY (1996) Dioxygen activation by enzymes with mononuclear non-heme iron active sites. Chem Rev 96: 2607–2624 [DOI] [PubMed] [Google Scholar]
- Ricciardolo FL, Sterk PJ, Gaston B, Folkerts G (2004) Nitric oxide in health and disease of the respiratory system. Physiol Rev 84: 731–765 [DOI] [PubMed] [Google Scholar]
- Rodríguez JM, Timm DE, Titus GP, Beltran-Valero De Bernabe D, Criado O, Mueller HA, Rodriguez De Cordoba S, Penalva MA (2000) Structural and functional analysis of mutations in alkaptonuria. Hum Mol Genet 9: 2341–2350 [DOI] [PubMed] [Google Scholar]
- Schopfer FJ, Baker PR, Freeman BA (2003) NO-dependent protein nitration: a cell signaling event or an oxidative inflammatory response? Trends Biochem Sci 28: 646–654 [DOI] [PubMed] [Google Scholar]
- Schweizer HP (1993) Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15: 831–833 [PubMed] [Google Scholar]
- Schweizer HP, Hoang TT (1995) An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158: 15–22 [DOI] [PubMed] [Google Scholar]
- Sias SR, Stouthamer AH, Ingraham JL (1980) The assimilatory and dissimilatory nitrate reductases of Pseudomonas aeruginosa are encoded by different genes. J Gen Microbiol 118: 229–234 [DOI] [PubMed] [Google Scholar]
- Silvestrini MC, Galeotti CL, Gervais M, Schinina E, Barra D, Bossa F, Brunori M (1989) Nitrite reductase from Pseudomonas aeruginosa: sequence of the gene and the protein. FEBS Lett 254: 33–38 [DOI] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP (2000) Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407: 762–764 [DOI] [PubMed] [Google Scholar]
- Smith KM, Bu Y, Suga H (2003) Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs. Chem Biol 10: 81–89 [DOI] [PubMed] [Google Scholar]
- Soum E, Drapier JC (2003) Nitric oxide and peroxynitrite promote complete disruption of the [4Fe–4S] cluster of recombinant human iron regulatory protein 1. J Biol Inorg Chem 8: 226–232 [DOI] [PubMed] [Google Scholar]
- Spallarossa A, Forlani F, Pagani S, Salvati L, Visca P, Ascenzi P, Bolognesi M, Bordo D (2003) Inhibition of Azotobacter vinelandii rhodanese by NO-donors. Biochem Biophys Res Commun 306: 1002–1007 [DOI] [PubMed] [Google Scholar]
- Suga H (2003) Quorum sensing in Pseudomonas aeruginosa: cell to-cell communication to sense the cell density of the same species. Tanpakushitsu Kakusan Koso 48: 1609–1615 [PubMed] [Google Scholar]
- Sutton VR, Kiley PJ (2003) Techniques for studying the oxygen-sensitive transcription factor FNR from Escherichia coli. Methods Enzymol 370: 300–312 [DOI] [PubMed] [Google Scholar]
- Vanin AF, Papina AA, Serezhenkov VA, Koppenol WH (2004) The mechanisms of S-nitrosothiol decomposition catalyzed by iron. Nitric Oxide 10: 60–73 [DOI] [PubMed] [Google Scholar]
- Vasil'eva SV, Stupakova MV, Lobysheva II, Mikoyan VD, Vanin AF (2001) Activation of the Escherichia coli SoxRS-regulon by nitric oxide and its physiological donors. Biochemistry (Moscow) 66: 984–988 [DOI] [PubMed] [Google Scholar]
- Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S (2003) Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol 185: 4585–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodmansee AN, Imlay JA (2003) A mechanism by which nitric oxide accelerates the rate of oxidative DNA damage in Escherichia coli. Mol Microbiol 49: 11–22 [DOI] [PubMed] [Google Scholar]
- Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Wei T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G (2002) Reduced oxygen concentrations in airway mucus contribute to the early and late pathogenesis of Pseudomonas aeruginosa cystic fibrosis airway infection. J Clin Invest 109: 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Cruz-Ramos H, Hill S, Green J, Sawers G, Poole RK (2000) Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr). Sensitivity to oxygen, reactive oxygen species, and nitric oxide. J Biol Chem 275: 4679–4686 [DOI] [PubMed] [Google Scholar]
- Wu M, Guina T, Brittnacher M, Nguyen H, Eng J, Miller SI (2005) The Pseudomonas aeruginosa proteome during anaerobic growth. J Bacteriol 187: 8185–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano Y, Nishikawa T, Komatsu Y (1998) Involvement of the RpoN protein in the transcription of the oprE gene in Pseudomonas aeruginosa. FEMS Microbiol Lett 162: 31–37 [DOI] [PubMed] [Google Scholar]
- Yoon SS, Coakley R, Lau GW, Lymar SV, Gaston B, Karabulut AC, Hennigan RF, Hwang SH, Buettner G, Schurr MJ, Mortensen JE, Burns JL, Speert D, Boucher RC, Hassett DJ (2006) Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J Clin Invest 116: 436–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock RE, Parsek MR, Noah TL, Boucher RC, Hassett DJ (2002) Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell 3: 593–603 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1
Supplementary Figure S1
Supplementary data