Abstract
Resistance to tamoxifen is observed in half of the recurrences in breast cancer, where the anti-estrogen tamoxifen acquires agonistic properties for transactivating estrogen receptor α (ERα). In a previous study, we showed that protein kinase A (PKA)-mediated phosphorylation of serine 305 (S305) of ERα results in resistance to tamoxifen. Now, we demonstrate that phosphorylation of S305 in ERα by PKA leads to an altered orientation between ERα and its coactivator SRC-1, which renders the transcription complex active in the presence of tamoxifen. This altered orientation involves the C-termini of ERα and SRC-1, which required a prolonged AF-1-mediated interaction. This intermolecular reorientation as a result of PKA-mediated phosphorylation of ERα-S305 and tamoxifen binding provides a unique model for resistance to the anticancer drug tamoxifen.
Keywords: estrogen receptor α, FRET, protein kinase A, SRC-1, tamoxifen resistance
Introduction
Tamoxifen is a highly effective anticancer drug in estrogen receptor α (ERα)-positive breast cancer patients. In recurrent disease however, still half of the patients develop resistance, where tamoxifen acquires agonistic properties for transactivation of ERα. Various mechanisms may account for insensitivity to tamoxifen, including activation of the mitogen-activated protein kinase (MAPK), protein kinase A (PKA) and p21-activated kinase-1 (PAK-1) signaling pathways that show enhanced activity in tamoxifen-resistant breast tumors (Michalides et al, 2004; Gutierrez et al, 2005; Holm et al, 2006). These kinases may directly target ERα. However, the molecular details of how these events contribute to tamoxifen resistance remain elusive. Antagonists of ERα act by altering the orientation of the C-terminally located α-helix 12 of the ligand binding domain (LBD) of ER (Brzozowski et al, 1997). In the agonist-bound state, cofactors bind to the pocket composed of helices 3, 4, 5 and 12 (Pike, 2006). Anti-estrogens induce a distortion in α-helix 12 covering this binding pocket, thereby preventing the association with the p160 family of coactivators (Shiau et al, 1998). These cofactors are essential to initiate transcription (Kamei et al, 1996) and include SRC-1 (or NcoA-1), SRC-2 (also known as TIF-2, GRIP1 or NcoA-2) and SRC-3 (also known as RAC3, ACTR, AIB1, P/CIP or TRAM) (Xu and Li, 2003). SRC-1 can interact with CREB binding protein (CBP), as well as with both the N-terminal AF-1 and the C-terminal AF-2 domains of ERα (Metivier et al, 2001). The AF-1 and AF-2 domains cooperate in transactivation of ERα (Metivier et al, 2002; Dutertre and Smith, 2003). Activity of SRC-1 is modified by phosphorylation at multiple sites, two of which are attributed to PKA activation (Rowan et al, 2000a). The SRCs can have different cellular properties: while SRC-1 and SRC-3 can be recruited to unliganded ERα, SRC-3 is more readily displaced from ERα in the presence of antagonists as compared to SRC-1 (Sharp et al, 2006). Overexpression of SRC-1 and SRC-3 is correlated with tamoxifen resistance in breast cancer patients (Osborne et al, 2003; Myers et al, 2004). Moreover, agonistic activity of tamoxifen is enhanced by overexpression of SRC-1 in normal uterus tissue (Shang and Brown, 2002).
Previously we reported that tamoxifen resistance mediated by PKA is caused by phosphorylation of serine 305 (S305) of ERα (Michalides et al, 2004). Recently, S305 of ERα was also reported to be the target of PAK-1 (Wang et al, 2002), and its overexpression correlates with resistance to tamoxifen in breast cancer patients (Holm et al, 2006). These findings identify S305 as a crucial site in ERα that upon phosphorylation by either PKA or PAK-1 is responsible for resistance to tamoxifen. This phosphorylation switches tamoxifen from an antagonist to an agonist of ERα, and affects conformational changes in ERα following binding to tamoxifen and other anti-estrogens (Michalides et al, 2004; Zwart et al, 2007). The mechanistic details of this process are, however, still unclear. Here, we report that PKA-mediated phosphorylation of ERα alters the orientation between ERα and coactivator SRC-1, without affecting the overall binding between ERα and SRC-1 in tamoxifen-treated cells. We applied fluorescent resonance energy transfer (FRET) in living cells to visualize changes in orientation between ERα and SRC-1 following tamoxifen binding, which was dependent on phosphorylation of the PKA target S305 in ERα. This affected the recruitment of RNA polymerase II and led to ER-mediated transcription in cells treated with tamoxifen. In summary, tamoxifen resistance via PKA or PAK-1 occurs through phosphorylation of S305 in ERα, which alters its orientation towards coactivator SRC-1 and recruitment of RNA polymerase II, thereby stimulating ER-driven transcription by the anti-estrogen tamoxifen.
Results
PKA phosphorylates ERα-S305
To follow the immediate interactions between ERα and its cofactor SRC-1 in cells transfected with tagged ERα and SRC-1 constructs, U2OS cells were used as a model cell line, since they are easily transfectable. However, the same results were also obtained in HeLa cells and in breast cancer cells T47D and MCF7 (Michalides et al, 2004; this study). To show that S305 is indeed phosphorylated by PKA, U2OS cells transfected with ERα were treated with 8-Br-cAMP, to activate PKA. This resulted in a specific phosphorylation of ERα-S305 (Figure 1A). The mutant ERαS305A, where the S305 was replaced with an alanine to prevent phosphorylation of that site by PKA, was not detected with the ERα-S305-specific antibody. The ERα-S305 phosphate-specific antibody did not detect the putative phosphomimics of ERα-S305, ERαS305E or ERαS305D, where S305 was replaced with glutamate or aspartate, respectively. We therefore preferred wild-type ERα treated with PKA activators 8-Br-cAMP or forskolin, with ERαS305A as a control. The concentration of (anti)-estrogens used in the study was similar to those in the previous study (Michalides et al, 2004), and were optimally effective for the levels of ERα expressed in the cells.
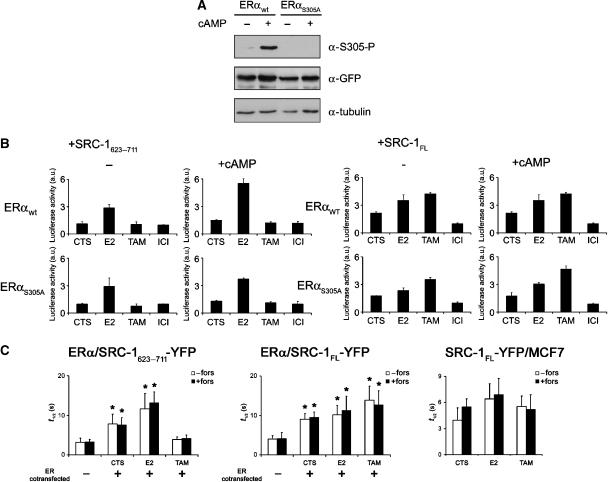
Figure 1.
Binding between ERα and SRC-1 is not affected by PKA activation. (A) Characterization of S305-ERα as PKA target. U2OS cells were transfected with the wild-type YFP-ERα-CFP construct or the S305A mutant thereof, cultured in the presence or absence of 8-Br-cAMP and analyzed for S305 phosphorylation of wild-type ERα or the phospho-mutant of ERα. Anti-tubulin staining was used as a loading control, anti-GFP as an expression control. Absence of the phosphorylated S305-ERα protein in the cells transfected with S305A, but its presence in the PKA-treated cells transfected with wild-type indicates the inability to phosphorylate mutant ERαS305A by PKA. (B) M2H analysis of ERα/SRC-1 interactions. U2OS cells transfected with DBD-SRC-1 (623–711 or full length), TA-ERα (wt or S305A), GAL4-luciferase and Renilla luciferase DNA were cultured in medium containing CTS only, or in the presence of 1 μM Estradiol (E2), 1 μM ICI 182,780 or 1 μM 4′OH-tamoxifen for 96 h. Twenty-four hours before analysis 100 μM 8-Br-cAMP was added, where indicated. Luciferase activity was measured and related to ICI 182,780 values without cAMP, for every transfectant, and set to 1. Bars indicate standard deviations from three independent experiments. (C) FRAP analysis of ERα/SRC-1 interactions. U2OS cells were transfected with SRC-1623−711-YFP (left panel) or SRC-1FL-YFP (middle panel), and ERα-CFP, MCF7 cells were transfected with SRC-1FL-YFP only (right panel). These were cultured in CTS containing medium, and 1 μM Estradiol (E2) or 1 μM 4′OH-tamoxifen was added 15 min before analysis, where indicated. Cells were pretreated with 10 μM forskolin for 15 min, where indicated. YFP was bleached and fluorescence intensities were followed in time in the bleach spot from which t1/2 (half-time to recovery) was calculated as described in Materials and methods. Bars indicate standard deviations from >10 cells per condition. Student t-test was performed for each bar, compared to SRC-1623−711-YFP or SRC-1FL-YFP expression alone. *P<0.05.
Phosphorylation of ERα-S305 by PKA does not influence overall binding to SRC-1
Since ligand-induced activation of ERα is generally followed by binding to a coactivator, we hypothesized that insensitivity to tamoxifen by PKA-mediated phosphorylation of S305 would affect the binding between ERα and cofactor SRC-1. This was first investigated in a mammalian two-hybrid (M2H) assay. To distinguish between the interaction of SRC-1 with AF-1 and/or AF-2 domains of ERα, the SRC-1 truncation mutant, aa 623–711 (SRC-1623−711) was used that only binds to the ligand binding AF-2 domain of ERα, as described previously (Llopis et al, 2000), as well as the full-length SRC-1 (SRC-1FL), which interacts with both AF-1 and AF-2 domains of ERα (Metivier et al, 2001). Expression of the fusion proteins was confirmed by Western blotting (Supplementary Figure S1). Chimeras of ERα and the transactivation domain of GAL4 (TA-ERα), as well as SRC-1623−711 or SRC-1FL fused to the DNA binding domain of GAL4 (DBD-SRC-1), were cotransfected with a GAL4-responsive luciferase expression construct and Renilla luciferase construct as a transfection control (Figure 1B). In this assay, luciferase activity is directly related to the binding between the two fusion proteins. SRC-1623−711 interacted with ERα in the presence of E2 and this was only slightly increased by preincubation of the cells with PKA activator 8-Br-cAMP (Figure 1B, left panel). No binding was observed under tamoxifen and ICI 182,780 conditions, also not in the presence of 8-Br-cAMP. The binding of full-length SRC-1 was increased by E2 and, surprisingly, also by the partial antagonist tamoxifen (Figure 1B, right panel). Treatment with the full ERα antagonist ICI 182,780 resulted in a loss of binding. ICI 182,780 not only inhibits ERα activity but also induces degradation of ERα, and defined the inactive state of ERα in our measurements. The interaction between ERα and SRC-1FL was not influenced by 8-Br-cAMP under all tested conditions. These results indicated that PKA activation does not affect the overall binding between ERα and SRC-1. In addition, whereas binding of SRC-1 to the AF-2 domain of ERα is induced by E2 and was abrogated by tamoxifen (Llopis et al, 2000), binding of SRC-1 to the AF-1 domain of ERα prevailed under hormone-depleted conditions and in the presence of tamoxifen, as was reported previously (Metivier et al, 2001).
In order to verify the M2H results in living cells and in a dynamic context, we visualized interactions between ERα and SRC-1 using fluorescence recovery after photobleaching (FRAP) in U2OS cells. In FRAP, fluorophores in the region of interest are bleached using a high-intensity laser beam and recurrence of fluorescence is followed in time, as described before (Stenoien et al, 2001), and illustrated in Supplementary Figure S2. The mobility of the fluorophore-tagged protein of interest, whether alone or in (transient) complex with interaction partners, can be monitored in this way. The statistics of the t1/2 recovery values are given in Figure 1C. SRC-1623−711 interacts with the AF-2 domain of ERα. Upon expressing SRC-1623−711 in the absence of ERα, the t1/2 value was low (indicating rapid diffusion in the nuclear compartments) and was not changed after addition of E2 or tamoxifen (data not shown). When SRC-1623−711 was coexpressed with ERα, introduction of the hormone E2 decreased the mobility of SRC-1623−711, suggesting interactions with ERα following binding to E2. The mobility of SRC-1623−711 under conditions of charcoal-treated serum (CTS) was slower than in the presence of tamoxifen, and was not affected by treatment with forskolin that activates PKA. The t1/2 recovery values for SRC-1FL indicated increased interactions between ERα and SRC-1FL under conditions of CTS, E2 and tamoxifen as well, since they were higher than the t1/2 recovery value of SRC-1FL-YFP alone (Figure 1C, middle column). The latter was not affected by E2 or tamoxifen (data not shown). The mobility of SRC-1FL-YFP in breast cancer MCF7 cells showed a similar pattern as was obtained in U2OS cells cotransfected with exogenous ERα (Figure 1C, right column), indicating that the FRAP results are not cell type specific and are in U2OS cells with exogenous ERα comparable to the breast cancer MCF7 cells with an endogenous ERα only.
The combined results of the M2H and FRAP experiments indicated that SRC-1FL can interact with ERα in hormone-depleted (CTS) as well as in E2 and tamoxifen-treated cells. They strongly suggested that the AF-1 domain of ERα is involved in binding under all three conditions, whereas the AF-2 domain is participating in binding only in the presence of E2. Importantly, PKA activation, which is known to induce tamoxifen resistance, did not influence the interaction between SRC-1 and ERα.
Phosphorylation of S305 by PKA alters the orientation between ERα and SRC-1 under tamoxifen conditions
If binding is not affected by PKA-mediated phosphorylation of ERα-S305, does this phosphorylation have an effect on the mode of interaction between ERα and SRC-1? We investigated this by FRET technology using confocal imaging in HeLa cells. FRET is the radiationless energy transfer from one fluorophore to the next, and occurs when two dipole moments of overlapping fluorophores couple within a distance of ∼80 Å. Intermolecular FRET thereby provides a most direct method to study interactions between two proteins, and is strictly dependent on the distance between the fluorophores as well as their relative orientations, allowing visualization of conformational changes within protein–protein complexes. In these experiments and in the FRAP experiments described above, we used cells with a 2- to 3-fold expression of exogenous ERα-CFP as compared to endogenous ERα expression in MCF7 cells (Figure 2A). FRET was detected between ERα-CFP and SRC-1623−711-YFP (Figure 2B); here, excitation of CFP at 430 nm yielded emission of YFP that, after correction of leak-through, only could have arisen from FRET (lower left panel). The observed FRET signal was normalized for the amount of CFP emission as is described in Materials and methods, yielding the corrected donor FRET efficiency (ED) that is presented in the lower right panel. ED represents the FRET per donor–acceptor fluorophore pair, and is independent of donor fluorescence. Under hormone-depleted conditions, a high ED was observed for ERα-CFP and SRC-1623−711-YFP, which was strongly reduced by tamoxifen (Figure 2B), indicating that the interaction in CTS between ERα and SRC-1623−711 was altered by tamoxifen. When FRET efficiency was quantified by determining the ED per pixel, as described in Materials and methods, we observed that the average efficiency of 16% FRET between ERα-CFP and SRC-1623−711-YFP under hormone-depleted conditions was reduced to an average of 5% after addition of tamoxifen to this particular cell (Figure 2D) and others (Figure 2E). Note that the spread in pixel ED values is due to contribution of noise. PKA activation by forskolin under conditions of CTS did not significantly influence FRET efficiency between ERα and SRC-1623−711 (Figure 2C, and quantified in Figure 2E). Subsequent treatment with tamoxifen still led to a substantial reduction in FRET efficiency. This indicated that PKA activation did not influence the binding between the AF-2 domain of ERα and SRC-1623−711-YFP, which agrees with the results obtained by FRAP and M2H (Figure 1).
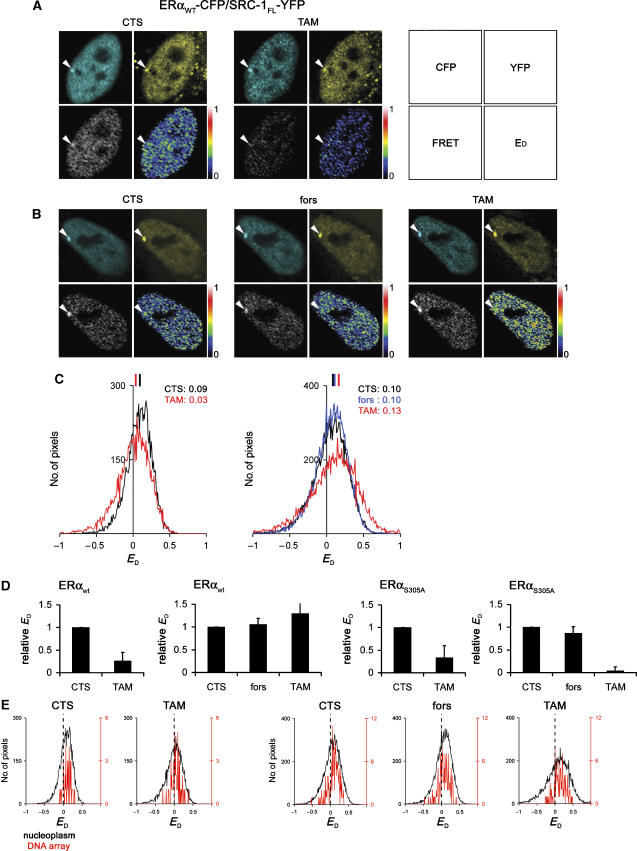
Figure 2.
Orientation between ERα and SRC-1623−711 is altered in the presence of tamoxifen and is independent of phosphorylation of S305 of ERα by PKA. (A) Expression levels of ERα-CFP related to endogenous expression. MCF7 cells were transfected with ERα-CFP, fixed and stained for ERα. Cells with CFP-positive nuclei were compared to exclusively endogenous ERα expressing cells. FRET measurements were performed on PRL array containing HeLa cells, transfected with ERαwt-CFP and SRC-1623−711-YFP. (B, C) FRET images were generated from cells cultured in hormone-depleted (CTS) medium only (B and C, left panels) or after treatment with 10 μM forskolin before FRET measurements (C, middle panel). Subsequently, 1 μM 4′OH-tamoxifen (TAM) was added and after 15 min cells were imaged (B and C, right panels). FRET and donor FRET efficiency (ED) were calculated as described in Materials and methods. For each condition described in panels B and C, the raw ED values from the whole nucleus were related to the total amount of pixels. (D) Mean ED is indicated and quantifications (E) are performed on >10 independent measurements, where the ED under CTS conditions is set to 1 for each experiment (relative ED). Bars indicate standard deviations. Bars at the right indicate relative intesity.
We next investigated whether PKA treatment had any effect on the orientation between full-length SRC-1 and ERα using FRET (Figure 3). FRET between ERα and SRC-1FL was less efficient as with SRC-1623−711, which may be attributed to the much larger protein size of the full-length SRC-1 and hence the larger distance between the fluorophores at the ends of the tagged proteins, but may also be the result of different orientation of the fluorophores. FRET between ERα-CFP and SRC-1FL-YFP was detected in hormone-deprived cells and was reduced by tamoxifen (Figure 3A), supporting the FRAP experiments, where these proteins could be found to interact under hormone-deprived conditions. The mean FRET efficiency of 9% in CTS was reduced to 3% when tamoxifen was added (Figure 3C). This is remarkable, since the M2H and FRAP experiments demonstrated that the interaction between ERα and SRC-1FL was not affected by tamoxifen (Figure 1). This suggested that a loss of FRET under conditions of tamoxifen indicated an altered orientation between ERα and SRC-1, where the C-termini of ERα and SRC-1FL that carry the fluorophores position differently in tamoxifen-treated cells than under CTS conditions. We next investigated whether activation of PKA influenced this reorientation upon tamoxifen binding. PKA activation through forskolin had no effect on FRET efficiency under hormone-deprived conditions (Figure 3B). However, subsequent treatment with tamoxifen did not show alterations in FRET efficiency analogous to cells not pretreated with forskolin, indicating that the orientation between ERα and SRC-1FL is stabilized by activation of PKA. The ERαS305A mutant behaved similar as wild-type ERα under non-PKA stimulated conditions: forskolin treatment followed by tamoxifen still resulted in a loss of FRET (Figure 3D, right panel and Supplementary Figure S3). This indicates that phosphorylation of wild-type ERαS305 by PKA affects the orientation between ERα and full-length SRC-1 (quantified in Figure 3D and E). These data were verified in U2OS (not shown) and MCF7 cells (Supplementary Figure S4D–F), indicating that the results of these FRET experiments were cell type independent. These results were further confirmed by fluorescence lifetime imaging microscopy (FLIM), which is an alternative FRET approach (Zwart et al, 2005) (Supplementary Figure S4A–C). Taken together, these data showed that the orientation between ERα-CFP and SRC-1FL-YFP is altered by tamoxifen, unless PKA phosphorylates S305 in ERα.
Figure 3.
Orientation between ERα and SRC-1FL is altered by tamoxifen alone, but is stabilized by activation of PKA. FRET experiments were performed analogous as described at Figure 2, now using SRC-1FL-YFP and wild-type ERα (A–D) or ERαS305A (D, right panel). Arrows indicate the PRL array (A, B), for which the raw ED values (including noise) were related to the total amount of pixels and presented for both the PRL array (in red) and entire nucleus (in black) for the cells shown in panels A and B (E). Bars at the right indicate relative intensity (A, B).
ERα interacts in the nucleus with SRC-1 irrespective of DNA promoter content
Given the limits of resolution of the light microscope and the relatively low number of specific estrogen response elements (ERE) in the genome (Carroll et al, 2006), it is not possible to directly relate high FRET efficiency between ERα and SRC-1 to ERE binding (Figures 3 and 4). Since the transcription function of ERα can be mediated by direct interaction with EREs at the promoters of target genes, we determined FRET signals in HeLa cells that contain a multicopy prolactin promoter/enhancer (PRL) DNA array containing natural ERα binding sites. Transcription from this visible locus is regulated by ligand-bound ER, and ERα and recruited cofactors can be readily identified (Sharp et al, 2006). This permitted a clear distinction between defined ERE-associated ERα and SRC-1 events on the PRL array and in the remainder of the nucleus, as shown in Figure 3A and B (indicated by arrowheads). Here, we compared the FRET efficiency on the PRL array with that throughout the remainder of the nucleus (Figure 3E). The levels of ERα-CFP, SRC-1-YFP proteins and FRET events were significantly higher on the PRL array than outside, as shown in Figure 3A and B. However, when FRET from these cells was related to the emission of the donor (ED) to render the readout fluorophore concentration independent, similar ED values were obtained on the array as compared to levels outside the array in the nucleus. Also, the effects on ED of tamoxifen alone or in combination with forskolin on the PRL array were comparable with those detected outside the array (Figure 3E). These results indicated that the interaction detected between ERα and SRC-1FL on a genuine ER binding domain, the PRL array, reflects the interaction at other sites in the nucleus.
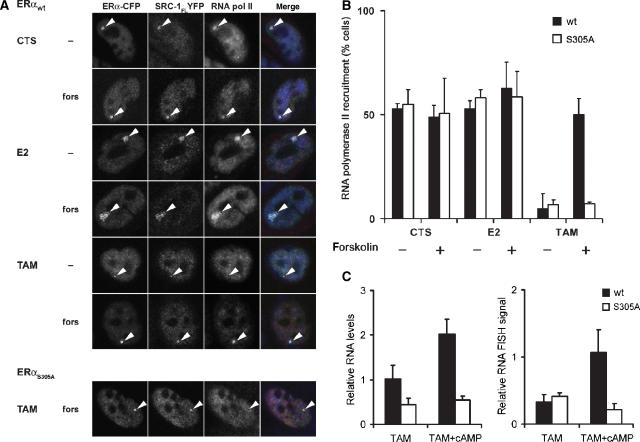
Figure 4.
PKA-mediated phosphorylation of S305 of ERα is responsible for RNA polymerase II recruitment on the PRL array and for enhanced transcription of a hormone-responsive reporter gene under conditions of tamoxifen. (A) Staining for RNA polymerase II was performed on PRL array containing HeLa cells that were transfected with ERαwt-CFP or ERαS305A-CFP and SRC1FL-YFP and cultured in hormone-depleted (CTS) medium. Where indicated, they were pretreated with 10 μM forskolin. Subsequently cells were treated with 1 μM Estradiol (E2) or 1 μM 4′OH-tamoxifen for 2 h and then fixed and stained for RNA polymerase II. Arrowheads indicate the PRL array that was analyzed for RNA polymerase II staining. (B) Quantification of the RNA polymerase II signals on the PRL array under conditions as described in Figure 6A. Quantifications are from three independent experiments, n>40 cells each. Bars indicate standard deviations. (C) PKA-mediated phosphorylation of S305 on ERα enhances transcription of the PRL array under conditions of tamoxifen. QPCR was performed on PRL array containing HeLa cells, transfected with ERαwt-CFP or ERαS305A-CFP, which were treated with 1 μM tamoxifen or left untreated (CTS). Sixteen hours before analysis, 100 μM 8-Br-cAMP was added, where indicated. mRNA levels were measured as described in Materials and methods, and related to CTS values, which was set to 1. Bars indicate standard deviation from two independent experiments. RNA FISH of the hormone-responsive DsRed2 reporter gene behind the PRL-based promoter in PRL-HeLa transfected with ERαwt-CFP or ERαS305A-CFP. Two hours before analysis, cells were treated with forskolin, where indicated, and subsequently with 1 μM 4′OH-tamoxifen. Signal intensity of the FISH DsRed2, as visualized by Alexa-546, is shown. Bars indicate standard deviation from two independent experiments.
Phosphorylation of serine-305 of ERα by PKA enables recruitment of RNA polymerase II to the PRL transcription locus, enhancing transcription under tamoxifen conditions
Genuine tamoxifen resistance would imply that the effect of PKA-induced phosphorylation of ERα-S305 on the positioning of SRC-1 and ERα following exposure to tamoxifen would lead to tamoxifen-mediated transactivation of ERα, with recruitment of RNA polymerase II to the ERα/SRC-1 complex as a hallmark. Therefore, we investigated the RNA polymerase II recruitment in the PRL array containing cells that were transfected with ERα-CFP and SRC-1FL-YFP, and examined colocalization of ERα and SRC-1 with RNA polymerase II on the PRL array. Approximately one-half of the cells expressing both ERαwt-CFP and SRC-1FL-YFP showed colocalization with RNA polymerase II on the PRL array under conditions of CTS and E2 (Figure 4A, and quantified in Figure 4B). The PRL array did show a more ‘open', decondensed structure due to Estradiol (E2) when compared to hormone-depleted conditions (CTS). This open chromatin structure was previously reported to be tightly linked with increased transcriptional activity (Sharp et al, 2006). In our experiments, activation of PKA by forskolin did not affect colocalization of the three proteins (ERα, SRC-1 and RNA polymerase II) under conditions of CTS and E2. As reported previously (Sharp et al, 2006), tamoxifen-treated cells showed a more condensed PRL array, on which both ERα and SRC-1, but no RNA polymerase II was recruited (Figure 4A). Strikingly, activation of PKA under tamoxifen conditions now resulted in recruitment of RNA polymerase II to the array, indicating that PKA activation influenced the orientation between ERα and SRC-1, such that RNA polymerase II recruitment to the PRL array is facilitated under tamoxifen conditions. The S305A mutant of ERα did not result in RNA polymerase II recruitment under identical conditions (Figure 4A and Supplementary Figure S5). This indicates that RNA polymerase II recruitment to the ERα complex in tamoxifen-treated cells requires phosphorylation of ERαS305 by PKA.
To directly investigate whether these events resulted in enhanced transcription, we measured transcription of the integrated DsRed2 reporter gene that is under control of the ER-responsive elements on the PRL array (Sharp et al, 2006) by quantitative RT–PCR and RNA FISH as described in Materials and methods (Figure 4C). PKA activation led in tamoxifen-treated cells transfected with ERαwt to a two-fold increase in transcription of the DsRed2 gene, as compared to tamoxifen alone, whereas no such effect was observed in cells transfected with ERαS305A. Tamoxifen exposure resulted in a similar PKA-induced increase in DsRed2 transcripts, which was specifically associated with the PRL array, as shown by RNA FISH in cells transfected with wild-type ERα, but not in ERαS305A-transfected cells.
Our results indicated that tamoxifen resistance is due to a change in the orientation between ERα and SRC-1, which was dependent on PKA-mediated phosphorylation of ERαS305. This reorientation between the C-termini of ERα and SRC-1FL induced RNA polymerase II recruitment, resulting in enhanced transcription.
Interaction between ERα and AF-1 binding domain of SRC-1 is maintained under tamoxifen conditions
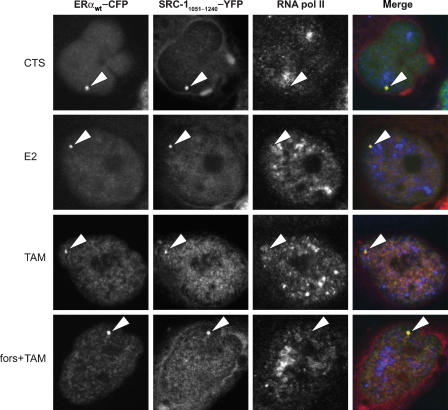
Which domain outside the AF-2 binding domain on SRC-1 is responsible for the sustained interaction between ERα and the full-length SRC-1 following tamoxifen exposure, as was observed in the FRET experiments? To study this, we determined colocalization of ERα-CFP, RNA polymerase II and an AF-1 binding domain of SRC-1 (Onate et al, 1998; Webb et al, 1998; Glass and Rosenfeld, 2000; Metivier et al, 2001). We reduced the SRC-1 fragment, which was previously reported to interact with the AF-1 of ERα by others (Metivier et al, 2001), to a SRC-11051−1240 fragment still showing interaction with the AF-1 of ERα (data not shown). We observed colocalization of ERα-CFP and SRC-11051−1240-YFP on the PRL array under conditions of CTS, E2 and tamoxifen, and also under tamoxifen conditions after PKA activation (Figure 5). No RNA polymerase II was recruited under these conditions, which is explained by a dominant-negative function of the SRC-11051−1240 mutant. This may also be indicated by the condensed shape of the PRL array in cells, also in the presence of E2, which is representative for the lack of transcriptional activity (Sharp et al, 2006).
Figure 5.
SRC-11051−1240-YFP recruitment to the PRL DNA array in the presence of tamoxifen. Staining of RNA polymerase II was performed on PRL array containing HeLa cells, transfected with ERαwt-CFP and SRC11051−1240-YFP and cultured in medium containing only CTS. Subsequently, cells were treated with 1 μM E2 or 1 μM 4′OH-tamoxifen for 2 h and then fixed and stained for RNA polymerase II. Arrowheads indicate the PRL DNA array that was analyzed for RNA polymerase II staining.
The condensed shape of the PRL array in the presence of E2 when ERα was coexpressed with the dominant-negative SRC-11051−1240 mutant also indicated that other endogenous SRCs do not compensate or otherwise influence our measurements. The combined results indicated that PKA-mediated phosphorylation of ERα-S305 in the presence of tamoxifen resulted in a reorientation between the C-termini of ERα and SRC-1, whereas the binding is sustained via an interaction between ERα and the AF-1 binding domain of SRC-1 in SRC-11051−1240. This altered orientation between the C-termini of ERα and SRC-1 resulted in RNA polymerase II recruitment and transcriptional activity under tamoxifen conditions, and explains how PKA activation can result in tamoxifen resistance.
Discussion
Regulated gene expression is achieved through the coordinated assembly of transcription factors, cofactors and the basal transcription machinery to transcription start sites, and demands a spatio-temporal coordination of interactions between these components. Traditional models of transcription tend to be static and depend on overall interactions measured by various kinds of binding assays, whereas the generation of transcription factor complexes requires a fine-tuned recruitment of components depending mainly on affinity of interfaces. The present study shows that the orientation of a tamoxifen-bound ERα transcription factor towards its coactivator SRC-1 is altered by PKA-mediated phosphorylation of S305 in the hinge region of ERα. This reorientation is responsible for RNA polymerase II recruitment and ER-dependent transcription in the presence of tamoxifen, as is illustrated in Figure 6. This reorientation was observed by a direct intermolecular FRET in living cells (Figures 2 and 3) that measures distance and/or orientation between the two fluorescently tagged proteins. Other methods, such as M2H and FRAP (Figure 1), measure protein–protein interactions, respectively, interactions with more static complexes and stability of these complexes. These M2H and FRAP approaches did not show an effect of PKA on overall binding between ERα and SRC-1 under tamoxifen conditions. However, this PKA-mediated reorientation of ERα leads to recruitment of RNA polymerase II and enhanced transcription from a hormone-responsive reporter gene (Figure 4). Our data indicate that the orientation of interfaces between ERα and SRC-1 in the transcription complex is crucial for efficacy of transcription and provides a molecular mechanism for tamoxifen resistance.
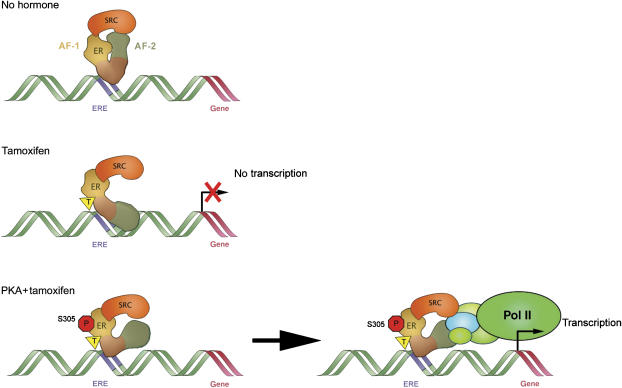
Figure 6.
A model of the molecular mechanism of the PKA- or PAK1-induced tamoxifen resistance. In the presence of tamoxifen, SRC-1 binds to the AF-1 but not to the AF-2 domain of ERα. Phosphorylation of ERα-S305 by PKA results in an altered orientation between the C-termini of ERα and SRC-1, which differs from the orientation under conditions of CTS. This ERα-S305-specific phosphorylation by PKA leads to recruitment of RNA polymerase II and to elevated transcription in the presence of tamoxifen.
We found no differences in the FRET efficiency resulting from the interactions of ERα with SRC-1 on the PRL array, as compared to FRET signals scattered in the nucleoplasm (Figure 3E). This indicates that ERα/SRC-1 interactions can be visualized throughout the nucleus, and that their relative orientations are unaltered by DNA binding. Possibly such complexes are not assembling at the transcription start sites, but exist as preformed complexes in the nucleoplasm, as also suggested by others (Zheng et al, 2005). At present, it is technically impossible to determine if the nucleoplasmic hot spots are sites of active genes containing EREs, or non-promoter-associated EREs.
The PKA-mediated phosphorylation of S305 in the hinge region of ERα is responsible for a reorientation of the C-terminus of ERα under tamoxifen conditions, which we measured by FRET using C-terminally tagged ERα-CFP and SRC-1-YFP constructs (Figures 2 and 3). This reorientation alters the interaction between the C-termini of ERα and SRC-1 and depends on prolonged association via AF-1 of ERα (Figure 5). This was inferred from the absence of interactions between the AF-2 binding SRC-1 fragment and ERα under tamoxifen conditions. In addition, this AF-2/SRC-1 interaction was also not induced by PKA activation (Figures 1B, C and 2), whereas the AF-1 binding SRC-1 fragment did interact with ERα under tamoxifen conditions (Figure 5). Our results thus support previous findings that AF-1 binding is a prerequisite for resistance to tamoxifen (Glaros et al, 2006), whereas functional synergy between AF-1 and AF-2 enhances SRC-1 recruitment and subsequent transcription (Metivier et al, 2001). Since the PKA effect on orientation is visualized only in the presence of tamoxifen, and not under conditions of CTS, the AF-1 domain should be regarded as a ligand dependent (in this case tamoxifen-dependent) transactivation domain that requires PKA activation. The AF-2 binding fragment of the SRC-1 construct that we used in this study as an AF-2 probe, encompasses aa 623–711 and contains two LXXLL motives that interact with the cofactor binding pocket in the AF-2 of the ERα-LBD (Llopis et al, 2000). The results with this AF-2 probe in our M2H and FRAP experiments indicate that it binds to ERα under conditions of E2 and CTS, but not in the presence of tamoxifen. This binding was not influenced by activation of PKA. The full-length SRC-1FL, however, binds to ERα under all three conditions when measured by M2H and FRAP, and was also observed on the PRL array in colocalization studies (Sharp et al, 2006) and FRET (Figure 3), as was also the case for the AF-1 binding SRC-11051−1240 fragment (Figure 5). This binding is also not affected by PKA activation. Since SRC-1FL binds to both AF-1 and AF-2 domains of ERα (Metivier et al, 2001), and SRC-623−711 only to the AF-2 domain, our results indicate that the AF-1 functional core in ERα, which is positioned at the start of the B-domain (aa 39–45 of ERα) (Metivier et al, 2001), is involved in the interaction between ERα and the 1051–1240 domain of SRC-1 in tamoxifen-treated cells. Indeed, an SRC-1 mutant, previously described to bind the AF-1 region of ER (Metivier et al, 2001), was still capable of interacting with ER on the PRL array in the presence of tamoxifen (Figure 5). Interaction between this AF-1 helical core and SRC-1 has been reported to be essential for ERα activity in the presence of tamoxifen (Glaros et al, 2006), and is most likely involved in the PKA-mediated reorientation between ERα and SRC-1 that we observed in our experiments under tamoxifen conditions. The outcome of our M2H and FRAP experiments, that measure interactions between ERα and SRC-1FL molecules, was not influenced by tamoxifen. The outcome of the FRET experiments that measure distance and orientation between them was, however, affected by tamoxifen. From these results, we concluded that the orientation between the C-termini of ERα and SRC-1FL, rather than the interaction between the two proteins, was altered by tamoxifen. This tamoxifen-associated alteration is prevented by phosphorylation of S305 of ERα by PKA, which stabilizes in the presence of tamoxifen the orientation towards SRC-1 (Figure 3 and Supplementary Figure S4). These findings led to a model where PKA-mediated phosphorylation of S305 results in a conformational state of ERα, by which the orientation between ERα and SRC-1 facilitates recruitment of RNA polymerase II under conditions of tamoxifen. The effect of PKA on the conformation of ERα involves most likely the reorientation between the N- and C-termini of ERα that we have observed by intramolecular FRET, as has been reported before (Michalides et al, 2004). This intramolecular change was dependent on PKA-mediated phosphorylation of S305 and resulted in enhanced transactivation of ERα- and tamoxifen-dependent proliferation after PKA activation. In tamoxifen-resistant ER-positive patients, PKA activity was found increased, underlining the clinical relevance of these findings. The results of the present study strongly indicate that the change of ERα conformation upon PKA activation results in a permissive orientation between ERα and SRC-1 that facilitates transcription under tamoxifen conditions. Besides the phosphorylation of direct target sites in ERα, the phosphorylation of SRC-1 (Rowan et al, 2000b) and reduced association between ERα and transcriptional corepressors NCoR and SMRT (Wagner et al, 1998) may contribute to PKA-mediated resistance to anti-estrogens. Since the PKA-induced reorientation, the RNA polymerase II recruitment and enhanced transcription were not observed when the ERαS305A mutant was used (Figure 3D and Supplementary Figure S3), we conclude that phosphorylation of S305 by PKA is the main target for PKA-induced tamoxifen resistance that may still act in conjunction with phosphorylation of SRC-1 and NCoR/SMRT.
The reorientation of the C-terminus of tamoxifen-bound ERα toward SRC-1 that is induced by either PKA- or PAK-1-mediated phosphorylation of S305 in the hinge region of ERα, provides a unique model for resistance to tamoxifen. It demonstrates that the effect of interacting agents can be nullified by activation of other signaling pathways, adding to the complexity of estrogen-mediated transcriptional events and complicating interfering strategies. This mechanism also provides a framework for selection and development of agents that are insensitive to these modifications on ERα, and contributes to identification of conditions for tamoxifen resistance in breast cancer patients.
Materials and methods
Plasmids, cell culture and transfections
Human osteosarcoma U2OS, MCF7 and HeLa cells were cultured in DMEM medium in the presence of 10% FCS and standard antibiotics. Cells containing ERα were cultured in phenol red-free DMEM medium containing 5% CTS (Hyclone), 48 h before analysis.
For characterization of the ERα-S305 phosphorylation of ERα by PKA, U2OS cells were transfected with the YFP-ERα-CFP construct or the S305A variant thereof (Michalides et al, 2004), treated with 100 μM 8-Br-cAMP (Sigma) and analyzed by Western blotting using antibodies against GFP (van Ham et al, 1997), tubulin (Sigma) or phospho-S305-ERα (Upstate, USA). Cloning strategies are described in the Supplementary data.
M2H assay
U2OS cells were cultured in 12-well plates, 96 h before analysis. Twenty-four hours later, cells were transfected using PEI (polyethylenimine, MW 25 kDa (Polysciences Inc.)) (Boussif et al, 1995) with TA-ERαwt or TA-ERαS305A (0.5 μg), DBD-SRC-1-YFP (full length or aa 623–711) (0.5 μg), GAL4-responsive Luciferase reporter construct (0.5 μg) and ER-insensitive Renilla luciferase construct (1 ng) as control for transfection efficiency (Michalides et al, 2004). Four hours after transfection, medium was replaced with Dulbecco's medium without phenol red, supplemented with 5% CTS (Hyclone) and cultured in the absence or presence of E2, ICI 182,780 (Tocris) or 4′OH-tamoxifen (Sigma), at a final concentration of 1 μM. Twenty-four hours before analysis, medium with (anti-) estrogens was replaced and 8-Br-cAMP (Sigma) was added where indicated, at a final concentration of 100 μM. Luciferase activity was determined as described previously (Bindels et al, 2002).
FRAP is described in the Supplementary data.
FRET imaging by sensitized emission
HeLa cells were cultured on coverslips for 48 h before imaging. Twenty-four hours before imaging, cells were transfected with the constructs indicated, and medium was replaced with Dulbecco's medium without phenol red, supplemented with CTS. Mel-JuSo cells, stably transfected with pcDNA3 constructs containing only CFP or YFP, were included to the culture for leak-through corrections and internal controls. Coverslips were placed in 2 ml bicarbonate/Hepes-buffered saline and analyzed in a heated tissue culture chamber at 37°C under 5% CO2. Where indicated, forskolin was added at a final concentration of 10 μM for 15 min. FRET between CFP and YFP molecules was determined by calculating the sensitized emission (the YFP emission upon CFP excitation) using separately acquired donor and acceptor images, as described previously (Zwart et al, 2005). In short, images were acquired on a TCS-SP2 confocal microscope (Leica). Three images were collected: CFP (excited at 430 nm and detected between 470 and 490 nm), indirect YFP (excited at 430 nm and detected between 528 and 603 nm) and direct YFP (excited at 514 nm and detected between 528 and 603 nm). Because of considerable overlap of CFP and YFP spectra, YFP emission was corrected for leak-through of CFP emission, and for direct excitation of YFP during CFP excitation. FRET was calculated using correction factors obtained from cells expressing either CFP or YFP alone, which were included for every image, as described before (van Rheenen et al, 2004). Then the apparent ED was calculated by relating the FRET to the total emission of the donor cell, after which the ED image was overlaid with a false color look up table. Using these methods, differences in FRET efficiency can be measured with an accuracy of 0.5% (Zwart et al, 2005). For graphic representation, the ED was calculated for each pixel from the raw data files of the represented cell, and was exported to Microsoft Excel. Here, the amount of pixels was related to the corresponding ED, and plotted as a histogram (Zwart et al, 2005).
RNA polymerase II recruitment assay
For CLSM analysis, prolactin promoter/enhancer (PRL) array containing HeLa cells (Sharp et al, 2006) were cultured in Dulbecco's medium containing CTS and were transfected with ERαwt-CFP or ERαS305A-CFP and SRC-1FL-YFP or the 1051–1240 truncation mutant thereof, using PEI. After 4 h, the medium was replaced with Dulbecco's medium without phenol red, supplemented with 5% CTS. Cells were cultured in CTS only or supplemented with 10 μM forskolin for 15 min. Subsequently, cells were treated for 2 h with 1 μM Estradiol, 1 μM 4′OH-tamoxifen or left untreated, thereafter fixed with 3.7% formaldehyde in PBS and subsequently stained with anti-RNA polymerase II antibody 8WG16 (Covanche Research Products Inc.) and secondary antibodies conjugated to Alexa 633 (Molecular Probes, Leiden, The Netherlands). Images were obtained with a Leica TCS SP2 System equipped with a × 63 oil-immersion objective. CFP was excited at 458 nm, and emission measured at 460–500 nm. YFP was excited at 514 nm, and emission measured at 528–600 nm. Alexa 633 was excited at 633 nm, and emission measured at 645–720 nm.
Quantitative RT–PCR and RNA FISH
Prolactin promoter/enhancer (PRL−) array containing HeLa cells (Sharp et al, 2006) were transfected with ERαwt-CFP or ERαS305A-CFP using electroporation, and subsequently cultured in Dulbecco's medium containing CTS. Immediately after seeding the cells, 1 μM 4′OH-tamoxifen was added or the cells were left untreated. After 6 h, 8-Br-cAMP was added, where indicated, at a final concentration of 100 μM. After 16 h, cells were lysed and RNA was extracted using Trizol (Invitrogen), according to the manufacturer's protocol. RNA was reverse transcribed using SuperScript(tm) III Reverse Transcriptase (Invitrogen), on which QPCR was performed using CYBR Green (Applied Biosystems), according to the manufacturer's protocols. The DsRed2 cDNA was amplified with the forward primer 5′ CCAGTTCCAGTACGGCTCCA and the reverse primer 5′ GCCGTCCTCGAAGTTCATCA. As a control, the observed DsRed2 signal was related to β-actin RNA levels, using a forward primer 5′ CCTGGCACCCAGCACAAT and reverse primer 5′ GGGCCGGACTCGTCATACT.
RNA FISH was performed as described previously (Sharp et al, 2006). PRL array containing HeLa cells were transfected with either CFP-ERαwt or CFP-ERαS305A and treated 2 h before analysis with 10 μM forskolin for 15 min, followed by treatment with 1 μM 4′OH-tamoxifen, or treated with 4′OH-tamoxifen only. The cells were stained with an antibody against ERα (Clone 60C, Upstate), and RNA FISH for DsRed2 transcripts was performed as described previously (Sharp et al, 2006). Informative arrays were analyzed for the Alexa-546 signal intensity of DsRed2 transcripts.
Supplementary Material
Supplementary Data
Acknowledgments
We thank Lennert Janssen for assistance in generating the chimeric mutants of ERα and SRC-1, Dr Rene Bernards (NKI) for kindly providing plasmids pPC86, pPC97 and pCMV-SRC-1, Anja Duursma for help with the Q-RT-PCR, Dr Jacco van Rheenen for advice on the FRET procedures, Coenraad Kuijl for help with FRAP analysis and Dr Mariska Rondaij for critically reading this manuscript, and Mrs Maureen G Mancini for technical advice on the PRL-HeLa cells. This study was supported by Grant NKI 2005-3388 of the Dutch Cancer Society.
References
- Bindels EM, Lallemand F, Balkenende A, Verwoerd D, Michalides R (2002) Involvement of G1/S cyclins in estrogen-independent proliferation of estrogen receptor-positive breast cancer cells. Oncogene 21: 8158–8165 [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA 92: 7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M (1997) Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389: 753–758 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M (2006) Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Dutertre M, Smith CL (2003) Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen receptor-alpha: regulation by phosphorylation sites in the A/B region depends on other receptor domains. Mol Endocrinol 17: 1296–1314 [DOI] [PubMed] [Google Scholar]
- Glaros S, Atanaskova N, Zhao C, Skafar DF, Reddy KB (2006) Activation function-1 domain of estrogen receptor regulates the agonistic and antagonistic actions of tamoxifen. Mol Endocrinol 20: 996–1008 [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14: 121–141 [PubMed] [Google Scholar]
- Gutierrez MC, Detre S, Johnston S, Mohsin SK, Shou J, Allred DC, Schiff R, Osborne CK, Dowsett M (2005) Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. J Clin Oncol 23: 2469–2476 [DOI] [PubMed] [Google Scholar]
- Holm C, Rayala S, Jirstrom K, Stal O, Kumar R, Landberg G (2006) Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst 98: 671–680 [DOI] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85: 403–414 [DOI] [PubMed] [Google Scholar]
- Llopis J, Westin S, Ricote M, Wang Z, Cho CY, Kurokawa R, Mullen TM, Rose DW, Rosenfeld MG, Tsien RY, Glass CK (2000) Ligand-dependent interactions of coactivators steroid receptor coactivator-1 and peroxisome proliferator-activated receptor binding protein with nuclear hormone receptors can be imaged in live cells and are required for transcription. Proc Natl Acad Sci USA 97: 4363–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Flouriot G, Pakdel F (2001) Synergism between ERalpha transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 alpha-helical core and for a direct interaction between the N- and C-terminal domains. Mol Endocrinol 15: 1953–1970 [DOI] [PubMed] [Google Scholar]
- Metivier R, Stark A, Flouriot G, Hubner MR, Brand H, Penot G, Manu D, Denger S, Reid G, Kos M, Russell RB, Kah O, Pakdel F, Gannon F (2002) A dynamic structural model for estrogen receptor-alpha activation by ligands, emphasizing the role of interactions between distant A and E domains. Mol Cell 10: 1019–1032 [DOI] [PubMed] [Google Scholar]
- Michalides R, Griekspoor A, Balkenende A, Verwoerd D, Janssen L, Jalink K, Floore A, Velds A, van't Veer L, Neefjes J (2004) Tamoxifen resistance by a conformational arrest of the estrogen receptor alpha after PKA activation in breast cancer. Cancer Cell 5: 597–605 [DOI] [PubMed] [Google Scholar]
- Myers E, Fleming FJ, Crotty TB, Kelly G, McDermott EW, O'higgins NJ, Hill AD, Young LS (2004) Inverse relationship between ER-beta and SRC-1 predicts outcome in endocrine-resistant breast cancer. Br J Cancer 91: 1687–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate SA, Boonyaratanakornkit V, Spencer TE, Tsai SY, Tsai MJ, Edwards DP, O'Malley BW (1998) The Steroid Receptor Coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the Activation Function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem 273: 12101–12108 [DOI] [PubMed] [Google Scholar]
- Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SA, Wong J, Allred DC, Clark GM, Schiff R (2003) Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst 95: 353–361 [DOI] [PubMed] [Google Scholar]
- Pike AC (2006) Lessons learnt from structural studies of the oestrogen receptor. Best Pract Res Clin Endocrinol Metab 20: 1–14 [DOI] [PubMed] [Google Scholar]
- Rowan BG, Garrison N, Weigel NL, O'Malley BW (2000a) 8-Bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein. Mol Cell Biol 20: 8720–8730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan BG, Weigel NL, O'Malley BW (2000b) Phosphorylation of steroid receptor coactivator-1. Identification of the phosphorylation sites and phosphorylation through the mitogen-activated protein kinase pathway. J Biol Chem 275: 4475–4483 [DOI] [PubMed] [Google Scholar]
- Shang Y, Brown M (2002) Molecular determinants for the tissue specificity of SERMs. Science 295: 2465–2468 [DOI] [PubMed] [Google Scholar]
- Sharp ZD, Mancini MG, Hinojos CA, Dai F, Berno V, Szafran AT, Smith KP, Lele TT, Ingber DE, Mancini MA (2006) Estrogen-receptor-{alpha} exchange and chromatin dynamics. J Cell Sci 119: 4101–4116 [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95: 927–937 [DOI] [PubMed] [Google Scholar]
- Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O'Malley BW, Mancini MA (2001) FRAP reveals that mobility of oestrogen receptor α is ligand- and proteasome-dependent. Nat Cell Biol 3: 15–23 [DOI] [PubMed] [Google Scholar]
- van Ham SM, Tjin EP, Lillemeier BF, Gruneberg U, van Meijgaarden KE, Pastoors L, Verwoerd D, Tulp A, Canas B, Rahman D, Ottenhoff TH, Pappin DJ, Trowsdale J, Neefjes J (1997) HLA-DO is a negative modulator of HLA-DM-mediated MHC class II peptide loading. Curr Biol 7: 950–957 [DOI] [PubMed] [Google Scholar]
- van Rheenen J, Langeslag M, Jalink K (2004) Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission. Biophys J 86: 2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner BL, Norris JD, Knotts TA, Weigel NL, McDonnell DP (1998) The nuclear corepressors NCoR and SMRT are key regulators of both ligand- and 8-bromo-cyclic AMP-dependent transcriptional activity of the human progesterone receptor. Mol Cell Biol 18: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RA, Mazumdar A, Vadlamudi RK, Kumar R (2002) P21-activated kinase-1 phosphorylates and transactivates estrogen receptor-alpha and promotes hyperplasia in mammary epithelium. EMBO J 21: 5437–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Shinsako P, Anderson C, Feng W, Nguyen MP, Chen D, Huang S, Subramanian S, McKinerney E, Katzenellenbogen BS, Stallcup MR, Kushner PJ (1998) Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocrinol 12: 1605–1618 [DOI] [PubMed] [Google Scholar]
- Xu J, Li Q (2003) Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol 17: 1681–1692 [DOI] [PubMed] [Google Scholar]
- Zheng FF, Wu RC, Smith CL, O'Malley BW (2005) Rapid estrogen-induced phosphorylation of the SRC-3 coactivator occurs in an extranuclear complex containing estrogen receptor. Mol Cell Biol 25: 8273–8284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart W, Griekspoor A, Kuijl C, Marsman M, van Rheenen J, Janssen H, Calafat J, van Ham M, Janssen L, van Lith M, Jalink K, Neefjes J (2005) Spatial separation of HLA-DM/HLA-DR interactions within MIIC and phagosome-induced immune escape. Immunity 22: 221–233 [DOI] [PubMed] [Google Scholar]
- Zwart W, Griekspoor A, Rondaij M, Verwoerd D, Neefjes J, Michalides R (2007) Classification of anti-estrogens according to intramolecular FRET effects on phospho-mutants of estrogen receptor a. Mol Cancer Ther 6: 1526–1533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data