Abstract
We isolated MED25, which associates with retinoic acid (RA)-bound retinoic acid receptor (RAR) through the C-terminal nuclear hormone receptor (NR) box/LxxLL motif, and increases RAR/RXR-mediated transcription. When tethered to a promoter, MED25 showed intrinsic transcriptional activity in its PTOV domain, which is likely accomplished by direct association with CBP. Reporter assays using dominant negatives of MED25 demonstrated the importance of the N-terminal Mediator-binding and C-terminal domains in CBP and RAR/RXR binding, which affect MED25 activity. Downregulation of MED25 specifically reduced RAR but not thyroid hormone receptor (TR) activity. Stimulation of RAR by MED25 was correlated with enhanced RA cytotoxicity in vivo. Chromatin immunoprecipitation (ChIP) assays revealed the RA-dependent recruitment of MED25 to the RARβ2 promoter. Recruitment of CBP and TRAP220 was diminished by the overexpression of a MED25 NR box deletion mutant, and by treatment with MED25 siRNA. Time-course ChIP assays indicated that CBP, together with RAR and MED25, is recruited early, whereas TRAP220 is recruited later to the promoter. Our data suggest that MED25, in cooperation with CBP and Mediators through its distinct domains, imposes a selective advantage on RAR/RXR activation.
Keywords: CBP, MED25, mediator, RAR, TRAP220/MED1
Introduction
Retinoic acid (RA) signaling is mediated by retinoic acid receptor (RAR) and retinoid X receptor (RXR), which are members of the nuclear hormone receptor (NR) superfamily (Tsai and O'Malley, 1994; Chambon, 1995; Mangelsdorf et al, 1995). RA-bound receptors activate gene transcription in association with cis-acting RA response elements (RARE) located in the regulatory regions of target genes. RARs and RXRs contain two activation functions (AFs) (reviewed in Chambon, 1996): ligand-independent N-terminal AF-1 and RA-induced AF-2 associated with the ligand-binding domain. Transcriptional regulation by NRs, including RARs and RXRs, involves the binding and recruitment of coactivators and/or Mediators to target gene promoters (McKenna and O'Malley, 2002).
The best-characterized NR coactivators identified thus far are members of the SRC/p160 family of proteins (McKenna et al, 1999; Leo and Chen, 2000). Although the exact mechanism of action of the SRC/p160 proteins is unclear, their ability to associate with histone acetyltransferases (HATs), such as CBP/p300 (Chakravarti et al, 1996; Hanstein et al, 1996; Torchia et al, 1997; Voegel et al, 1998) and pCAF (Blanco et al, 1998), the intrinsic HAT activity of some family members (Chen et al, 1997; Spencer et al, 1997) suggest a role in chromatin remodeling. A second type of NR coactivator complex is the multimeric thyroid hormone receptor (TR)-associated protein (TRAP)–Mediator complex, which copurifies with ligand-activated TR from HeLa cells. It is composed of at least 16 different polypeptides ranging in size from 15 to 240 kDa (Fondell et al, 1996; Ito and Roeder, 2001). Most TRAP–Mediator subunits have been identified in other metazoan holocomplexes, including NAT, DRIP, ARC, and CRSP (reviewed in Malik and Roeder, 2000). The ability of the TRAP complex to stimulate TR-mediated transcription in vitro on naked DNA templates in the absence of TATA-binding protein-associated factors suggests that TRAPs mediate a novel NR coactivator pathway or an activation step distinct from those mediated by the SRC/p160 proteins and CBP/p300, possibly involving a more direct influence on the basal transcription machinery (Fondell et al, 1999). One component of the TRAP–Mediator complex, TRAP220 (also called ARC/DRIP205, PBP, CRSP200, and MED1), directly associates with TR, VDR, and PPAR through its NR box. Although TRAP220 can interact with other NRs, including RAR and RXR, data derived from TRAP220-null mouse embryo fibroblasts indicate that TRAP220 is dispensable for the transcriptional activation of these receptors, but is critical for the function of TR (Malik et al, 2004). These data suggest that Mediator component(s) other than TRAP220 may be present for the activation of RAR/RXR.
Using a yeast two-hybrid screen, we identified MED25 (also called p78, PTOV2, ARC92, and ACID1), which interacts with RAR in the presence of all-trans retinoic acid (AtRA). Here, we report the isolation, characterization, and function of MED25. Our results suggest a role for MED25 in chromatin modification and preinitiation complex assembly through the recruitment of the histone acetyltransferase CBP and a Mediator complex to RA-responsive promoters. Our results further indicate that MED25 may efficiently coordinate the transcriptional activation of RAR/RXR in higher eukaryotic cells.
Results
Isolation of MED25 and its interaction with the ligand-binding domain of RAR
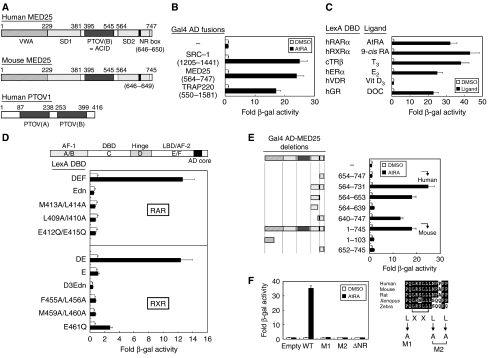
From a yeast two-hybrid screen of a HeLa cDNA library, using the ligand-binding domain (or DEF) of RARα as bait, we isolated the known NR coregulators SRC-1, RIP140, and PNRC, as well as p78 (or PTOV2), which is also known as ACID1 or ARC92 (Benedit et al, 2001; Wang et al, 2002; Mittler et al, 2003; Yang et al, 2004). These proteins were recently designated as MED25 of the Mediator complex (Bourbon et al, 2004) and will hereafter be referred to as MED25. A conserved domain database search using the primary sequence of MED25 revealed several notable domains (Figure 1A). A PTOV domain is located between the SD1 and SD2 domains, whereas the PTOV1 protein contains two conserved PTOV domains (Benedit et al, 2001). Similar to SRC-1 and TRAP220, MED25 showed a strong ligand-dependent interaction with RARα (Figure 1B). In addition to RAR, MED25 also interacts with RXR, TR, ER, and glucocorticoid receptor (GR), but not with vitamin D nuclear receptor (VDR), in the presence of their cognate ligands (Figure 1C). The absence of an interaction with VDR seems to be specific, as VDR was active in the interaction with SRC-1 (data not shown). No RAR or RXR isotype-specific interactions were found in our yeast assays (data not shown).
Figure 1.
Characterization of the interaction between MED25 and RAR in yeast. (A) Structural features of MED25 (human and mouse homologs) and PTOV1 (human). Notable domains include a von Willebrand factor type A (VWA) domain, a synapsin I domain 1 (SD1), a PTOV (a conserved region found in prostate tumor overexpressed protein 1) domain, a VP16 activator-interacting domain (ACID), an SD2 domain, and an NR box (LRSLL). (B) Isolation of MED25, which interacts with RAR DEF in the presence of retinoid, by yeast two-hybrid screening. Interactions between other known RAR cofactors (SRC-1 and TRAP220) and RAR are shown for comparison. (C) Interaction of MED25 with other nuclear receptors. The assays were performed using the indicated NRs in the presence of their cognate ligands. (D) The AF-2 AD core of RAR/RXR is critical for its ligand-dependent interaction with MED25. The location of the AF-2 AD core is shown (Edn, E region with deleted AF-2 AD, residues 212–403 of hRARα; D3Edn, D3E region with deleted AF-2 AD, residues 237–448 of mRXRα), and the point mutations are indicated. (E) Mapping of the MED25 domain responsible for the interaction with RAR. Additional deletions were constructed using the original human MED25 (residues 564–747) clone isolated by yeast-two hybrid screening. For full-length MED25, the mouse homolog was used. Schematic representations of full-length MED25 and its deletion mutants. (F) The NR box (LRSLL) of MED25 is critical for the interaction. Four NR box mutants (L → A) were designed on the basis of an alignment of MED25 from various mammalian species, as indicated. In all experiments, yeast two-hybrid and β-galactosidase assays were used to determine the interaction between LexA-fused nuclear receptors, including RAR, and Gal4 AD-fused RAR cofactors, including MED25 (and its mutants). Fold activity indicates the value relative to that of the Gal4 AD empty control.
Mapping of the domains involved in the RAR/MED25 interaction
The AF-2 activation domain (AD) core of NRs is critical for coactivator binding and coactivator-mediated transcriptional activation; therefore, we tested whether the AF-2 AD core of RAR/RXR is vital for its interaction with MED25. As indicated in Figure 1D, wild-type RAR and RXR showed ligand-dependent interactions with MED25, whereas no interaction was observed with dominant-negative (dn) MED25 (i.e., AF-2 AD core deletions) or with most of the AD core point mutants, except the E → Q mutant. None of the mutants except E → Q showed transcriptional activity when fused to Gal4 DNA-binding domain (DBD); similar results were obtained with SRC-1 (data not shown).
The human MED25 clone isolated in our screen codes for amino acids 564–747. A conserved motif search revealed one LxxLL motif (NR box, LRSLL; residues 646–650). To demonstrate that the NR box is responsible for the RAR interaction, several deletion mutants of MED25 were generated, and their interactions with RAR were determined by yeast two-hybrid assay. The MED25 derivatives containing the NR box (residues 564–731, 564–653, and 640–747) retained the ability to interact with RAR, whereas those derivatives that lacked the NR box (residues 654–747 and 564–639) did not (Figure 1E). Full-length mouse MED25 (mMED25) interacted with RAR to the same extent as the human homolog. In addition, mMED25 derivatives in which the NR box had been deleted showed no interaction. Subsequently, four NR box point mutants (L → A) were generated by PCR and subcloned into pGAD10, which harbors the Gal4 AD fusion. As shown in Figure 1F, all three leucines were required for the binding of MED25 to RAR. Overall, these data suggest that the C-terminal NR box in MED25 and the AF-2 AD core in RAR (or RXR) are important for the ligand-dependent interaction between the two proteins.
In vitro and in vivo interactions
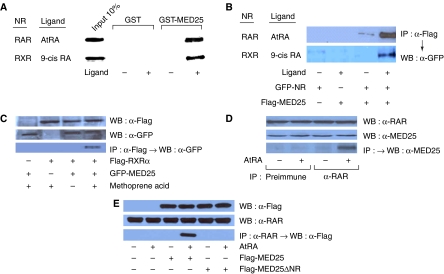
To confirm the physical interaction between RAR (or RXR) and MED25 in vitro and in vivo, we performed GST pull-down and immunoprecipitation (IP) assays, respectively. Our GST pull-down assays, performed using GST-fused human MED25 (residues 564–747) and in vitro translated, [35S]-labeled retinoid receptors, confirmed the ligand-dependent interaction of MED25 with RAR (and RXR) (Figure 2A).
Figure 2.
MED25 interaction with RAR in vitro and in mammalian cells. (A) In vitro translated [35S]-labeled NR (hRAR and mRXR) was incubated with 2 μg of GST-fused MED25 (residues 564–747) in the presence of the cognate ligand (2 μM). Bound proteins were visualized by SDS–PAGE and autoradiography. (B) NIH3T3 cells were cotransfected with 2 μg of GFP-NR (hRAR and mRXR) and 2 μg of the Flag-mMED25 (mouse, full length) expression vectors. IP was carried out using anti-Flag antibody. The precipitated proteins were visualized by WB using anti-GFP antibody. (C) HeLa cells were cotransfected with 2 μg each of the GFP-MED25 and Flag-RXRα expression vectors in the presence of the RXR-specific agonist methoprene acid. IP was carried out using anti-Flag antibody. The precipitated proteins were visualized by WB using anti-GFP antibody. (D) Interaction between endogenous proteins. HeLa cells were cultured in the absence and presence of AtRA. Cell extracts were subjected to IP with preimmune serum or anti-RAR antibody. Interactions were detected by WB using polyclonal anti-MED25 antibody raised against residues 48–53 of human MED25 in rabbit. The inputs were 10% of the IP samples, and are shown by WB using anti-RAR and -MED25 antibodies. (E) Deletion of the NR box abrogates the interaction. Flag-mMED25 or Flag-mMED25 ΔNR (NR box deletion) was expressed in HEK293 cells in the presence of AtRA. The interaction was determined by IP using anti-RARα antibody followed by WB using anti-Flag antibody. The inputs were 10% of the IP samples.
For the IP assays, NIH3T3 cells were cotransfected with GFP-fused RAR and Flag-tagged mMED25 constructs. IP with an anti-Flag antibody and subsequent Western blotting (WB) with an anti-GFP antibody demonstrated the ligand-dependent interaction of MED25 with mRARα and mRXRα (Figure 2B). The interaction with RXRα was confirmed using an RXR-specific agonist, methoprene acid (BIOMOL, Plymouth Meeting, PA) (Figure 2C). The endogenous interaction was further verified by IP using anti-RAR and WB with anti-MED25 antibodies (Figure 2D). The NR box deletion mutant of MED25 (ΔNR) exhibited defective RAR binding in HEK293 cells (Figure 2E), confirming that the interaction of MED25 with RAR occurs via its NR box.
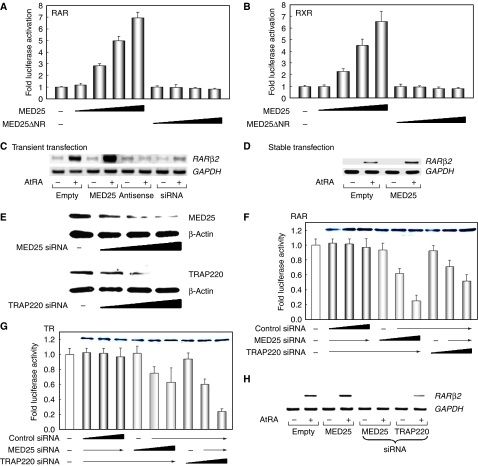
Effect of MED25 on the transcriptional activity of RAR/RXR
To determine whether the RAR/MED25 interaction is functionally relevant, the effect of MED25 on RAR (or RXR) activity was investigated using a tk-luciferase reporter containing the RARE (or RXRE). As shown in Figure 3A and B, MED25 strongly enhanced RAR/RXR-mediated transcription in a ligand- and dose-dependent manner. However, no stimulatory effect was observed with the NR box deletion mutant. Instead, increased MED25 ΔNR expression slightly decreased the ligand-dependent transcriptional activity of RAR/RXR, suggesting a dominant-negative interaction between MED25 ΔNR and endogenous MED25.
Figure 3.
MED25 is a coactivator of RAR as determined using overexpression (A–D) and reduced expression (E–H) conditions. (A, B) Effect of MED25 on the transcriptional activity of RAR and RXR. NIH3T3 cells were transfected with expression vectors for RAR (or RXR) and MED25 (or ΔNR), with RARE- or RXRE-tk-luciferase, in the absence or presence of 1 μM ligand (AtRA for RAR; 9-cis RA for RXR). Relative luciferase activity was determined by luciferase assay after normalizing to the observed β-galactosidase activity. The fold activation is a comparison between the luciferase activity in the presence and absence of ligand. The data shown are averages from three independent experiments. (C, D) Effect of MED25 on endogenous RARβ2 expression. (C) HeLa cells were transiently transfected with a MED25 sense or antisense expression vector and treated with MED25 siRNA. (D) MCF-7 cells stably expressing MED25 were selected by G-418 treatment. Total RNA was extracted from each cell and subjected to RT–PCR using primer pairs specific for the RARβ2 coding sequence. GAPDH was used as an internal control. (E–H) Downregulation of MED25 resulted in a significant and specific reduction in RAR activity. (E) Increasing amounts (0, 80, 160, and 240 pmol) of MED25 or TRAP220 siRNA were added to HeLa cells. The extent of knockdown was monitored by WB. β-Actin was used as an internal control. (F) HeLa cells were transfected with control, MED25, or TRAP220 siRNA (0, 40, 80, and 120 pmol) and the RARE-tk-luciferase reporter in the presence of 1 μM of AtRA. The level of endogenous RARα was determined by WB (inset). Luciferase activity was determined as described above. (G) The DR4-tk-luciferase reporter was used instead, and the level of TR is shown (inset). Each graph shows the average data obtained from three independent experiments. (H) The knockdown effect was analyzed by RT–PCR using primer pairs specific for the RARβ2 coding sequence.
We next assessed the effect of the level of MED25 on endogenous AtRA-regulated gene expression using transient and stable transfection assays. Transfected cells were treated with vehicle or AtRA; thereafter, the transcription of a known RAR target gene, RARβ2, was monitored by RT–PCR. MED25 expression was determined by WB using either anti-Flag or anti-MED25 antibody (data not shown). As shown in Figure 3C and D, the mRNA expression of RARβ2 was greatly increased by both transient (in HeLa cells) and stable (in MCF-7 cells) overexpression of MED25 in the presence of AtRA compared with the level of the control GAPDH. To avoid nonphysiological effects such as squelching, which are sometimes associated with overexpression studies, the effect of MED25 downregulation was monitored using antisense MED25 DNA or siRNA treatment. As shown in Figure 3C, RARβ2 expression was greatly reduced under these conditions, while the control siRNA had little effect on RA-dependent RARβ2 expression (Supplementary Figure S1). The expression of p21WAF1, another RA- and growth arrest-responsive gene (Liu et al, 1996), was also significantly increased by MED25 overexpression and reduced by treatment with MED25 siRNA (Supplementary Figure S1).
To compare the NR specificity of MED25 with that of TRAP220, assays were performed using specific siRNAs; Mediator expression was determined by WB (Figure 3E). MED25 siRNA, but not the control siRNA, effectively decreased RA-dependent luciferase activity without affecting RAR expression (Figure 3F); however, the repressive effect of TRAP220 siRNA on RAR activity was marginal compared with that of the MED25 siRNA. In contrast, the MED25 and TRAP220 siRNAs had opposite effects on TR activity (Figure 3G), suggesting that MED25 may be a candidate mediator for RAR, whereas TRAP220 may mediate TR activity. Neither the MED25 nor TRAP220 siRNA had an effect on p53-mediated transcriptional activity (data not shown). The effect of these Mediators on RAR activity was further confirmed by RT–PCR analysis of endogenous RARβ2 expression (Figure 3H). Additional confirmation was provided by RT–PCR of the control siRNA (Supplementary Figure S1B). Overall, these results suggest that MED25 mediates RAR rather than TR activity.
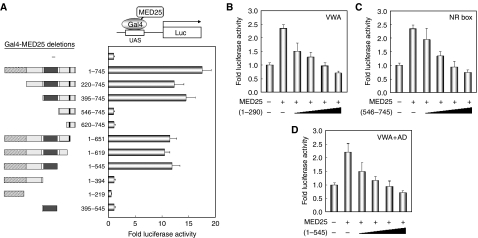
The MED25 domains responsible for transcriptional activation of RAR/RXR
To examine the role of MED25 as a coactivator and to establish the domains required for this function, Gal4 DBD-fused MED25 deletions were generated, and the expression of each construct was monitored with anti-Gal4 antibody (data not shown). Upon transfection with the Gal4 DBD-responsive reporter into COS-1 cells, full-length MED25 showed strong transcriptional activity. Analysis of a series of N- and C-terminal truncations revealed that MED25 residues 395–545 are required for autonomous activation (Figure 4A); however, this region was insufficient for activation, probably due to instability (Mittler et al, 2003). Structurally, this region corresponds to the PTOV (or ACID) domain (Benedit et al, 2001; Yang et al, 2004). Interestingly, PTOV1, which contains two PTOV domains in tandem, was transcriptionally active when tethered to the promoter (Supplementary Figure S2A). Given these findings and our in vitro and yeast interaction data (Supplementary Figure S2C and D), MED25 may be a coactivator of RAR/RXR. As PTOV1 harbors autonomous transcriptional activity but does not interact with RAR/RXR, it may exert a dominant-negative effect on MED25 (Supplementary Figure S2B).
Figure 4.
The MED25 domain responsible for transcriptional activation of RAR. (A) Mapping of the domain required for autonomous transcriptional activity. Gal4-DBD full-length mouse MED25 and -deletion mutant expression vectors (0.25 μg each) were transfected into HeLa cells together with a Gal4-responsive 17-mer tk-luciferase reporter. A schematic representation of the MED25 fragments. Relative luciferase activity was determined by luciferase assay after normalizing to the observed β-galactosidase activity. (B–D) Dominant-negative effect of the MED25 mutants. HeLa cells were transfected with expression vectors for RARα and MED25 (0.4 μg) with RARE-tk-luciferase in the presence of AtRA. The cells were further transfected with increasing amounts (0, 0.1, 0.2, 0.3, and 0.4 μg) of each MED25 deletion mutant. The residues included in each fragment are indicated: (1–290), containing the VWA domain; (546–745), the NR box; (1–545), the VWA and AD, as defined above. Luciferase activity was determined as described above. Each graph includes the data obtained from three independent experiments.
To analyze the biological relevance of the MED25 domains required for enhanced RAR activity, a series of transcription–competition assays were performed using MED25 fragments (residues 1–290, 546–745, and 1–545). In the presence of MED25, the level of AtRA-induced RAR activity increased. At a fixed amount of MED25, the level of RAR activity gradually decreased as the amount of the MED25 fragments increased (Figure 4B–D). Similar interference, albeit not as great, was observed in the absence of MED25 overexpression (data not shown). These results indicate that all of the MED25 mutants tested exerted a dominant-negative effect on endogenous MED25. Thus, it is likely that the Mediator-binding domain (residues 1–290), the activation or PTOV domain (residues 395–545), and the NR box (residues 646–650) are functionally important for mediating the ligand-induced transcriptional activity of RAR.
Effect of MED25 on RA cytotoxicity
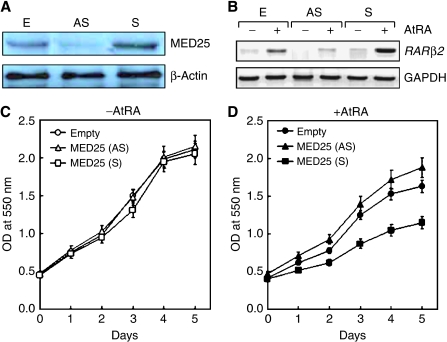
The cytotoxic role of RA has been well documented in various cancer cell lines (Sun and Lotan, 2002); thus, the effect of MED25 on RA cytotoxicity was investigated in MCF-7 cells stably overexpressing or silencing MED25. The expression of MED25 protein and RARβ2 gene was monitored by WB (Figure 5A) and RT–PCR (Figure 5B), respectively. The growth of the MCF-7 cells was only marginally affected by either condition in the absence of AtRA (Figure 5C); however, the growth of cells overexpressing MED25 was greatly retarded in the presence of 5 μM AtRA compared with that of the parental cells stably transfected with the empty expression vector (Figure 5D). Moreover, the suppression of endogenous MED25 expression by a MED25 antisense expression vector increased cellular proliferation. These results suggest that MED25 may cause RA cytotoxicity by activating RAR in vivo. In support of this hypothesis, we determined the effect of MED25 on the expression of p21WAF1, an RAR target responsible for cell cycle arrest, and found that RA-dependent p21WAF1 expression was greatly increased by MED25 overexpression and completely abrogated by MED25 downregulation (Supplementary Figure S1).
Figure 5.
Biological role of MED25 in RA signaling. MCF-7 cells stably transfected with empty, MED25 sense (S), or MED25 antisense (AS) expression vector were selected by G-418 treatment. (A) Cellular levels of MED25 were monitored by WB, using polyclonal anti-MED25 antibody. (B) RARβ2 expression was monitored by RT–PCR as described. (C, D) MTT assays (Cho et al, 2006) were performed to monitor cell growth in the absence (C) and presence of 5 μM AtRA (D).
MED25 cooperates with CBP through its PTOV domain
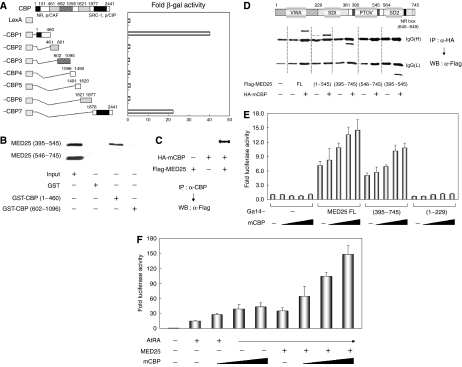
To investigate how the activation function of MED25 is transduced to RAR, we tested for an association between MED25 and the candidate coactivator CBP, which exhibits HAT activity. Yeast two-hybrid assays using LexA-fused CBP fragments and Gal4-fused MED25 indicated that the N-terminal (residues 1–460) and, to a lesser extent, the C-terminal (residues 1878–2441) fragments of CBP could interact with MED25 (Figure 6A). These regions are responsible for the HAT activity of CBP, SRC-1 and p/CIP binding, and NR binding, suggesting that MED25 serves those functions. GST pull-down assays also indicated that residues 395–545 of MED25, but not residues 546–745, interact in vitro with CBP residues 1–460, but not with CBP residues 602–1096 (Figure 6B). The CBP/MED25 interaction was further confirmed by IP using cells transfected with hemagglutinin (HA)-tagged CBP and Flag-tagged MED25 (Figure 6C). To map the region of MED25 required for its interaction with CBP, NIH3T3 cells were cotransfected with HA-fused CBP and Flag-tagged MED25 deletion constructs. IP with anti-HA antibody and subsequent WB with anti-Flag antibody revealed that the interaction of MED25 with CBP requires residues 395–545, which correspond to the PTOV, or ACID, domain (Figure 6D). Of note, an interaction was also observed between CBP and PTOV1, which contains two PTOV domains and shows marked transcriptional activation when targeted to a promoter (Supplementary Figure S2).
Figure 6.
Cooperation of MED25 with CBP. (A) The interaction between MED25 and CBP in yeast. Yeast two-hybrid assays were performed using LexA DBD fused to CBP deletions and Gal4 AD fused to MED25. Interactions were monitored by β-galactosidase assay. (B) In vitro interactions between MED25 and CBP. [35S]-labeled MED25 (residues 395–545) or MED25 (residues 546–745) was mixed with GST, GST-CBP (residues 1–460), or GST-CBP (residues 602–1096). The GST pull-down results were visualized by autoradiography. (C) Interaction between MED25 and CBP in mammalian cells. HA-tagged CBP and Flag-tagged MED25 expression vectors were transfected alone or together into NIH3T3 cells. IP of the cellular extracts with anti-CBP antiserum was followed by WB with anti-Flag antibody. (D) Mapping of the MED25 domain needed to bind CBP. NIH3T3 cells were transfected with HA-tagged CBP and Flag-tagged MED25 (or its deletions). WB using anti-Flag antibody was performed using precipitates obtained from cellular extracts and anti-HA antibody. The locations of the IgG heavy and light chains are indicated. (E) Effect of CBP on the intrinsic transcriptional activity of MED25. NIH3T3 cells were transfected with Gal4 DBD-responsive luciferase reporter, 0.1 μg of Gal4 DBD-fused MED25 (or its deletions), and increasing amounts (0, 0.01, 0.05, 0.1, and 0.2 μg) of CBP expression vector. (F) Effect of CBP on MED25-mediated transcriptional enhancement of RAR. NIH3T3 cells were transfected with RARE-tk-luciferase reporter, RARα, and increasing amounts (0, 0.1, 0.2, and 0.4 μg) of CBP expression vector in the absence or presence of MED25. Extracts were subjected to luciferase assays as described (E, F).
The role of the CBP/MED25 interaction was determined by two sets of transfection assays. First, NIH3T3 cells were cotransfected with a Gal4-responsive luciferase reporter and Gal4-MED25, -MED25 (residues 395–745), or -MED25 (residues 1–229) plus increasing amounts of CBP. As shown in Figure 6E, CBP gradually increased the autonomous transcriptional activity of MED25 and MED25 (395–745); CBP-binding-defective MED25 (1–229) was unresponsive. In the second assay, increased CBP expression enhanced the transcriptional activity of RAR and further stimulated RAR activity in the presence of MED25 (Figure 6F). MED25 residues 395–545 were also required for autonomous transcriptional activity and the transcriptional enhancement of RAR. Therefore, MED25, acting through the region comprising residues 395–545, may activate RAR by recruiting CBP for histone acetylation.
Sequential recruitment of MED25, CBP, and TRAP220 to RA-responsive chromatin
To confirm that the interaction between MED25 and RAR, CBP, or TRAP220 occurs in vivo, HEK293 cells were transfected with Flag-MED25 in the presence of AtRA. IP demonstrated that MED25 associates with RAR in a ligand-dependent manner (Figure 7A). In addition, overexpressed MED25 could interact with endogenous CBP. Further IP analysis showed that MED25 associates with the Mediator TRAP220. No ligand was required for the interaction between MED25 and CBP or TRAP220.
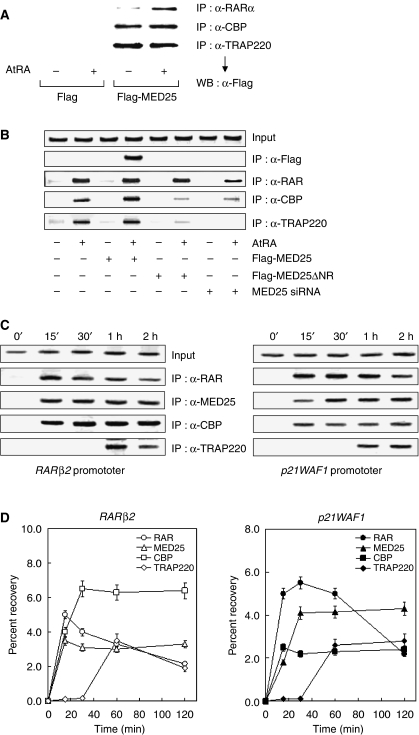
Figure 7.
Chromatin association of MED25 with RAR, CBP, and TRAP220. (A) Binding of MED25 to endogenous RAR, CBP, and TRAP220. HEK293 cells were transfected with Flag-MED25 expression vector in the absence or presence of 2 μM AtRA. RAR and the indicated cofactors were immunoprecipitated from whole extracts of transfected cells using the indicated antibodies. MED25 was detected in precipitates by WB with anti-Flag antibody. (B) AtRA-dependent recruitment of MED25 and other RAR cofactors. HEK293 cells were transfected with Flag-MED25 or MED25 ΔNR expression vector or MED25 siRNA and then treated with or without 2 μM AtRA for 2 h. Crosslinked chromatin was prepared and immunoprecipitated with the antibodies indicated on the right. The precipitates were subjected to PCR analysis using primer pairs spanning the human RARβ2 promoter. The control represents the PCR product obtained before IP. (C) Sequential recruitment of MED25 and other RAR cofactors. ChIP assays were performed as in Figure 7B, except that HeLa cells were treated with AtRA for varying lengths of time (shown above the lanes) and another RA-responsive promoter was included (p21WAF1). The primers used for the p21WAF1 promoter were obtained from Zeng et al (2002). The antibodies used are indicated on the right. (D) Real-time quantitative PCR was performed using the double-stranded DNA binding dye SYBR Green in an iCycler (Bio-Rad). All of the data (the average of three independent amplifications) represent the recovery of each DNA fragment relative to the total input DNA.
Chromatin immunoprecipitation (ChIP) assays performed at a steady state indicated that Flag-tagged MED25, but not Flag alone, associates with the AtRA-responsive RARβ2 promoter in the presence of AtRA (Figure 7B). The defective association of MED25 ΔNR with the promoter implies that the NR box of MED25 is critical for its recruitment to the promoter, probably through an interaction with prebound RAR. The association of RAR with the promoter was unaffected by the presence of MED25; however, MED25 expression slightly increased the binding of CBP and TRAP220 to the promoter. Interestingly, chromatin binding by CBP and TRAP220 was greatly impaired by the expression of MED25 ΔNR, which is defective in RAR binding but retains the domains responsible for binding CBP and TRAP220. Similar results were obtained using MED25 siRNA. These effects were further confirmed by a separate ChIP assay that included control siRNA (Supplementary Figure S3). Thus, MED25 is critical for the recruitment of CBP and TRAP220 to RAR-responsive promoters in vivo.
To determine the recruitment order of the RAR cofactors, HeLa cells were cultured with AtRA for varying lengths of time and then used for ChIP assays (Figure 7C). Most RAR was recruited to the RARβ2 promoter within 15 min of AtRA treatment; however, after 30 min, additional RAR was recruited, which remained at the promoter for the duration of the experiment. MED25 reached maximal binding at 15 min and persisted until the end of the experiment. Interestingly, CBP reached its maximal level of chromatin binding within 30 min and subsequently decreased. In contrast to the rapid recruitment of RAR, MED25, and CBP, the recruitment of TRAP220 to the RARβ2 promoter displayed slower kinetics, reaching its maximal level at 2 h. The p21WAF1 promoter was used to confirm the sequential recruitment of MED25, CBP, and TRAP220. As indicated, the time-dependent order of recruitment of these factors to the p21WAF1 promoter was the same as that to the RARβ2 promoter. Consistent with these observations, quantitative real-time PCR showed a similar pattern of recruitment (Figure 7D). Overall, these results suggest the following sequential association of RAR cofactors to AtRA-responsive promoters: RAR and MED25 → CBP → TRAP220.
The role of Mediator binding in MED25 function
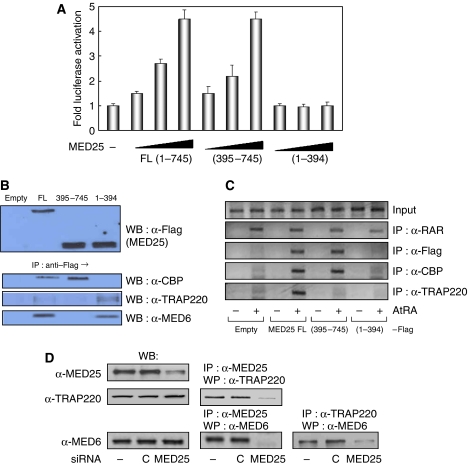
Luciferase reporter assays indicated that full-length MED25 and MED25 (395–745), which harbors the CBP-binding domain, stimulate RAR activity to similar levels, whereas MED25 (1–394), which contains the Mediator-binding domain, does not (Figure 8A). The interactions among the MED25 fragments, CBP, and the Mediators (TRAP220 and MED6) were confirmed by IP analysis (Figure 8B). ChIP assays also demonstrated that these protein–protein interactions are required for recruitment to the chromatin-associated RARβ2 promoter (Figure 8C). These results suggest that the Mediator-binding domain of MED25 does not stimulate RAR activity, although it is required for Mediator binding and recruitment to chromatin. However, an endogenous experiment using siRNA indicated that TRAP220 recruitment absolutely depends on the presence of MED25 (Figure 7B). In addition, knocking down MED25 impaired its ability to interact with TRAP220 and with MED6, and it further reduced the association of TRAP220 with MED6, which may indicate Mediator complex formation (Figure 8D). These studies suggest that MED25 is required for recruitment of the Mediator complex to the chromatin-associated RAR target promoter in vivo, although its role in RAR stimulation is unclear.
Figure 8.
Role of Mediator binding in MED25 function. (A) HeLa cells were transfected with the expression vectors for full-length MED25 (FL) or a MED25 fragment (residues 395–745 or 1–394) with RARE-tk-luciferase in the absence or presence of ligand (1 μM). The fold activation is a comparison between the luciferase activity in the presence and absence of ligand. (B) Flag-MED25 FL or a MED25 fragment (residues 395–745 or 1–394) was expressed in HeLa cells. The interaction was determined by IP using anti-Flag antibody and WB using anti-CBP, anti-TRAP220, and anti-MED6 antibodies. (C) After the transfections described above, crosslinked chromatin was prepared and immunoprecipitated with the antibodies indicated on the right. The precipitates were subjected to PCR analysis using primer pairs spanning the human RARβ2 promoter. (D) HeLa cells were treated with MED25 siRNA (−, untreated; C, control siRNA) and the extracts were subjected to WB with the indicated antibodies. The interaction was determined by IP followed by WB using the indicated antibodies.
Discussion
The Mediator complex, which is highly conserved from yeast to humans, is responsible for diverse transcriptional activation, including basal transcription, by recruiting RNA polymerase II to the target promoter (Boube et al, 2000; Borggrefe et al, 2002; Sato et al, 2004). Documented examples of transcriptional activation indicate that TRAP80 (MED17) targets p53 and VP16 for activation (Ito et al, 1999), while MED23 (TRAP150β or SUR-2) targets E1A and phosphorylated Elk1 (Stevens et al, 2002), MED15 targets Smad2/3 (Kato et al, 2002), and MED16 targets differentiation-inducing factor (Kim et al, 2004). TRAP220/DRIP205 was originally identified as a component of the TRAP/DRIP complexes that interact with TR and VDR in the presence of their cognate ligands (Yuan et al, 1998; Rachez et al, 1999). Other studies have isolated similar or identical complexes, including ARC (Naar et al, 1999) and CRSP (Ryu and Tjian, 1999). TRAP220/DRIP205 was also identified as a PPAR-binding protein and was thus named PBP (Zhu et al, 1997). TRAP220/DRIP205/PBP was recently renamed MED1 (Bourbon et al, 2004). It is well known that TRAP220 mediates the transcriptional activation of TR, VDR, and PPAR via a direct interaction between its two NR boxes and the AF-2 AD core of the NR. MED25 was originally identified as a PTOV1 relative (Benedit et al, 2001; Wang et al, 2002) and was later found to be identical to ACID1 or ARC92, which is a direct target of the activator VP16 (Mittler et al, 2003; Yang et al, 2004). Interestingly, MED25 is present in humans and Drosophila, and a GenBank search indicated that MED25 is present in the genomes of Xenopus and zebrafish, but neither MED25 nor the related PTOV1 has been found in Caenorhabditis elegans or yeast (Benedit et al, 2001). As most Mediator components are present in lower eukaryotes such as yeast, the presence of MED25 in vertebrates alone may indicate an evolutionary advantage for transcriptional activation in higher eukaryotes. We found that MED25 interacts with RAR and RXR in a ligand-dependent manner. Further studies demonstrated that MED25 associates with the AF-2 AD core in RAR (or RXR) through its C-terminal NR box, causing the transcriptional activation of RAR/RXR.
Although MED25 shares several features with TRAP220, it has a distinct structure and function in NR activation. Structurally, MED25 contains several notable features: an N-terminal VWA domain (residues 1–228; responsible for Mediator binding), a central ACID domain (residues 395–545; for binding to the VP16 activation domain or PTOV box), and a C-terminal NR box (residues 646–650; for NR binding). In addition, two SD1 domains (residues 229–381) and an SD2 domain (residues 554–731) are present, although their functions are unclear. Except for the NR box, these domains are not present in TRAP220. MED25 is also functionally distinct from TRAP220 in that MED25 shows autonomous transcriptional activity when tethered to a promoter through fusion to the Gal4 DBD. Furthermore, only TRAP220 links ligand-activated NRs to the preinitiation complex through its association with RNA polymerase II. Generally, transcriptional activation by NR requires not only histone acetylation by the HAT complex containing CBP/p300, pCAF, and SRC-1/p160 but also NR linkage to the preinitiation complex. These two events could occur sequentially or independently. TRAP220 plays a role in recruiting the preinitiation complex to NR-specific promoters and thus requires additional HAT activity for full NR activation. In this regard, HAT activity can be delivered by a direct interaction between the NR and the HAT complex. However, these processes would be more efficient if one factor were to recruit both the preinitiation and HAT complexes. Such is the case for MED25, which can interact with both CBP and the Mediator complex through its central PTOV and N-terminal VWA domains, respectively.
The RAR-binding motif, or NR box, is absolutely required for the recruitment of MED25 to the endogenous RAR-responsive RARβ2 promoter. Overexpression of MED25 ΔNR greatly impaired the recruitment of both CBP and TRAP220. As MED25 ΔNR is defective in chromatin binding but still capable of associating with CBP and TRAP220, it may prevent the binding of CBP and TRAP220 to the RARβ2 promoter. Downregulation of endogenous MED25 also significantly reduced the association of CBP and TRAP220 with the promoter. Under the same conditions, less RAR was recruited. Thus, it is plausible that the promoter association of MED25, CBP, and TRAP220 is important for the function of RAR on the chromatin template. RAR and MED25 were recruited to the promoter at roughly the same time after AtRA treatment (15 min), and they remained largely unchanged during the 2 h that followed. In contrast, CBP and TRAP220 were recruited to the RARβ2 and p21WAF1 promoters at different times (maximum at 30 min and 2 h, respectively; Figure 7C and D). Overall, these results suggest the following sequential association of RAR cofactors at AtRA-responsive chromatin: RAR and MED25 → CBP → TRAP220. First, MED25 is recruited to an RAR-bound promoter in an AtRA-dependent manner. Next, CBP associates with the chromatin via an AtRA-dependent interaction with RAR. During this step, more CBP is recruited to the chromatin, probably through a direct interaction with MED25. Subsequently, RAR- and CBP-bound MED25 recruits TRAP220 to the chromatin, probably through its N-terminal VWA domain.
What is the consequence of Mediator binding to MED25? Our Gal4- and RAR-responsive luciferase assays (Figures 4A and 8A) indicate that the CBP-binding domain of MED25, and not the Mediator-binding domain, is critical for transcriptional activation in the case of overexpression. The Mediator-binding domain appears to contribute to basal transcription (Figure 4B). However, our endogenous experiments using siRNA indicate that MED25 is not only responsive to RA signaling in vivo (Figure 3C, H and F, and Supplementary Figure S1A and B) but also able to recruit TRAP220 to RA-responsive promoters (Figure 7B and Supplementary Figure S3), probably through its N-terminal Mediator-binding domain (Figure 8C). In addition, siRNA-mediated MED25 knockdown impaired the interaction of MED25 with TRAP220 and MED6, and it further reduced the association of TRAP220 with MED6, which may indicate Mediator complex formation (Figure 8D). These data thus suggest that Mediator binding by MED25 may be critical for RA signaling in vivo, although this was not observed during MED25 overexpression.
What is the purpose of the NR coactivators MED25 and TRAP220 in the Mediator complex? Transcription assays followed by transient transfections have shown that TRAP220 stimulates the ligand-activated transcription of TR, VDR, and PPAR, in addition to that of RAR and RXR. However, assays using TRAP220-null fibroblasts derived from a TRAP220–/– mouse indicated that TRAP220 is required for TR activity but not for the transcriptional activity of RAR/RXR (Malik et al, 2004). Furthermore, the phenotype of the TRAP220–/– mouse was distinct from that of the RAR–/– mouse. These data suggest that factors other than TRAP220 may mediate retinoid signaling in TRAP220-null cells. Given that it enhances the activity of RAR and RXR, MED25 may be a physiological candidate for RAR/RXR signaling in vivo, although this hypothesis should be tested using MED25-null cells. Our knockdown assays using MED25 and TRAP220 siRNA support this hypothesis. In this respect, it is possible that the two Mediators target different sets of NRs for activation: TRAP220 for TR, VDR, and PPAR and MED25 for RAR, RXR, ER, and GR.
MED25 is also called PTOV2 because it possesses one of the two conserved PTOV domains present in PTOV1. PTOV1 is overexpressed in certain prostate cancer cell lines and tissues compared with normal tissue; therefore, it is implicated in prostate tumorigenesis (Benedit et al, 2001). Our study demonstrated that the PTOV domain of MED25 is responsible for autonomous transcriptional activation, probably through its interaction with CBP, and thus contributes to the transcriptional activation of RAR/RXR. Further assays revealed that PTOV1 harbors autonomous transcriptional activation, as does MED25, exerts a dominant-negative effect on MED25, and associates with CBP (Supplementary Figure S3). It was recently reported that PTOV1 shuttles between the cytoplasm and the nucleus in a cell cycle-dependent manner, and that its overexpression markedly induces cell proliferation (Santamaria et al, 2005). Considering that ligand-activated RAR/RXR functions in antiproliferation and tumor cell apoptosis (Sun and Lotan, 2002), the transcriptional interference by PTOV1 may help tumor cells escape growth inhibition and thus grow more efficiently. However, it remains to be determined whether the cross-association of MED25 and PTOV1 with CBP affects the proliferation regulated by RA-activated RAR/RXR.
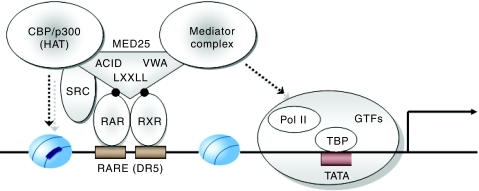
As summarized in Figure 9, MED25 associates with RAR/RXR in the presence of RA through its NR box (LRSLL) and AF-2 AD core, respectively. The N-terminal VWA domain (responsible for Mediator binding), the central ACID domain (responsible for CBP binding), and the C-terminal NR box (responsible for RAR/RXR binding) are required for the transcriptional enhancement of RAR/RXR. These results demonstrate that MED25 functions in chromatin modification and preinitiation complex assembly by recruiting CBP and the Mediator complex, respectively, to RAR/RXR-responsive promoters. These results further suggest that MED25 plays a key role in efficiently coordinating these events in higher eukaryotic cells.
Figure 9.
Model for the transcriptional activation of RAR/RXR by MED25. For details, see the Discussion section.
Materials and methods
Cell lines and cell culture
HeLa, NIH3T3, and HEK293 cell were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% heat-inactivated fetal bovine serum (FBS) and Antibiotic-Antimycotic (all from Invitrogen, Carlsbad, CA) in a 5% CO2 atmosphere at 37°C. For transcription assay, FBS was pretreated with charcoal.
Plasmids and cloning
All cDNAs were constructed according to standard methods and verified by sequencing. The multicopy yeast expression plasmids used in the two-hybrid assays were described previously (Um et al, 1998). Deletion mutants of the desired genes were created by PCR amplification and subcloned into the pBTM116 or pGAD10 (BD Biosciences, Palo Alto, CA). Flag (2 ×)-tagged cTRβ, mRARβ, mRXR, and MED25 genes were placed on pcDNA3 vector (Invitrogen, Carlsbad, CA). Gal4-tagged MED25 deletions were constructed with pG4MpolyII vector (Um et al, 1998). Other MED25 mutants with point mutations or deletion of NR box were created by recombinant PCR and subcloned into Flag-pcDNA3 vector. GFP- and HcRed-tagged constructs were generated using pEGFP-C3 and pHcRed1 (BD Biosciences), respectively. For GST-fused proteins, either pGEX2T or pGEX4T-1 (Amersham Pharmacia Biotech, Piscataway, NJ) was used.
Yeast two-hybrid screening and assays
A Hela cDNA library (BD Biosciences) was screened for proteins that interact with RAR using yeast reporter strain L40. Experimental procedures were same as previously reported (Kim et al, 2002), except LexA-fused hRARα DEF was used as bait in this study. To map the RAR/RXR interaction domain in MED25, deletion derivatives of MED25 were fused to Gal4 AD by subcloning into pGAD10 vector. To localize the MED25 interaction motif in RAR/RXR, AF-2 AD core mutants of RAR/RXR were fused to LexA DBD by subcloning into pBTM116 vector. The level of interaction was determined by β-gal assays.
Glutathione-S-transferase pull-down assays
GST fusions of MED25 and CBP fragments were purified on glutathione–Sepharose beads (Amersham) by standard methods. Indicated NR proteins and MED25 fragments were in vitro translated in rabbit reticulocyte lysate (Promega, Madison, WI) supplemented with [35S]methionine (Amersham). Detailed experimental procedures were previously described (Kim et al, 2002), except that assays were performed in presence of a ligand in this study.
Co-IP assays
After transfection with indicated plasmid DNA using Lipofectamine plus reagent (Invitrogen), either NIH3T3, HeLa, or HEK293, cells were washed in phosphate-buffered saline (PBS), and cell lysates were prepared by adding 1 ml of TEN-modified buffer (50 mM Tris–Cl, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA, 1 mM PMSF) supplemented with protease inhibitors (Roche Molecular Biochemicals, Mannheim, Germany). Lysates were precleared by preincubation with protein A/G-Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) for 15 min, and incubated at 4°C for 2 h with beads and a 1:200 dilution of indicated antibodies: mouse monoclonal anti-Flag M2 antibody (F-3165; Sigma, St Louis, MO); rabbit polyclonal anti-RARα antibody (sc-551; Santa Cruz); rabbit polyclonal anti-CBP antibody (sc-1211; Santa Cruz); mouse monoclonal anti-HA antibody (12CA5; Roche); goat polyclonal anti-TRAP220 antibody (sc-5334; Santa Cruz). Beads were then washed once with TEN and twice with PBS, and the immune complexes were released from the beads by boiling in sample buffer for 5 min. Following electrophoresis on 10% SDS–polyacrylamide gels, immunoprecipitates were analyzed by Western blotting using either rabbit polyclonal anti-GFP antibody (sc-8334; Santa Cruz), anti-Flag antibody (1:1000), or anti-MED25 antibody (polyclonal serum against amino acids 48–53 of human MED25 were generated in rabbits).
Transient transfection and luciferase assay
NIH3T3 or HeLa cells were seeded in 12-well culture plate at a density of 1.5 × 105 cells per well. Cells were then transiently transfected with Gal4-tk-luciferase reporter (or RARE-tk-luciferase) and SV40-driven β-gal expression vector as an internal control. Depending on the experimental conditions, Gal4-MED25, MED25, RAR, RXR, or CBP expression vector was cotransfected. Luciferase activity was measured by adding 20 μl Luciferin into 30 μl of lysates using Analytical Luminescenceluminometer according to the manufacturer's instructions (Promega). β-Galactosidase activity was determined in 96-well plates that were read at 405 nm using ELISA reader. The luciferase activities were normalized to the β-galactosidase activity.
Stable transfection and RT–PCR
MCF-7 cells were transfected with either MED25 subcloned into the G418-resistant vector pcDNA3 (2 × Flag-tagged) or empty vector for 48 h and treated with 1 mg/ml of G418. Every 5–7 days cells were incubated with fresh medium containing G418 for a month. Cells stably expressing Flag-MED25 were verified using anti-Flag antibody.
For RT–PCR, transiently transfected HeLa or stably transfected MCF-7 cells with Fag-MED25, grown in DMEM with 5% charcoal-stripped FBS, were treated with 2 μM AtRA. Total RNA was extracted using TRIzol Reagent (Invitrogen), and 5 μg RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen) and random oligo(dT) primers (New England Biolabs, Beverly, MA). RT products were amplified by PCR using the following primer pairs: for the RARβ2 coding sequence (375 bp), forward, 5′-GGAACGCATTCGGAAGGCTT-3′ and reverse, 5′-AGCACTTCTGGAGTCGACAG-3′; for control GAPDH coding sequence (212 bp), forward, 5′-GTGGATATTGTTGCCATCA-3′ and reverse, 5′-GACTCCACGACGTACTCA-3′.
RNA interference
The sequences of the custom siRNA duplex (Stealth system, Invitrogen) were as follows: for MED25, sense, 5′-UUGGUGAAAUGGAACUGGACCAUCC-3′ and antisense, 5′-GGAUGGUCCAGUUCCAUUUCACCAA-3′; for TRAP220/MED1, sense, 5′-AUGGCUGGAAGAAUUUGUGCUCACC-3′ and antisense, 5′-GGUGAGCACAAAUUCUUCCAGCCAU-3′. The siRNA duplex control used was the Stealth RNAi Negative Control with medium GC content from Invitrogen. Transfection of siRNA was performed with Lipofectamine plus reagent in HeLa cells according to the manufacturer's instructions. In independent experiments, transfection efficiency was assessed to be greater than 90% using fluorescein-labeled siRNA (Invitrogen). Knockdown of MED25 and TRAP220 was verified by Western blotting.
ChIP
Either HEK293 cells transfected with MED25 (or ΔNR or siRNA) or HeLa cells were treated with AtRA (2 μM) for indicated times. Cells were then treated with the crosslinking reagent formaldehyde (1% final concentration) for 10 min at 37°C, rinsed twice with cold PBS, and swollen on ice in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl, pH 8.1) for 10 min. Nuclei were collected and sonicated on ice. Supernatants were obtained by centrifugation for 10 min and diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl, pH 8.1, 167 mM NaCl). The mixture (i.e., fragmented chromatin) was then incubated with 2 μl of anti-Flag, anti-RARα, anti-CBP, or anti-TRAP220 antibody on a rotator at 4°C for 4 h. Then 20 μl of protein A/G PLUS-agarose were added and incubated for 1 h at 4oC with rotation to collect the antibody/chromatin complex. Crosslinked, precipitated chromatin complexes were recovered and reversed according to Upstate's protocol (Upstate, Chicago, IL). Final DNA pellets were recovered and analyzed by PCR, using a pair of primers that encompass the RARβ2 promoter region (288 bp). The primers used were forward 5′-AAGCTCTGTGAGAATCCTG-3′ and reverse 5′-GGATCCTACCCCGACGGTG-3′.
Supplementary Material
Supplementary Figures
Supplementary Material and Figure Legends
Acknowledgments
This work was supported by a grant to S-JU from the Ministry of Science and Technology (2004-01348), Republic of Korea. S-JU, E-JK, and U-HP were supported by BK21 project from Ministry of Education & Human Resources Development.
References
- Benedit P, Paciucci R, Thomson TM, Valeri M, Nadal M, Caceres C, de Torres I, Estivill X, Lozano JJ, Morote J, Reventos J (2001) PTOV1, a novel protein overexpressed in prostate cancer containing a new class of protein homology blocks. Oncogene 20: 1455–1464 [DOI] [PubMed] [Google Scholar]
- Blanco JC, Minucci S, Lu J, Yang XJ, Walker KK, Chen H, Evans RM, Nakatani Y, Ozato K (1998) The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev 12: 1638–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Davis R, Erdjument-Bromage H, Tempst P, Kornberg RD (2002) A complex of the Srb8, -9, -10 and -11 transcriptional regulatory proteins from yeast. J Biol Chem 277: 44202–44207 [DOI] [PubMed] [Google Scholar]
- Boube M, Faucher C, Joulia L, Cribbs DL, Bourbon HM (2000) Drosophila homologs of transcriptional mediator complex subunits are required for adult cell and segment identity specification. Genes Dev 14: 2906–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ (2004) A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell 14: 553–557 [DOI] [PubMed] [Google Scholar]
- Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM (1996) Role of CBP/P300 in nuclear receptor signalling. Nature 383: 99–103 [DOI] [PubMed] [Google Scholar]
- Chambon P (1995) The molecular and genetic dissection of the retinoid signaling pathway. Recent Prog Horm Res 5: 317–332 [DOI] [PubMed] [Google Scholar]
- Chambon P (1996) A decade of molecular biology of retinoic acid receptors. FASEB J 10: 940–954 [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM (1997) Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90: 569–580 [DOI] [PubMed] [Google Scholar]
- Cho YS, Kim EJ, Park UH, Sin HS, Um SJ. (2006) Additional sex comb-like 1 (ASXL1), in cooperation with SRC-1, acts as a ligand-dependent coactivator for retinoic acid receptor. J Biol Chem 281: 17588–17598 [DOI] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG (1996) Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA 9: 8329–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell JD, Guermah M, Malik S, Roeder RG (1999) Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA boxbinding protein-associated factors of TFIID. Proc Natl Acad Sci USA 96: 1959–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M (1996) p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA 93: 11540–11545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Roeder RG (2001) The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab 12: 127–134 [DOI] [PubMed] [Google Scholar]
- Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG (1999) Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell 3: 361–370 [DOI] [PubMed] [Google Scholar]
- Kato Y, Habas R, Katsuyama Y, Naar AM, He X (2002) A component of the ARC/Mediator complex required for TGF beta/Nodal signalling. Nature 418: 641–646 [DOI] [PubMed] [Google Scholar]
- Kim EJ, Park JS, Um SJ (2002) Identification and characterization of HIPK2 interacting with p73 and modulating functions of the p53 family in vivo. J Biol Chem 277: 32020–32028 [DOI] [PubMed] [Google Scholar]
- Kim TW, Kwon YJ, Kim JM, Song YH, Kim SN, Kim YJ (2004) MED16 and MED23 of Mediator are coactivators of lipopolysaccharide- and heat-shock-induced transcriptional activators. Proc Natl Acad Sci USA 101: 12153–12158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo C, Chen JD (2000) The SRC family of nuclear receptor coactivators. Gene 245: 1–11 [DOI] [PubMed] [Google Scholar]
- Liu M, Iavarone A, Freedman LP (1996) Transcriptional activation of the human p21WAF1/CIP1 gene by retinoic acid receptor. J Biol Chem 271: 31723–31728 [DOI] [PubMed] [Google Scholar]
- Malik S, Guermah M, Yuan CX, Wu W, Yamamura S, Roeder RG (2004) Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol Cell Biol 24: 8244–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG (2000) Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci 25: 277–283 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM (1995) The nuclear receptor superfamily: the second decade. Cell 83: 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O'Malley BW (1999) Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20: 321–344 [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108: 465–474 [DOI] [PubMed] [Google Scholar]
- Mittler G, Stuhler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, Meisterernst M (2003) A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J 22: 6494–6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R (1999) Composite co-activator ARC mediates chromatindirected transcriptional activation. Nature 398: 828–832 [DOI] [PubMed] [Google Scholar]
- Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP (1999) Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398: 824–828 [DOI] [PubMed] [Google Scholar]
- Ryu S, Tjian R (1999) Purification of transcription cofactor complex CRSP. Proc Natl Acad Sci USA 96: 7137–7142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A, Castellanos E, Gomez V, Benedit P, Renau-Piqueras J, Morote J, Reventos J, Thomson TM, Paciucci R (2005) PTOV1 enables the nuclear translocation and mitogenic activity of flotillin-1, a major protein of lipid rafts. Mol Cell Biol 25: 1900–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, Conaway JW, Conaway RC (2004) A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell 14: 685–691 [DOI] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O'Malley BW (1997) Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389: 194–198 [DOI] [PubMed] [Google Scholar]
- Stevens JL, Cantin GT, Wang G, Shevchenko A, Berk AJ (2002) Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296: 755–758 [DOI] [PubMed] [Google Scholar]
- Sun SY, Lotan R (2002) Retinoids and their receptors in cancer development and chemoprevention. Crit Rev Oncol Hematol 41: 41–55 [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza Kamei JY, Westin S, Glass CK, Rosenfeld MG (1997) The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387: 677–6849192892 [Google Scholar]
- Tsai MJ, O'Malley BW (1994) Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem 63: 451–486 [DOI] [PubMed] [Google Scholar]
- Um SJ, Harbers M, Benecke A, Pierrat B, Losson R, Chambon P (1998) Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J Biol Chem 273: 20728–20736 [DOI] [PubMed] [Google Scholar]
- Voegel JJ, Heine MJ, Tini M, Vivat V, Chambon P, Gronemeyer H (1998) The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J 17: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, McCarty IM, Balazs L, Li Y, Steiner MS (2002) A prostate-derived cDNA that is mapped to human chromosome 19 encodes a novel protein. Biochem Biophys Res Commun 296: 281–287 [DOI] [PubMed] [Google Scholar]
- Yang F, DeBeaumont R, Zhou S, Naar AM (2004) The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci USA 101: 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG (1998) The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA 95: 7939–7944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Kumar A, Meng G, Gao Q, Dimri G, Wazer D, Band H, Band V (2002) Human papilloma virus 16 E6 oncoprotein inhibits retinoic X receptor-mediated transactivation by targeting human ADA3 coactivator. J Biol Chem 277: 45611–45618 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qi C, Jain S, Rao MS, Reddy JK (1997) Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem 272: 25500–25506 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Material and Figure Legends