Abstract
Epigenetically silent transposons and repeats constitute a substantial proportion of eukaryotic genomes, but their impact on cellular gene function remains largely unexplored. In Arabidopsis, transposons are silenced by DNA methylation, and this methylation is often abolished by mutations in a chromatin-remodeling gene DDM1 (DECREASE IN DNA METHYLATION 1). The ddm1 mutation induces various types of developmental abnormalities through de-repression of transposons and repeats. Here, we report a novel mechanism for a ddm1-induced syndrome, called bonsai (bns). We identified the gene responsible for the bns phenotypes by genetic linkage analysis and subsequent transcriptional analysis. The bns phenotypes are due to silencing of a putative Anaphase-Promoting Complex (APC) 13 gene. The BNS gene silencing was associated with DNA hypermethylation, which is in contrast to the ddm1-induced hypomethylation in the other genomic regions. This paradoxical BNS hypermethylation was reproducibly induced during self-pollination of the ddm1 mutant, and it was mediated by a long interspersed nuclear element (LINE) retrotransposon flanking the BNS gene. We discuss possible molecular mechanisms and the evolutionary implications of transposon-mediated epigenetic changes in the BNS locus.
Keywords: anaphase promoting complex, BONSAI, epigenetic inheritance, heterochromatin, small RNA
Introduction
Methylation of cytosine is a heritable epigenetic mark involved in several important biological processes, including genomic imprinting and transposon silencing (Jaenisch and Bird, 2003; Rangwala and Richards, 2004; Chan et al, 2005; Zilberman and Henikoff, 2005). Transposons are methylated in diverse organisms, and loss of cytosine methylation leads to activation of transposons (Yoder et al, 1997; Walsh et al, 1998; Miura et al, 2001; Singer et al, 2001; Kato et al, 2003; Selker et al, 2003). Genome-wide mapping of DNA methylation in the flowering plant Arabidopsis demonstrated that a majority of cytosine methylation is concentrated in heterochromatic regions, where transposons and repetitive sequences accumulate (Lippman et al, 2004; Zhang et al, 2006; Zilberman et al, 2007). Unexpectedly, however, recent high-resolution mapping studies revealed that ∼20–30% of expressed genes have methylation within their transcribed regions, although the methylation level is generally lower than that in transposons (Zhang et al, 2006; Zilberman et al, 2007). Interestingly, the proportion of those methylated genes increases toward heterochromatic pericentromeric regions, possibly reflecting direct or indirect interaction(s) of epigenetic states between euchromatic genes and heterochromatic sequences (Zilberman et al, 2007). In the large genomes of plants and vertebrates, transposons and repeats are also scattered among and within genes. However, the impact of such local heterochromatin on activity of cellular genes remained largely unexplored.
The impact of epigenetic changes on transposon activity can be directly examined using Arabidopsis mutants with defective genomic DNA methylation. In plants, cytosine methylation is found in both CG and non-CG contexts. In Arabidopsis, methylation at CG sites is maintained by DNA methyltransferase MET1, an ortholog of Dnmt1 in mammals, while methylation at non-CG sites depends on DNA methyltransferase genes, CMT3 and DRM2 (Finnegan et al, 1996; Ronemus et al, 1996; Bartee et al, 2001; Lindroth et al, 2001; Cao et al, 2003; Kankel et al, 2003). Another gene involved in maintenance of methylation and silencing of heterochromatin loci is a chromatin-remodeling ATPase gene DDM1 (DECREASE IN DNA METHYLATION 1), which is involved in both CG and non-CG methylation (Vongs et al, 1993; Jeddeloh et al, 1998). In addition, chromatin and RNAi components involved in de novo DNA methylation have recently been identified using several reporter transgene systems (Aufsatz et al, 2002; Kanno et al, 2004, 2005; Chan et al, 2005; Herr et al, 2005; Onodera et al, 2005; Pontier et al, 2005; Pontes et al, 2006). Notably, many of the putative endogenous targets of this pathway are located near transposon sequences, which might epigenetically regulate adjacent genes (Huettel et al, 2006).
Several examples of developmental variants were recovered in both met1 and ddm1 mutant lines (Finnegan et al 1996; Kakutani et al, 1996, 2004; Ronemus et al, 1996; Kankel et al, 2003; Saze et al, 2003). Genetic analysis of some of these ddm1-induced developmental variants revealed that each of the abnormalities is due to a heritable change in a locus other than DDM1 (Kakutani et al, 1996). For example, a ddm1-induced dwarf phenotype named bal is produced by the overexpression of a cluster of disease resistance genes (Stokes et al, 2002). Another ddm1-induced developmental variation, characterized by a delay in flowering onset, is due to ectopic expression of the imprinted homeobox gene FWA (Kakutani, 1997; Soppe et al, 2000; Kinoshita et al, 2004). Although these abnormalities behave as dominant traits, some of the ddm1-induced abnormalities behave as heritable recessive traits, suggesting that a different mechanism is responsible (Kakutani et al, 2004).
Here, we report the identification of the target gene of a ddm1-induced loss-of-function epigenetic abnormality called bns (Kakutani, 1997; Kakutani et al, 2004). The loss of BONSAI gene function was due to gene silencing associated with DNA hypermethylation and small RNA accumulation. The de novo methylation of the BONSAI gene was induced reproducibly in independent ddm1 mutant lines. This ectopic methylation depends on the presence of a long interspersed nuclear element (LINE) retrotransposon insertion within the 3′ non-coding region. The LINE insertion, which is found in the majority of natural accessions, generates a potential trigger for epigenetic variation with strong developmental effects.
Results
Repeated self-pollination of a ddm1 mutant induced a combination of phenotypes named bns
Repeated self-pollination of the DNA hypomethylation mutant ddm1 results in a variety of developmental abnormalities (Kakutani et al, 1996). Genetic analyses of some of the phenotypes have revealed that they are caused by gain-of-function alleles, which reflect overexpression of the responsible genes (Soppe et al, 2000; Stokes et al, 2002). However, not all of the developmental abnormalities are gain-of-function alleles. An example is a ddm1-induced developmental syndrome that we named bns.
The bns phenotypes were characterized by short, compact inflorescence, resulting in reduced plant height (Figure 1A and B). The bns variant showed disrupted phyllotaxis, reduced apical dominance and production of clusters of bracts and flowers at the apex of the inflorescence (Figure 1C and D). These phenotypes seem to reflect the inhibition of internode elongation and the termination of shoot growth at the apical meristems (Figure 1; Kakutani, 1997; Kakutani et al, 2004).
Figure 1.
The bns phenotypes. (A) WT Col plants (two on the left) and bns plants in a DDM1/DDM1 background (two on the right; hereafter referred to as bns). Both are six weeks old. (B) A close-up image of bns. (C) Inflorescences of bns. (D) A cluster of flowers produced in bns.
After backcrossing to the parental wild-type (WT) Columbia (Col), the bns phenotype was not detectable in the F1 population, suggesting that the abnormal phenotypes are not due to a gain-of-function mutation. In the self-pollinated progeny of an F1 plant, we recovered F2 plants showing the bns phenotype. The phenotypic plants included both ddm1/ddm1 and DDM1/- genotypes. This observation suggests that the bns phenotypes are produced by a heritable change in a locus (or loci) other than DDM1.
Identification of the BNS gene
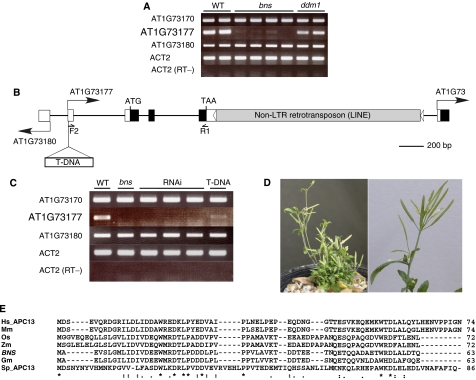
To further understand the basis of the heritable bns phenotypes, we examined their inheritance in the F2 progeny from a cross of a ddm1 plant with bns phenotypes (Col) to a WT Landsberg erecta (Ler) plant. The genotype was determined for 531 F2 plants with clear bns phenotypes, which comprised about 10% of the F2 population. Characterization of Col/Ler polymorphisms throughout the genome revealed that all of the phenotypic plants were homozygous for the Col haplotype in one locus in the bottom arm of chromosome 1, suggesting that a loss-of-function allele in this locus is responsible for the bns trait. This locus was narrowed to an interval between genetic markers NGA111 and BW54 (five recombinants and two recombinants, respectively, out of the 1062 chromosomes examined). We compared the transcript levels of 54 predicted genes in this genetically defined BNS region between WT and bns plants (backcrossed to DDM1/DDM1), using a reverse transcription (RT)–PCR assay. We found that one gene (AT1G73177) showed a severe reduction in its expression in bns DDM1 compared to WT plants (Figure 2A). The AT1G73177 transcript was also reduced in the self-pollinated ddm1 plants with the bns phenotypes (data not shown). The identified gene consists of four exons and encodes a predicted 63-amino acid (aa) protein (Figure 2B), and this annotation is supported by the presence of full-length cDNA in nucleotide sequence databases (GenBank: AY088589). A truncated non-LTR-type retrotransposon (LINE, long interspersed nuclear elements) sequence (AT1G73175) was found in the 3′UTR in the WT Col genome (Figure 2B and see below). Two flanking genes, AT1G73170 and AT1G73180, did not show a detectable reduction in their transcript level in bns plants (Figure 2A).
Figure 2.
Identification of the BNS gene. (A) RT–PCR for the BNS gene (AT1G73177) and neighboring genes (AT1G73170 and AT1G73180). Total RNA isolated from wild-type Col (WT), bns (backcrossed to DDM1) and ddm1 plants (before repeated self-pollination) was used. The BNS transcript was also reduced in ddm1 lines after repeated self-pollination (not shown). Actin2 (ACT2) was used as a control. (B) A schematic representation of the BNS locus. Boxes represent exons (coding sequences in black and UTRs in white for BNS and the neighboring genes, and in gray for the LINE sequence). Black arrows indicate the annotated transcription start sites and transcript orientation (www.arabidopsis.org). Horizontal white arrowheads represent the target site duplications of the LINE insertion. The position of the T-DNA insertion in the first exon of BNS in SALK_027397 line is also indicated. The positions of primer pair, F2 and R3, used for RT–PCR of AT1G73177, are also shown. (C) Knockdown of BNS transcripts in the RNAi lines and in the T-DNA insertion line. RT–PCR was performed with total RNA from wild-type Col (WT), bns, transgenic plants expressing dsRNA of BNS gene (RNAi) and SALK_027397 line homozygous for the T-DNA insertion (T-DNA). (D) Phenotypes of a BNS RNAi line (left panel), and an inflorescence in a plant homozygous for the T-DNA insertion (right panel). (E) Multiple aa sequence alignment of BNS (Arabidopsis thaliana; AT1G73177) and APC13 homologs in Homo sapiens (Hs, NP_056206), Mus musculus (Mm, NP_852059), Oryza sativa (Os, NP_001060376), Zea mays (Zm, AY105005), Glycine max (Gm, CX701269) and Schizosaccharomyces pombe (Sp, NP_595754). The sequences were aligned using the ClustalW program that highlights the identical and conserved aa with asterisks and dots, respectively.
To test whether the bns phenotypes are due to the repression of AT1G73177, this gene was knocked down by RNAi in WT Col plants by transformation with a transgenic construct producing double-stranded RNA (dsRNA) of the gene sequence (Figure 2C). The transgenic lines showed the bns-like phenotypes (i.e., reduced plant height and clustered flowers) associated with a reduction in AT1G73177 transcript abundance (Figure 2C and D). In addition, we analyzed the effect of a T-DNA insertion in the upstream non-coding region in the first exon (SALK_027397). In the insertion mutant, a transcript was still detectable by RT–PCR, but the level was less than that observed in WT plants (Figure 2B and C). The plants homozygous for the T-DNA insertion showed similar phenotypes, although they were much milder (Figure 2D), further confirming that the reduction in AT1G73177 transcript induces the bns phenotypes. From these results, together with the recessive nature of the bns mutation, we concluded that the loss or reduction in AT1G73177 function is most likely to be responsible for the bns phenotypes.
The BNS gene product has similarity to a subunit of the Anaphase-Promoting Complex/Cyclosome (APC/C)
The predicted BNS protein has a high similarity to the mammalian Swm1/Apc13, a subunit of Anaphase-Promoting Complex/Cyclosome (APC/C) (Figure 2E). The APC/C is a large ubiquitin–protein ligase complex that regulates cell cycle progression in eukaryotic cells (Castro et al, 2005). Swm1/Apc13 was originally identified for its role in spore wall assembly in Saccharomyces cerevisiae (Ufano et al, 1999), and was later found to be a core subunit of the APC/C (Yoon et al, 2002; Hall et al, 2003). The protein is evolutionarily conserved in a wide range of organisms (Schwickart et al, 2004) (Figure 2E). We detected BNS expression in all tissues examined in WT plants (Supplementary Figure 1).
bns is an epigenetic mutation associated with DNA hypermethylation
Despite the marked reduction in BNS expression in the bns line, the nucleotide sequence of the BNS gene in the bns line was identical to that in the WT progenitor strain, Col (from −825 to +946; data not shown). These results suggested that the silencing of the BNS gene has an epigenetic basis. We therefore examined the level of DNA methylation in this region.
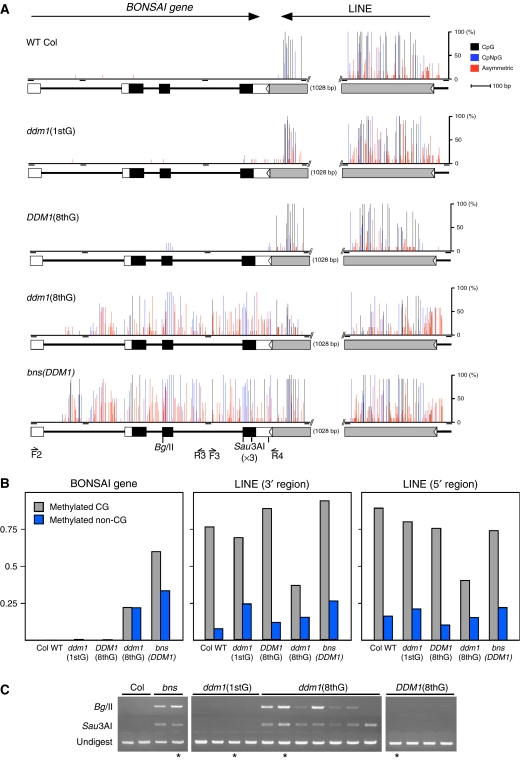
To detect DNA methylation, we used the following two methods: digestibility by methylation-sensitive restriction enzymes and bisulfite genomic sequencing. Bisulfite genomic sequencing revealed that the BNS gene in the WT Col genome was almost free of DNA methylation (WT Col in Figure 3A). On the other hand, the flanking LINE sequence was heavily methylated, especially at CG sites (Figure 3A and B). In the bns line, the BNS gene region was also heavily methylated (bottom diagram in Figure 3A), which is in contrast to the situation in WT Col plants. The hypermethylation at the BNS locus was found at both CG and non-CG sites.
Figure 3.
DNA methylation pattern in the BNS locus. (A) Schematic representations of the BNS locus and cytosine methylation level analyzed by bisulfite sequencing. After treatment with bisulfite, DNA fragments were amplified using four pairs of primers separately (the positions indicated as short horizontal black bars), and cloned for sequencing (12 clones for each amplified region). The percentage of methylated cytosine is indicated by vertical bars (black, CG; blue, CNG; red, asymmetric cytosine). Boxes below represent exons (coding sequences in black and UTRs in white for BNS, and in gray for the LINE sequence). (B) Proportion of methylated cytosines in the BNS locus, which is based on the results shown in panel A. (C) Methylation of the BNS region detected by restriction digestion. Genomic DNA was digested by methylation-sensitive restriction enzymes BglII (5′-AGATCT-3′) or Sau3AI (5′-GATC-3′) (http://rebase.neb.com), and was subsequently used as template for PCR amplification. The positions of the restriction sites and primers used for the PCR are indicated in the bottom of panel A; primer pairs F2+R3 and F3+R4 were used after BglII and Sau3AI digestion, respectively. Asterisks (*) indicate the samples used for bisulfite sequencing in panels A and B.
The methylation status of the BNS region was confirmed by digestion with methylation-sensitive restriction enzymes and subsequent PCR. WT Col samples did not show a PCR signal, reflecting the complete digestion of the genomic DNA (left panel in Figure 3C). Samples derived from bns plants showed bands reflecting incomplete digestion due to methylation. These results are consistent with the results of the bisulfite sequencing.
Repeated self-pollination of ddm1 mutant reproducibly induced de novo DNA methylation in the BNS gene
The hypermethylation in the BNS gene contrasts with the global DNA hypomethylation induced by the ddm1. We examined whether this paradoxical DNA hypermethylation in the BNS locus reflected one single purely stochastic event, or BNS hypermethylation could be reproducibly induced in a ddm1 mutant background.
In order to see the initial effect of the ddm1 mutation, ddm1 homozygotes were selected from progeny derived by self-pollination of a DDM1/ddm1 heterozygote. This DDM1/ddm1 parent was generated by backcrossing a ddm1 mutant six times to WT Col parent, in order to remove heritable effects from the original ddm1/ddm1 mutant (Kakutani et al, 1996, 1999). ddm1/ddm1 plants segregated in the self-pollinated progeny of the backcrossed DDM1/ddm1 parent did not show signs of BNS gene hypermethylation (ddm1(1stG) in Figure 3). In order to see the effect of repeated self-pollination of the ddm1 mutant, seven ddm1 homozygotes in the segregating family were independently self-pollinated seven times. The hypermethylation of the BNS gene was detected in all seven independent ddm1 lines (ddm1(8thG) in Figure 3). The ddm1 mutation reproducibly induced BNS methylation, but this process was slow and required multiple generations. As a control, BNS methylation was also examined in four DDM1/DDM1 sibling lines segregated from the same DDM1/ddm1 parent and self-pollinated seven times in parallel (DDM1 (8thG) in Figure 3). BNS methylation was not detected in any of the four DDM1 control lines. The lack of methylation in the DDM1 sibling families further confirmed that the ddm1 mutation was responsible for the de novo methylation of the BNS gene.
The flanking LINE sequence showed a reduction in DNA methylation in the self-pollinated ddm1 plants (ddm1 (8thG) in Figure 3A and B). This result is consistent with the previous observations that DDM1 activity is required for the maintenance of DNA methylation and silencing of endogenous transposons (Miura et al, 2001; Singer et al, 2001; Lippman et al, 2004). The hypomethylation of the LINE was found only after repeated self-pollination, which is similar to the situation for the SINE-related sequence in the FWA promoter, which remains methylated in the initial generations of ddm1 inbreeding, but loses methylation stochastically in subsequent inbred generations (Soppe et al, 2000). Interestingly, the LINE sequence was methylated to the WT level in the bns mutant line backcrossed into a DDM1/DDM1 background (Figure 3A and B), suggesting de novo methylation in the DDM1 background.
The hypermethylation and silencing of BNS gene is associated with small RNAs
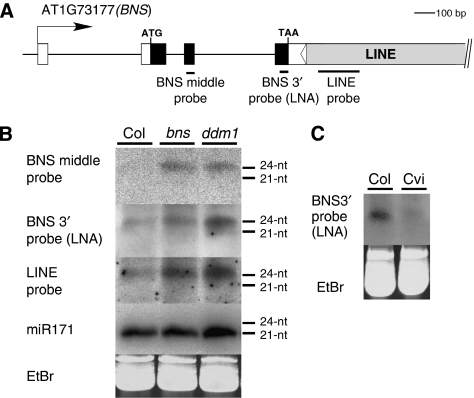
Epigenetic silencing of transposons and repeats are frequently associated with the production of small RNAs, which could be involved in RNA-directed DNA methylation (Zilberman et al, 2003; Chan et al, 2004; Matzke and Birchler, 2005). Because the BNS gene was methylated de novo in a ddm1 background, we examined small RNAs corresponding to this region. As shown in Figure 4, BNS gene silencing was associated with the accumulation of small RNA in the size of 24–25 nt, the length of small RNA species often detected for heterochromatic sequences (Hamilton et al, 2002; Xie et al, 2004; Henderson et al, 2006; Pontes et al, 2006). Small RNAs (24–25 nt) were also detected in the ddm1 mutant sample. A hybridization probe covering the 3′ region of the BNS gene near the boundary with the LINE (BNS 3′ probe) detected a weak but significant signal in the WT Col sample (Figure 4B and C), although the signal increased in the sample carrying a silent BNS allele.
Figure 4.
Small RNA northern analysis of the BNS locus. (A) The positions of three hybridization probes used are indicated. (B) Small RNA was examined in WT Col, bns in DDM1 background and self-pollinated ddm1 plants with reduced BNS expression. The same membrane was used for hybridization with each of the three probes and for the control miR171 probe. Ethidium bromide staining of the major RNA is shown as a control (EtBr). (C) WT Col and Cvi samples on a different membrane.
As is the case for many other silent transposons, small RNA was also detected for the LINE sequence (Figure 4B) in the WT Col sample. Interestingly, the amount of the small RNA for this family of LINE increased in the ddm1 and bns plants. The increase in the small RNA signal might mediate the de novo methylation of this LINE element, and that may explain the partial methylation of the LINE in the ddm1 mutant, and the de novo methylation of the element after introduction into a background with a WT DDM1 allele (Figure 3A and B).
The LINE insertion was found at the BNS locus in majority of Arabidopsis natural accessions
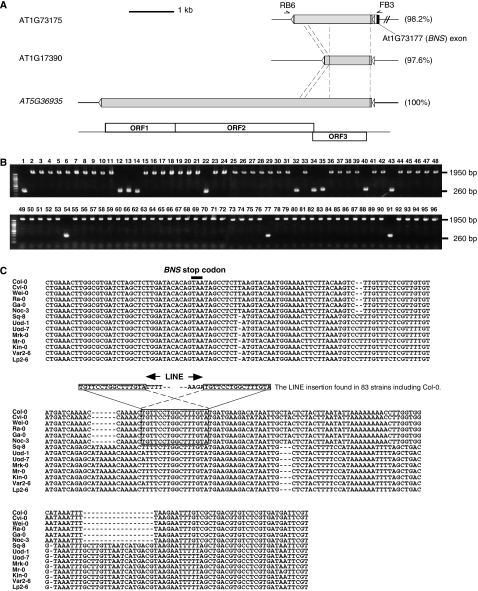
The LINE (AT1G73175) at the BNS locus belongs to a previously uncharacterized subfamily of LINE sequences in Arabidopsis (Wright et al, 1996; Noma et al, 2000, 2001). The presence of 16-bp target site duplication (TSD) followed by a 9-bp poly(A) sequence proximal to BNS indicates that the LINE sequence is inserted in a tail-to-tail orientation relative to the BNS gene (Figure 3A). In WT Col, BNS mRNA extends into the LINE sequence over the TSD and poly(A) sequences (Supplementary Figure 2). The Col genome contains two other members of this LINE subfamily, which share more than 97% nucleotide sequence identity (AT1G17390 and AT5G36935; Figure 5A). The copy on chromosome 5 (AT5G36935) is likely to be the full-length copy (Figure 5A). This copy encodes three open reading frames, with a structure similar to ATLN-L class LINEs (Noma et al, 2001). The presence of these three copies in the Col genome was confirmed by Southern blot analysis (Supplementary Figure 3).
Figure 5.
The AT5G36935 LINE family in the Arabidopsis genome. (A) Schematic representations of the LINE sequence at the BNS locus, and two other related sequences in the Col genome. Sequence identities are indicated in the parentheses. Exons of LINEs are shown as gray boxes (coding sequences in black and UTRs in white for the BNS gene). White triangles indicate target site duplications of the LINE insertion. AT1G17390 lacks a part of the internal sequence, as indicated by dashed lines. Predicted ORFs in AT5G36935 are shown at the bottom. (B) The LINE insertion at the 3′UTR of BNS in 96 natural strains of Arabidopsis thaliana. Presence of the LINE insertion was examined by PCR using BNS FB3 and LINE RB6 primers indicated in panel A. The PCR using genomic DNA isolated from 96 Arabidopsis strains amplified either ∼1950 bp (LINE+) or ∼260 bp (LINE−) fragment. Names of the strains used are shown in Supplementary data. (C) An alignment of the nucleotide sequences of the BNS 3′UTR in 13 strains that do not have the LINE insertion. Col sequence is also shown. Note that the 13 strains are classified into two groups based on similarity, and only one group has an identical target site sequence to that found in the Col genome (indicated by box). The stop codon of the BNS gene is indicated by the black bar.
In order to evaluate the impact of the LINE insertion in natural populations, we examined the presence of the LINE insertion at the BNS locus in 96 natural accessions of Arabidopsis thaliana. Among them, 83 accessions have the LINE insertion in the BNS 3′UTR, while 13 did not have the insertion (Figure 5B). This was confirmed by Southern analysis of the 96 natural accessions (data not shown). Among the 13 accessions without the LINE insertion, five contain sequences almost identical to Col apart from the LINE insertion (Figure 5C). In those accessions, the TSD sequence remained intact, suggesting that these are ancestral alleles before the LINE insertion. Presence of the LINE in the majority of natural accessions suggests that the LINE insertion per se does not have deleterious effects in natural populations.
Dependence of the BNS hypermethylation on the flanking LINE sequence
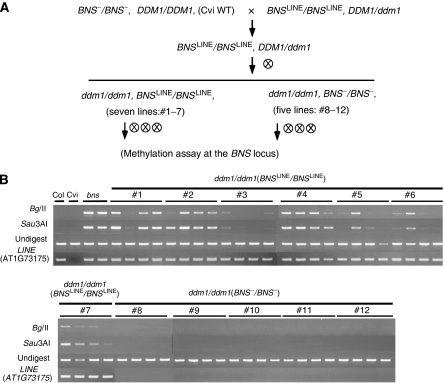
Using Cvi, which does not have the LINE insertion at the BNS locus (Figure 5C), we tested whether the LINE sequence is necessary for the ddm1-induced de novo methylation at the BNS locus. WT Cvi was crossed to a ddm1 heterozygote, which had already been backcrossed six times in the heterozygous state (Kakutani et al, 1996). A DDM1/ddm1 heterozygote originating from this cross was self-pollinated, and from the progeny, we selected ddm1 homozygotes with the BNS allele from genome of Col (BNSLINE/BNSLINE; homozygous for the LINE insertion) or Cvi (BNS−/BNS−; without the LINE insertion) (Figure 6A). After three rounds of self-pollination, DNA methylation of the BNS locus in these ddm1 plants was examined using methylation-sensitive restriction enzymes. All of seven independent ddm1 lines homozygous for the BNSLINE allele (from Col) showed de novo DNA methylation of the BNS locus, whereas none of the five ddm1 lines homozygous for the BNS− allele (from Cvi) showed ectopic DNA methylation (Figure 6B). This result suggests that ddm1-induced de novo methylation at the BNS gene depends on the presence of the LINE insertion in the 3′UTR.
Figure 6.
The ddm1-induced de novo methylation at the BNS locus depends on the presence of the LINE insertion. (A) Genetic scheme to generate ddm1 plants with or without the LINE insertion at the BNS locus. X and encircled X indicate outcross and self-pollination, respectively. BNSLINE: BNS allele from Col with LINE insertion. BNS-: BNS allele from Cvi without the LINE insertion. DNA methylation was examined after three generations of self-pollination. (B) DNA methylation at the BNS gene in twelve independent lines. Methylation was analyzed as described in Figure 3C.
Discussion
Mechanism for BNS gene hypermethylation triggered by the ddm1 mutation
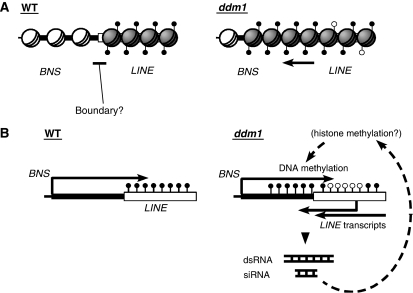
Here, we report the identification and characterization of a loss-of-function epigenetic developmental abnormality bns. The most striking feature of the bns trait is that the local hypermethylation of the BNS gene was induced in a background of global DNA hypomethylation. The hypermethylation of the BNS gene was not evident in newly segregated ddm1 homozygous plants (1stG in Figure 3), but it was reproducibly induced in the self-pollinated progeny of ddm1 mutants (8thG in Figure 3). These observations suggest that BNS hypermethylation may be due to an indirect effect of the globally hypomethylated ddm1 background. Similar observations have been previously reported for SUPERMAN (SUP) and AGAMOUS (AG) sequences; these sequences are stochastically hypermethylated in the absence of DDM1 or MET1 activity (Jacobsen and Meyerowitz, 1997; Jacobsen et al, 2000). In both SUP and AG, pyrimidine-rich sequences such as CT dinucleotide repeats are found in the hypermethylated target sequences, and the possible involvement of this simple-sequence motif has been proposed (Jacobsen et al, 2000). However, a pyrimidine-rich sequence was not found in the BNS locus, suggesting that it is not the basis for BNS hypermethylation (data not shown). Instead, our results suggest that BNS hypermethylation is mediated by the pre-existing LINE insertion in non-coding region of the BNS gene (Figure 6).
The ectopic hypermethylation at BNS in ddm1 background occurred in a spreading manner from the LINE into the BNS region (Figure 3A). Spread of heterochromatin into genic regions is also known in position-effect-variegation in Drosophila (Talbert and Henikoff, 2006) and telomeric silencing in budding yeast (Grunstein, 1997). Although the BNS locus resides in a euchromatic chromosomal arm, the dense DNA methylation on the LINE sequence at this locus suggests that the LINE sequence can function as local heterochromatin, which is maintained without affecting adjacent genes in the WT background. The DDM1 gene is necessary for the maintenance of the heterochromatic characteristics of LINE and other transposons (Gendrel et al, 2002; Lippman et al, 2003, 2004). DDM1 might also be necessary to define a heterochromatin boundary (Figure 7A). In mammals, the chromatin insulator CTCF has a barrier function that blocks the extension of heterochromatin. The CTCF-dependent insulator activity was abolished by loss of an SNF2-like chromodomain helicase/ATPase protein, leading to a decrease in euchromatic histone modifications and DNA hypermethylation around the boundary sequences (Ishihara et al, 2006).
Figure 7.
Two models for ddm1-induced de novo methylation at the BNS locus. (A) Heterochromatin spreading model. (B) RNA-directed DNA methylation model. Filled lollipops indicate DNA methylation. See Discussion for details.
Interestingly, the spreading of DNA methylation was not found in the other side (opposite from the BNS gene) of the LINE sequence. On that side, the expression of the gene AT1G73170, which has the transcription start site approximately 150 bp away from the TSD of the LINE, was not affected in bns and self-pollinated ddm1 lines (Figure 2A and B and data not shown). These results suggest that the spreading of cytosine methylation from the LINE was unidirectional. In addition, we could detect small RNA from the BNS coding sequence in the ddm1 mutant plants (Figure 4), which correlates with the ectopic methylation. These features raise the possibility that LINE transcripts induced in the ddm1 background form dsRNA with the BNS mRNA through the complementary sequence in the 3′UTR of BNS mRNA (Figure 7B; Supplementary Figures 2 and 3). It is also possible that the LINE at the BNS locus has a cryptic promoter within the element. Readout transcripts originating from cryptic promoters in LTR-retrotransposons and LINEs can affect transcription of adjacent genes in both mammals and plants (Michaud et al, 1994; Nigumann et al, 2002; Kashkush et al, 2003). Indeed, the plant promoter database (PlantProm; Shahmuradov et al, 2003) predicted the presence of a putative bidirectional promoter sequence at the 3′ end of the LINE sequence (data not shown). The promoter could be activated through the loss of DNA methylation in self-pollinated ddm1 lines (8thG in Figure 3A), and produce antisense transcripts of BNS mRNA, which subsequently result in the formation of dsRNA of BNS mRNA.
A low level of small RNA was also found in WT Col plants at the 3′ region of the BNS gene (Figure 4B and C). This situation might have parallels with the maize B paramutation system, in which small RNA are detected for the region controlling the epigenetic state of the B gene, even when this gene is expressed (Chandler, 2007). Similarly, small RNA was detected for the Arabidopsis FWA promoter even when it is unmethylated (Chan et al, 2006). Thus, the presence of small RNA correlates with the potential for the epigenetic silencing, which, in the case of the BNS gene, also correlates with presence of the flanking LINE sequence (Figure 4C).
Phenotypic effects of BNS gene silencing
The predicted product of the BNS gene shares similarity with APC13, a subunit of the Anaphase-Promoting Complex APC/C that regulates the metaphase–anaphase transition and exit from mitosis through the degradation of cell cycle regulators (Castro et al, 2005). In budding yeast, loss of Swm1/APC13 leads to slow growth and an accumulation of G2/M cells (Hall et al, 2003; Schwickart et al, 2004). The observed bns phenotypes—inhibition of internode elongation and termination of shoot growth at shoot apical meristems (Figure 1)—might result from defects in APC-dependent cell cycle events. Null mutants of other single-copy Arabidopsis APC/C components, APC2 and APC6, exhibit female gametophytic lethality due to cell cycle arrests at an early stage of embryo sac development (Capron et al, 2003; Kwee and Sundaresan, 2003). Although BNS is present as a single-copy gene, bns phenotypes (Figure 1) are distinct from that of apc2 or apc6. It is possible that the bns epiallele does not cause a complete loss-of-function, and the remaining activity circumvents the cell cycle arrest that occurs in apc2 or apc6 gametophytes.
Once the bns phenotypes were induced in the ddm1 background, these phenotypes were inherited by their progeny even in the presence of a WT DDM1 allele. However, phenotypic variability was observed in bns DDM1 lines (data not shown), suggesting that the epigenetically silent state may be unstable in a DDM1 background. Similarly, the loss-of-function epigenetic alleles of sup and ag are unstable (Jacobsen and Meyerowitz, 1997; Jacobsen et al, 2000).
The role of transposons as potential triggers for heritable epigenetic developmental variation
When a transposon is inserted near a cellular gene in maize, expression of that gene is often affected by the epigenetic state of the transposon (McClintock, 1965; Fedoroff, 1989). Similarly, in mouse, insertion of a retroviral element within a gene can form an allele that shows epigenetic variation, which is heritable over multiple generations (Whitelaw and Martin, 2001; Rakyan et al, 2003). In an evolutionary context, an important question is whether the newly generated allele, which is under the transposon control, survives within natural populations or not.
The presence of the LINE insertion at the BNS locus in a majority of Arabidopsis natural accessions suggests that the insertion of the LINE per se did not have a deleterious effect in natural populations. Considering that, it is striking that the LINE mediates changes in epigenetic state of the BNS gene that lead to strong developmental variation. It is often the case that alleles with transposon insertions are indistinguishable from the original allele in term of expression pattern, as long as the transposon is silent (Fedoroff, 1989; Martienssen, 1996). Such hidden phenotypic variability, generated by a transposon insertion, may broaden the potential for evolution, as implied from ‘canalization' phenomena in Drosophila (Waddington, 1959; Flatt, 2005). A systematic survey of polymorphisms in transposon insertion sites in natural populations might reveal beneficial impact of transposon insertions as a source for epigenetic variability, which is heritable but reversible.
Materials and methods
Plant materials and growth conditions
The identification and isolation of the bns strain was described in the previous study (Kakutani, 1997). Sources of the 96 natural accessions (Figure 5B) are described in Nordborg et al (2005). Seeds of these strains (CS22660) and the BNS T-DNA insertion line (SALK_027397) (Figure 2) were obtained from the Arabidopsis Biological Resource Center. Plant seeds were allowed to germinate and grow on a medium containing 0.5 × MS salts (SIGMA), 2% sucrose and 0.8% agar (pH 5.7), for 2 weeks under long-day conditions (16 h, light; 8 h, dark) at 22°C. The seedlings were subsequently transferred and grown on vermiculite under the conditions described above. The ddm1-1 mutant and the WT DDM1 alleles were distinguished by PCR, as described by Kato et al (2003). The LINE insertion in the BNS locus (Figure 5B) was detected by PCR with primers BNS FB3 (5′-CAG GAA ACT CAG CAA GCA GAT G-3′) and LINE RB6 (5′-GAG CCG TTT GCC AAC CAC GTG G-3′).
RT–PCR
Total RNA from Arabidopsis leaves was isolated with the RNeasy Plant Mini kit (QIAGEN) and was treated with DNase I (TAKARA). cDNA was synthesized using the TAKARA RNA PCR kit (AMV) Ver.3.0 (TAKARA) and an oligo-(dT) primer. A total of 500 ng RNA in the RT reaction mixture (total 10 μl) was reverse transcribed at 42–50°C for 1 h, followed by heat inactivation at 95°C for 5 min. A one-fifth portion of the RT reaction was used as a template for PCR (total 20 μl). PCR conditions were as follows: 94°C for 2 min, 26–30 cycles at 94°C for 15 s, 60°C for 30 s, 72°C for 45 s and 72°C for 5 min. Control reactions without RT were carried out as described above. The primer pairs used for RT–PCR were as follows: the BNS gene (BNS F2: 5′-GCT AGA GGT TTT TAG TTC TCT G-3′ and BNS R1: 5′-TGT ACT TAA GAG GCT ATT ACT G-3′); AT1G73170 (AT1G73170 F1: 5′-GCG ATA CGG GCA TTA CTA ACA G-3′ and AT1G73170 R1: 5′-TAA TCA GG CAA TAG AGG TAA CC-3′); AT1G73180 (AT1G73180 F1: 5′-GGC GAA GGT CCT TAT AAC ACT C-3′ and AT1G73180 R1: 5′-TGA TTT CTT CAA TCA GGC GTT G-3′); LINE (LINE FB5: 5′-AAA TTA CAC TTG AAC GTT CCG G-3′ and LINE RD: 5′-AGT GGG GAG GAG ACA ATT CTA CAC-3′) and Actin2 (ACT2F: 5′-CTA AGC TCT CAA GAT CAA AGG C-3′ and ACT2R: 5′-AAC ATT GCA AAG AGT TTC AAG G-3′). cDNA synthesized from total RNA isolated from WT Col plants was used for 3′ RACE of the BNS gene. The initial PCR was performed using the BNS F2 primer and the M13 Primer M4 supplied with the kit, and then nested with BNS F1 primer (5′-TGT GTG GAG TAC GGC TGC ATT G-3′) and M13 primer M4. Amplified fragments were cloned and sequenced.
Transgenic plants
To prepare the RNAi construct (Figure 2), the BNS genomic sequence was amplified from genomic DNA, using the 177 12attB1 F (5′-AAA AAG CAG GCT TGT GTG GAG TAC GGC TGC AT-3′) and 177 12attB2 R (5′-AGA AAG CTG GGT AGA GGC TAT TAC TGT GTA TC-3′) primers, and cloned by a GATEWAY BP reaction (Invitrogen) into the binary vector pHELLSGATE 2 (Wesley et al, 2001). Plants were transformed by the standard floral dip method (Clough and Bent 1998).
DNA methylation analysis
Genomic bisulfite sequencing was performed as described by Paulin et al (1998). Detail is shown in Supplementary data.
DNA methylation was also analyzed by restriction enzymes BglII and Sau3AI (Figure 3C). A 100 ng weight of genomic DNA was digested with BglII and EcoRI, or Sau3AI and EcoRI, in 40 μl reaction mix. Control ‘undigest' sample was digested with EcoRI alone. After digestion, PCR was performed by using 1 μl of the digested sample as a template. BNS F2 and BNS R3 (5′-TTC CTT ATG ACA TTT CAA GGT C-3′) primers were used for BglII-digested DNA, and BNS F3 (5′-GTA ATG GAG ACA CAT ACG TCA C-3′) and BNS R4 (5′-TAC AAA GCC AGG AAC AGT TTT G-3′) were used for Sau3AI-digested DNA.
Small RNA Northern analysis
Small RNA was isolated from mature leaves using the mirVana miRNA isolation kit (Ambion). RNA (25∼30 μg) was resolved on denaturing polyacrylamide/urea gels (15%). Electroblotting and hybridization were performed as described (Llave et al, 2002). Hybridization was performed overnight at 38°C using PerfectHyb Plus buffer (Sigma). Blots were washed at 42°C in 2 × SSC, 0.2% SDS for 10 min, and in 0.5 × SSC, 0.1% SDS for 60 min, and analyzed by BAS-2500 (Fuji film). BNS middle probe was 5′-TGG TTT CTT CAG TAT CAT CAG TTT TAA CAG CAA GCA CTG G-3′ and BNS 3′ probe was 5′-A+AG A+GC T+AG A+TC A+CG C+CA A+GT T+TC A+GC A+TC T+GC T+TG C+TG A-3′ (+ indicates LNA-modified bases). For the LINE probe, a PCR fragment amplified from genomic DNA by using LINE FB5 and LINE RD primers was used.
Supplementary Material
Supplementary Data
Acknowledgments
We thank Kyoko Munakata, Akiko Shiraishi, Kazuya Takashima and Akiko Terui for technical assistance, Hiromi Higo for help in the initial stage of mapping the BNS gene, Hidetaka Ito and Asuka Miura for sharing plant and DNA materials, Tatsuo Kanno and Marjori Matzke for advice for the small RNA detection, Yasushi Hiromi, Hiroyuki Sasaki, and Eric Richards for critical comments on the manuscript. We acknowledge Arabidopsis Biological Stock Center at Ohio State University for the seed stocks, and CSIRO for the pHELLSGATE vector. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (17027024) to TK and Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists to HS.
References
- Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ (2002) HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J 21: 6832–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L, Malagnac F, Bender J (2001) Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev 15: 1753–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltrasferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Capron A, Serralbo O, Fulop K, Frugier F, Parmentier Y, Dong A, Lecureuil A, Guerche P, Kondorosi E, Scheres B, Genschik P (2003) The Arabidopsis anaphase-promoting complex or cyclosome: molecular and genetic characterization of the APC2 subunit. Plant Cell 15: 2370–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A, Bernis C, Vigneron S, Labbe JC, Lorca T (2005) The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene 24: 314–325 [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE (2006) Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol 4: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE (2004) RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Chandler VL (2007) Paramutation: from maize to mouse. Cell 128: 641–645 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Fedoroff NV (1989) Maize transposable elements. In Mobile DNA, Howe M, Berg D (eds), pp 375–411. Washington, DC: American Society for Microbiology [Google Scholar]
- Finnegan E, Peacock J, Dennis E (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T (2005) The evolutionary genetics of canalization. Q Rev Biol 80: 287–316 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Grunstein M (1997) Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol 9: 383–387 [DOI] [PubMed] [Google Scholar]
- Hall MC, Torres MP, Schroeder GK, Borchers CH (2003) Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J Biol Chem 278: 16698–16705 [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, Matzke M (2006) Endogenous targets of RNA-directed DNA methylation and PolIV in Arabidopsis. EMBO J 25: 2828–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M (2006) CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell 23: 733–742 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Meyerowitz EM (1997) Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277: 1100–1103 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Sakai H, Finnegan EJ, Cao X, Meyerowitz EM (2000) Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr Biol 10: 179–186 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33 (Suppl): 245–254 [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Bender J, Richards EJ (1998) The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev 12: 1714–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T (1997) Genetic characterization of late-flowering traits induced by DNA hypomethylation mutation in Arabidopsis thaliana. Plant J 12: 1447–1451 [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ (1996) Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA 93: 12406–12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Kato M, Kinoshita T, Miura A (2004) Control of development and transposon movement by DNA methylation in Arabidopsis thaliana. Cold Spring Harbor Symp Quant Biol 69: 139–143 [DOI] [PubMed] [Google Scholar]
- Kakutani T, Munakata K, Richards EJ, Hirochika H (1999) Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics 151: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ (2004) Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol 14: 801–805 [DOI] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33: 102–106 [DOI] [PubMed] [Google Scholar]
- Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T (2003) Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr Biol 13: 421–426 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523 [DOI] [PubMed] [Google Scholar]
- Kwee HS, Sundaresan V (2003) The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the Anaphase Promoting Complex in Arabidopsis. Plant J 36: 853–866 [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, Carrington JC, Doerge RW, Colot V, Martienssen R (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476 [DOI] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R (2003) Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1: E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R (1996) Epigenetic silencing of Mu transposable elements in maize. In Epigenetic Mechanisms of Gene Regulation, Russo V, Martienssen R, Riggs AD (eds), pp 593–608. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Matzke MA, Birchler JA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- McClintock B (1965) The control of gene action in maize. Brookhaven Symp Biol 18: 162–184 [Google Scholar]
- Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP (1994) Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev 8: 1463–1472 [DOI] [PubMed] [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T (2001) Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214 [DOI] [PubMed] [Google Scholar]
- Nigumann P, Redik K, Matlik K, Speek M (2002) Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79: 628–634 [DOI] [PubMed] [Google Scholar]
- Noma K, Ohtsubo H, Ohtsubo E (2000) ATLN elements, LINEs from Arabidopsis thaliana: identification and characterization. DNA Res 7: 291–303 [DOI] [PubMed] [Google Scholar]
- Noma K, Ohtsubo H, Ohtsubo E (2001) A new class of LINEs (ATLN-L) from Arabidopsis thaliana with extraordinary structural features. DNA Res 8: 291–299 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, Jakobsson M, Kim S, Morozov Y, Padhukasahasram B, Plagnol V, Rosenberg NA, Shah C, Wall JD, Wang J, Zhao K, Kalbfleisch T, Schulz V, Kreitman M, Bergelson J (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Paulin R, Grigg GW, Davey MW, Piper AA (1998) Urea improves efficiency of bisulphite-mediated sequencing of 5′-methylcytosine in genomic DNA. Nucleic Acids Res 26: 5009–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS (2006) The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126: 79–92 [DOI] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E (2003) Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA 100: 2538–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala SH, Richards EJ (2004) The value-added genome: building and maintaining genomic cytosine methylation landscapes. Curr Opin Genet Dev 14: 686–691 [DOI] [PubMed] [Google Scholar]
- Ronemus M, Galbiati M, Ticknor C, Chen J, Dellaporta S (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657 [DOI] [PubMed] [Google Scholar]
- Saze H, Scheid OM, Paszkowski J (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Schwickart M, Havlis J, Habermann B, Bogdanova A, Camasses A, Oelschlaegel T, Shevchenko A, Zachariae W (2004) Swm1/Apc13 is an evolutionarily conserved subunit of the anaphase-promoting complex stabilizing the association of Cdc16 and Cdc27. Mol Cell Biol 24: 3562–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU, Tountas NA, Cross SH, Margolin BS, Murphy JG, Bird AP, Freitag M (2003) The methylated component of the Neurospora crassa genome. Nature 422: 893–897 [DOI] [PubMed] [Google Scholar]
- Shahmuradov IA, Gammerman AJ, Hancock JM, Bramley PM, Solovyev VV (2003) PlantProm: a database of plant promoter sequences. Nucleic Acids Res 31: 114–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, Yordan C, Martienssen RA (2001) Robertson's mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev 15: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802 [DOI] [PubMed] [Google Scholar]
- Stokes TL, Kunkel BN, Richards EJ (2002) Epigenetic variation in Arabidopsis disease resistance. Genes Dev 16: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S (2006) Spreading of silent chromatin: inaction at a distance. Nat Rev Genet 7: 793–803 [DOI] [PubMed] [Google Scholar]
- Ufano S, San-Segundo P, del Rey F, Vazquez de Aldana CR (1999) SWM1, a developmentally regulated gene, is required for spore wall assembly in Saccharomyces cerevisiae. Mol Cell Biol 19: 2118–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ (1993) Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Waddington CH (1959) Canalization of development and genetic assimilation of acquired characters. Nature 183: 1654–1655 [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH (1998) Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Whitelaw E, Martin DI (2001) Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat Genet 27: 361–365 [DOI] [PubMed] [Google Scholar]
- Wright DA, Ke N, Smalle J, Hauge BM, Goodman HM, Voytas DF (1996) Multiple non-LTR retrotransposons in the genome of Arabidopsis thaliana. Genetics 142: 569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13: 335–340 [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Feoktistova A, Wolfe BA, Jennings JL, Link AJ, Gould KL (2002) Proteomics analysis identifies new components of the fission and budding yeast anaphase-promoting complexes. Curr Biol 12: 2048–2054 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S (2005) Epigenetic inheritance in Arabidopsis: selective silence. Curr Opin Genet Dev 15: 557–562 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data