Abstract
During T-cell development, thymocytes with intermediate avidity for antigen–MHC complexes are positively selected and then differentiate into functional cytotoxic and helper T cells. This process is controlled by signalling from the T-cell receptor (TCR). Here, we show that the c-Myb transcription factor is a critical downstream regulator of positive selection, promoting the development of helper T cells and blocking the development of cytotoxic T cells. A gain-of-function c-Myb transgene stops development of cytotoxic T cells, instead causing accumulation of a precursor population. Conversely, loss of c-Myb in selecting cells results in significantly fewer helper T cells. In c-Myb-null thymocytes, Gata3, a critical inducer of T-helper cell fate, is not upregulated in response to T-cell receptor signaling, following selection. We show that Gata3 is a direct target of c-Myb, and propose that c-Myb is an important regulator of Gata3, required for transduction of the T-cell receptor signal for subsequent helper cell lineage differentiation.
Keywords: CD4, Gata3, Myb, T cell, transcription
Introduction
The question of how the same extracellular stimulus can in different contexts lead to a multiplicity of cell fates is central to all developing biological systems. In T-cell development, this is exemplified by the processes of positive and negative selection and the subsequent maturation of functional helper and cytotoxic T cells in the thymus. This developmental sequence is fundamental to the generation of an acquired immune system diverse enough to recognise and respond to any foreign antigen, yet stringently controlled to prevent the potentially disastrous recognition of self antigen (Rothenberg and Taghon, 2005).
Positive selection and commitment to either a helper or cytotoxic fate take place after immature thymocytes become double positive (DP) for the cell surface molecules CD4 and CD8, whose eventual function is to act as co-receptors for the T-cell receptor (TCR). Cells entering the DP compartment have already generated and expressed a productively rearranged TCRβ chain, and now start to rearrange the TCRα locus. Whereas the DP cells unable to productively rearrange TCRα, or whose TCR is not signalling-competent, die by neglect, those in which the rearranged TCRα and TCRβ chains form a fully functional TCR upregulate the activation marker CD69, indicating that selection has begun. In this process, DP thymocytes sample antigenic peptides presented to them in the context of MHC Class I or Class II. Those cells whose TCRs bind with very high affinity to peptide–MHC generally die by negative selection (Palmer, 2003), as they are dangerously self-reactive, whereas those bearing lower-affinity TCRs are positively selected (Germain, 2002; Starr et al, 2003), and depending on whether the TCR has been selected on MHC Class I or II, differentiate to become, respectively, CD4−CD8+ single-positive cytotoxic T cells (CD8 SP) or CD4+CD8− single-positive helper T cells (CD4 SP) (Bosselut, 2004; Singer and Bosselut, 2004; Kappes et al, 2005).
The two processes of positive selection and lineage commitment are dependent on the same proximal signalling events post-TCR ligation with antigen–MHC. However, this same initial signal can lead in the first instance to survival and positive selection, but in the second to differentiation down either the CD4 or CD8 lineages; presumably, cells can enforce divergent downstream signalling events, leading to different outcomes. The ‘strength of signal' model for this process holds that for DP cells recognising peptide bound to MHC Class II, co-engagement of the CD4 co-receptor and the TCR leads to a strong signal and CD4 SP fate, whereas for cells encountering peptide in the context of MHC Class I, the weaker CD8 co-receptor-TCR signal leads to CD8 SP fate (reviewed by Kappes et al, 2005). However, recent experiments have led to the ‘kinetic signalling' model, by which the duration and developmental timing of the proximal TCR signal dictates outcome (reviewed by Bosselut, 2004). In this model, any strength of TCR signal below the negative selection threshold induces differentiation of DP thymocytes to an intermediate CD4+CD8lo subset, where the decision to mature down a particular lineage takes place. Cells with an MHC Class II-specific TCR continue to transduce a TCR signal, as the CD4 co-receptor is still present, leading to differentiation down the CD4 lineage. However, cells with an MHC Class I-specific TCR can no longer signal adequately due to the downregulation of the CD8 co-receptor, and loss of the TCR signal results in differentiation down the CD8 lineage.
The nature of the proximal TCR signal has been investigated intensively and is thought to be transduced in two principal ways, via the mitogen-associated protein kinase (MAPK; Alberola-Ila and Hernandez-Hoyos, 2003) and the calcineurin (Neilson et al, 2004) signalling pathways. Experiments to determine how signalling down these pathways leads to commitment to a particular cell fate have identified a number of transcription factors able to dictate or modulate lineage choice. Runx3 and Runx1 are able to repress CD4 transcription and are important for specification of CD8 lineage fate (Taniuchi et al, 2002a; Ehlers et al, 2003; Sato et al, 2005), and the calcineurin-induced Tox protein can induce CD8 differentiation (Wilkinson et al, 2002; Aliahmad et al, 2004). ThPOK acts as a master regulator of CD4 differentiation, able to subvert and dictate cell fate as long as an initiating TCR signal has been received (He et al, 2005; Sun et al, 2005). Gata3 is also required for the development of CD4 SP cells (Hernandez-Hoyos et al, 2003; Pai et al, 2003), and when overexpressed, inhibits CD8 SP development (Nawijn et al, 2001). Gata3 is upregulated in response to TCR ligation following positive selection (Hernandez-Hoyos et al, 2003), but how this is achieved is unknown.
The c-Myb transcription factor has previously been shown to be important at multiple points during T-cell development, being involved in the development and maturation of the most immature thymocytes (Badiani et al, 1994; Allen et al, 1999; Pearson and Weston, 2000; Emambokus et al, 2003; Bender et al, 2004; Lieu et al, 2004) rearrangement of the TCRα, TCRβ, and TCRδ chains (Hernandez-Munain et al, 1996; Hernandez-Munain and Krangel, 2002; Bender et al, 2004), protection from apoptosis (Taylor et al, 1996; Bender et al, 2004), and survival and activation of peripheral T cells (Badiani et al, 1994; Lieu et al, 2004). Here, we show that, in addition to these roles, it is critical for positive selection and the induction of commitment to the CD4 lineage. Inactivation of c-Myb by homologous recombination at the DP stage causes a marked reduction in CD4 SP cell numbers. Conversely, expression of an active form of c-Myb during thymopoiesis results in suppression of the CD8 lineage even in the presence of transgenic TCRs, which normally lead to exclusive development of CD8 SP cells. Expression of activated c-Myb can enhance CD4 SP development, and a CD4+CD8lo intermediate population is observed even in the absence of a positive selection signal. c-Myb's effect on the positive selection signal for CD4 lineage commitment is at least partly due to its regulation of Gata3. Gata3 expression in response to TCR signalling is an early event, occurring shortly after an elevation in c-Myb expression, and it is not induced in the absence of c-Myb. We further define Gata3 as a direct transcriptional target of c-Myb. These data show that c-Myb plays an important role in transducing the post-selection TCR signal.
Results
c-Myb affects lineage choice following positive selection
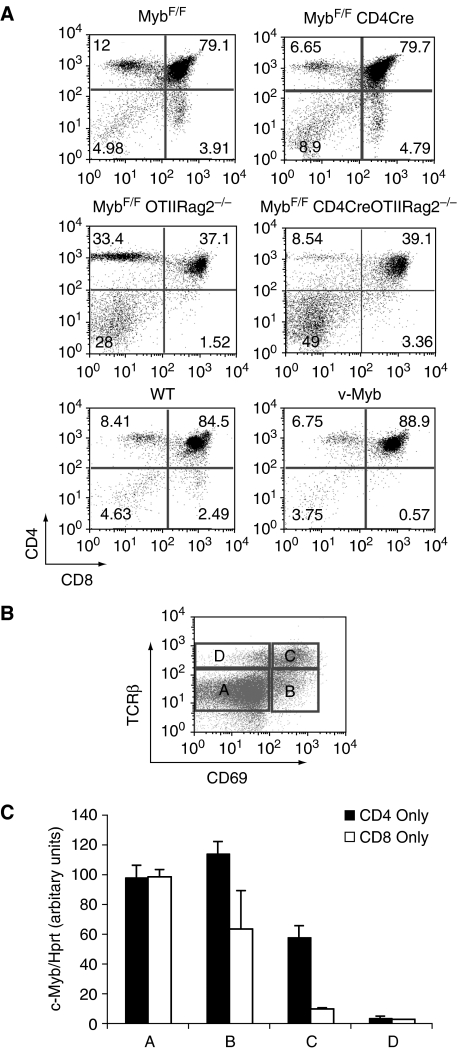
c-Myb activity can be abolished from the DP stage of thymocyte development onwards, by crossing mice homozygous for a floxed Myb allele (MybF/F; Emambokus et al, 2003) to CD4Cre transgenic mice, in which Cre recombinase is under the control of the murine cd4 promoter, enhancer and silencer (Lee et al, 2001). As assessed by qPCR of genomic DNA, the Myb allele is approximately 80% deleted, and Western blotting shows very little residual full-length protein (Supplementary Figure 1). Flow cytometry using CD4 and CD8 as markers of thymocyte subsets showed that c-Myb-knockout thymocytes from MybF/FCD4Cre animals had decreased numbers of CD4 SP cells and an increase in the CD8 SP population (Figure 1A, top two panels and Table I), such that the ratio of CD4 SP:CD8 SP decreased from an average 2.6:1 to 1:1. This effect was magnified when MybF/FCD4Cre animals were crossed onto an OTII:RAG2−/− background (Barnden et al, 1998). In OTII:RAG2−/− mice, exclusively CD4 SP thymocytes develop, as the MHC Class II-specific OTII transgenic TCR directs development of CD4 SP rather than CD8 SP cells, and there is no endogenous TCR rearrangement due to the absence of the rag2 gene. On this background, in the absence of c-Myb, CD4 SP development was reduced by 58% (Figure 1A, centre panels and Table I).
Figure 1.
Phenotypes of loss or gain of c-Myb during positive selection coincide with high levels of c-Myb expression. (A) Percentage distribution of thymocyte subsets measured by expression of CD8 (x-axis) and CD4 (y-axis) in: a MybF/F littermate control versus a MybF/F:CD4Cre mouse (top panels); a MybF/FOTIIRAG2−/− control versus a MybF/F:CD4Cre OTIIRAG2−/− mouse (centre panels), and an age-matched normal control versus a vMyb4 transgenic (lower panels). Data are representative plots. (B) Dot plot showing expression of CD69 (x-axis) and TCRβ (y-axis) on thymocyte subpopulations used in this study. Subsets boxed as (A–D) are successively more mature (C) q-RT–PCR quantitating c-Myb mRNA levels relative to hprt in thymocyte subsets (A–D) from MHC Class I (black bars; CD4 only) and MHC Class II (white bars; CD8 only) mice. Data are the mean of ⩾5 separate experiments. Error bars: standard deviation (s.d.).
Table 1.
T-cell populations
| Cell count × 10−6 (mean±s.d) | DN (%) | DP (%) | CD4 SP (%) | CD8SP (%) | |
|---|---|---|---|---|---|
| Thymus | |||||
| Wild type (n=5) | 68.75 (28.3) | 4.98 (1.12) | 85.25 (0.54) | 7.35 (0.95) | 2.19 (0.23) |
| vMyb4 (n=5) | 122.5 (35.9) | 3.49 (0.29) | 89.62 (0.49) | 6.4 (0.27) | 0.5 (0.05) |
| P-value, H0=identical mean | 0.059 | 0.072 | 0.00005 | 0.14 | 0.0004 |
| MybF/F (n=13) | 120.98 (54.3) | 6.85 (1.98) | 77.67 (3.51) | 11.10 (1.98) | 4.34 (0.49) |
| MybF/FCD4Cre (n=19) | 120.03 (57.76) | 10.76 (3.86) | 76.22 (6.23) | 6.42 (1.57) | 6.57 (1.84) |
| P-value, H0=identical mean | 0.909 | 0.0002 | 0.368 | 1.83E-08 | 1.29E-05 |
| MybF/FOTIIRag2−/− (n=3) | 4.83 (1.79) | 32 (5.29) | 38.36 (3.19) | 27.46 (5.35) | |
| MybF/FCD4Cre OTII Rag2−/−(n=3) | 2.96 (0.45) | 38.66 (9.07) | 48.7 (8.32) | 11.51 (2.76) | |
| P-value, H0=identical mean | 0.208 | 0.346 | 0.153 | 0.019 | |
| HYRag2−/− (n=7) | 57 (28.6) | 21.65 (4.31) | 59.25 (10.21) | 18 (8.17) | |
| vMyb HYRag2−/− (n=9) | 86.88 (67.7) | 11.58 (1.83) | 84.36 (2.62) | 3.35 (2.20) | |
| P-value, H0=identical mean | 0.257 | 0.0004 | 0.0004 | 0.002 | |
| NSHYRag2−/− (n=3) | 53 (7.5) | 25.66 (7.49) | 66.33 (3.51) | ||
| vMybNSHYRag2−/− (n=3) | 70.34 (15.4) | 13.76 (1.87) | 83.5 (2.29) | ||
| P-value, H0=identical mean | 0.083 | 0.102 | 0.003 | ||
| F5 (n=3) | 91.3 (27.2) | 9.20 (5.42) | 71.93 (6.72) | 4.52 (1.22 | 14.56 (1.98) |
| vMybF5 (n=5) | 171.6 (59.9) | 4.86 (1.19) | 75.96 (3.20) | 16.96 (1.59) | 2.33 (0.59) |
| P-value, H0=identical mean | 0.042 | 0.299 | 0.41 | 3.77E-05 | 0.006 |
| Spleen | |||||
| MybF/F (n=5) | 75.8 (3.19) | 17.3 (7.21) | 9.34 (4.82) | ||
| MybF/FCD4Cre (n=5) | 54.8 (11.9) | 10.75 (2.98) | 8.078 (1.93) | ||
| P-value, H0=identical mean | 0.478 | 0.116 | 0.608 | ||
| HYRag2−/− | |||||
| (n=3) | 10.5 (4.27) | 23.66 (7.76) | |||
| vMyb HYRag2−/− | |||||
| (n=4) | 8.75 (2.75) | 0.9 (0.26) | |||
| P-value, H0=identical mean | 0.57 | 0.03 | |||
| F5 (n=3) | 84.6 (32.57) | 9.26 (2.13) | 37.23 (2.65) | ||
| vMybF5 (n=5) | 78 (23.4) | 15.42 (1.49) | 1.45 (0.38) | ||
| P-value, H0=identical mean | 0.77 | 0.019 | 0.001 | ||
To determine whether gain of c-Myb would result in the reciprocal phenotype, namely loss of CD8 SP cells, we analysed thymocytes from vMyb4 transgenic mice. These animals express v-Myb, the oncogenic form of c-Myb, under the control of the human CD2 promoter and LCR, such that there is three- to fivefold overexpression (relative to wt) of v-Myb specifically in T cells (Badiani et al, 1996). Flow cytometry using CD4 and CD8 as markers of thymocyte subsets showed that, in 4–6 week old vMyb4 animals, numbers of mature CD8 SP cells were greatly reduced, whereas CD4 SP cell numbers were slightly higher than wild-type (wt) levels (Figure 1A, bottom panels and Table I). This effect was not due to any oncogenic effect of the v-Myb transgene, as TCR rearrangement in CD4 SP cells was polyclonal, and the mean age of tumour onset in these animals is 15 months (Badiani et al, 1996). These data suggest that c-Myb may play a role in controlling the ratio of CD4 SP to CD8 SP T cells, potentially through regulating positive selection or lineage commitment signals.
c-Myb is highly expressed in selecting DP thymocytes
To establish whether the expression pattern of c-Myb was consistent with its having a role during positive selection and commitment, we examined c-Myb expression during selection to either the CD4 SP or CD8 SP lineage. For this, we used MHC Class I null (β2M°) mice (Koller et al, 1990), whose DP thymocytes can only be selected on MHC Class II, and which therefore make exclusively CD4 SP cells, or MHC Class II null (H2-Ab1°) mice (Cosgrove et al, 1991), whose DP thymocytes can only be selected on MHC Class I, generating only CD8 SP cells. DP and SP thymocytes from each strain were sorted into four populations on the basis of expression levels of the TCRβ chain and of the activation marker CD69. These populations, shown in Figure 1B, were: A (TCRloCD69−); B (TCRintCD69+); C (TCRhiCD69+) and D (TCRhiCD69−), which correspond to: pre-selection (naïve) DP; post-selection DP and CD4+CD8lo; immature SP, and mature SP, respectively (Hernandez-Hoyos et al, 2003). c-Myb expression, assessed by qRT–PCR relative to Hprt, was high in pre-selection DP (Figure 1C, population A), was increased in post-selection DP committing to the CD4 lineage, but dropped if the post-selection DP were destined to become CD8 SP (Figure 1C, population B). In population C, a later stage of commitment, levels of c-Myb mRNA declined in both CD4- and CD8-committed cells, but the decline was again more extreme in the latter (Figure 1C, population C). Population D, comprising mature SP cells, expressed very little c-Myb. Therefore, expression of c-Myb during thymopoiesis is highest at the point of positive selection, and is strongly associated with commitment to the CD4 rather than the CD8 lineage.
c-Myb can force CD4+CD8lo differentiation
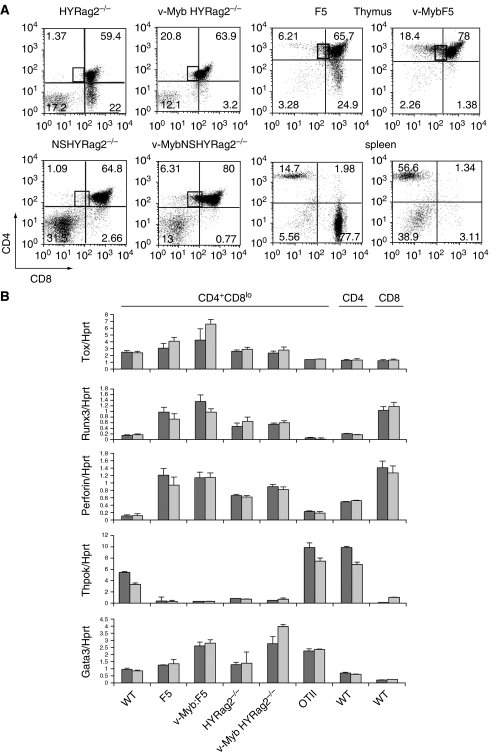
As manipulation of the levels of c-Myb could severely perturb the fate of positively selected thymocytes, we examined whether overexpression of activated c-Myb was able to act as a lineage switcher, subverting DP cells destined to become CD8 SP into the CD4 lineage. To do this, we bred the activated c-Myb transgenic line vMyb4 with two separate mouse models in which CD8 SP development is forced by the expression of an MHC Class I-specific transgenic TCR. HYRAG2−/− mice carry a transgenic TCR recognising the male-specific HY antigen (Kisielow et al, 1988) expressed on a Rag2 null background. In males, there are no mature SP cells, due to negative selection, and in females, only CD8 SP cells develop, as the HY TCR is specifically selected on MHC Class I (Figure 2A, upper left panel). The F5 transgenic TCR (Mamalaki et al, 1993) recognises an influenza virus antigen in the context of MHC Class I, selecting predominantly CD8 SP cells, which express the F5-specific TCRβ chain Vβ11 (Figure 2A, centre right panels). In vMyb:HYRAG2−/− crosses, negative selection in males was unaffected (data not shown) but in females, no CD8 SP cells were produced (Figure 2A, upper centre left panel, and Table I), showing that overexpression of v-Myb was able to block a CD8-specific differentiation signal. This was also true in the F5 model, where equally few CD8 SP cells were produced (Figure 2A, top right panel and Table I). Although there was no significant increase in transgene-specific CD4SP cells in vMybHYRAG2−/− cells, we did observe more Vβ11+CD4SP in vMybF5 thymuses and spleens (Figure 2A, right panels). Although it is possible that these are lineage-switched CD8 SP cells, it is more likely they are a result of the F5 β chain associating with endogenous TCRα chains, as is seen with vMybHYRAG2+/− animals (Supplementary Figure 2), indicating that on a recombination permissive background, there is a strong bias towards the CD4SP lineage even in the presence of CD8-specific transgenic TCRs.
Figure 2.
Activated c-Myb blocks CD8 lineage commitment. (A) Comparison of thymocyte development in vMyb transgenic mice crossed to CD8-specific transgenic TCR mice. x-axis: CD8, y-axis: CD4. CD4+CD8lo cells are boxed. Top left panels: crossed to HYRAG2−/− animals on a selecting background. Bottom left panels: to HYRAG2−/− animals on a nonselecting H2d background. Right panels: crossed to F5 transgenic animals, with plots gated on F5-specific Vβ11 TCR; top=thymus, bottom=spleen. Percentages of cells in each quadrant are shown. Plots are representative of between three and nine mice per experiment (see Table I). (B) qRT–PCR for expression of Tox, Runx3, Prf1, ThPOK and Gata3 versus an hprt control from CD4+CD8lo and SP cells sorted from thymuses of the genotypes shown on the x-axis. Two independent sorts are shown for each genotype. Error bars: s.d.
In both models, we observed an increase in the proportion of CD4+CD8lo cells, the transitional population lying between the DP and SP stages (Figure 2A, upper panels, boxed). To validate this population, we analysed mRNA from sorted vMyb:HYRAG2−/− and vMyb:F5 CD4+CD8lo cells relative to a wt control and to TCR transgenics lacking vMyb. For comparison, we also included CD4+CD8lo cells from OTIIRAG2−/− mice, which commit exclusively to the CD4 lineage, and also CD4 and CD8 SP cells from wt animals. Figure 2B shows that, in both CD8-specific models, in the presence or absence of vMyb, transcripts had been induced for Tox, Prf1 and Runx3. These CD8-specific genes are all upregulated upon positive selection and are markers of CD8 lineage commitment in the CD4+CD8lo subset (Liu et al, 2005), as exemplified by their lack of induction in the CD4-committed OTIIRAG2−/− cells. Gata3, a CD4 lineage-specific factor, was also found in the CD4+CD8lo subset in all lines of mice examined, and notably, appeared increased in the presence of the vMyb transgene. However, ThPOK, the CD4 master regulator, was not induced either in the presence or absence of vMyb, as compared to its induction in either wt or OTIIRAG2−/− CD4+CD8lo cells.
We also tested whether c-Myb might promote positive selection in the absence of a transgenic TCR-stimulatory signal. vMyb4 transgenics were bred to HYRAG2−/− animals on a nonselecting B10.D2 (H2d) background (NSHYRAG2−/−); as there is no cognate MHC, thymocytes in these animals are arrested at the preselection naïve DP stage (Figure 2A, lower left panel). Again, the vMyb4 transgene promoted the development of CD4+CD8lo transitional cells, but could not force further maturation (Figure 2A, lower left panels; CD4+CD8lo cells are boxed). From these data, we concluded that in the presence of a strong CD8 selection signal, overexpression of v-Myb results in inhibition of CD8 SP maturation, can enhance production of CD4 SP cells if endogenous recombination is permitted, and can force DP cells through the early stages of positive selection even in the absence of TCR signalling, such that there is an accumulation of CD4+CD8lo intermediate cells. As judged by expression of CD8 lineage-specific markers, many of the cells in the CD4+CD8lo subset may still be CD8-committed, but are unable to mature. Of note, the absence of ThPOK mRNA in the CD4+CD8lo cells in the F5 transgenic model suggests that the CD4SP cells produced are either aberrant, or may bypass the CD4+CD8lo stage.
c-Myb is required for correct induction post-selection of Gata3
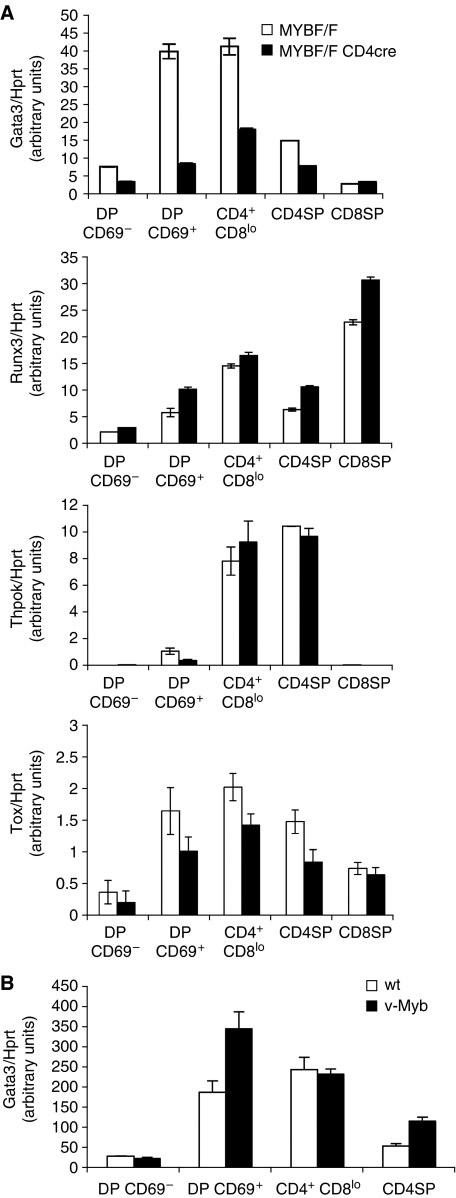
To further examine the effects of c-Myb on lineage-associated genes, mRNA was made from knockout MybF/F:CD4Cre and control MybF/F DP thymocytes flow sorted into DP CD69− (pre-selection naïve), DP CD69+ (post-selection), CD4+CD8lo (transitional) and SP subsets, and expression of Gata3, Runx3, ThPOK and Tox was quantitated by qRT–PCR relative to hprt. Expression of Gata3 was reduced in all Myb-knockout DP subsets examined, and also in CD4 SP cells (Figure 3A, top panel). Levels were reduced between two- and fourfold, with the greatest differences seen in post-selection DP CD69+ and CD4+CD8lo subsets. Expression of Tox was also slightly reduced (Figure 3A, fourth panel). The CD8 lineage regulator Runx3 was expressed at slightly higher levels at all points after positive selection had occurred (Figure 3A, second panel), and ThPOK, although repressed in DP CD69+ cells, was induced normally at the CD4+CD8lo stage in a lineage-specific fashion (Figure 3A, lower panel). Having noted that overexpression of v-Myb could upregulate Gata3 mRNA in the presence of transgenic TCRs, we also sorted DP and SP subsets from vMyb4 mice on a normal C57Bl/10 background. We found that in post-selection DP CD69+ and CD4 SP thymocytes from these mice, Gata3 mRNA levels were approximately doubled (Figure 3B). However, in contrast to the data in Figure 2B showing that Gata3 mRNA was increased in the CD4+CD8lo subsets of vMyb:HYRAG2−/− and vMyb:F5 animals, no such increase was detected in vMyb4 transgenics on a wt background. Notwithstanding this discrepancy, we conclude that in thymocytes lacking c-Myb, Gata3 is downregulated and its induction following positive selection is severely inhibited. Conversely, in thymocytes with excessive c-Myb activity, Gata3 is upregulated. These data suggest that c-Myb is an important modulator of Gata3 expression.
Figure 3.
Expression of lineage-specific genes in sorted thymocytes from c-Myb mutant mice. (A) qRT–PCR of mRNA from thymocyte subsets from MybF/F (white bars) or MybF/FCD4Cre (black bars) mice sorted as indicated on the x-axes. Expression of Gata3 (top panel), Runx3 (second panel), ThPOK (third panel) and Tox (bottom panel) is shown relative to hprt. Data are the mean of at least three experiments performed in triplicate; error bars indicate s.d. (B) Expression of Gata3 relative to hprt in vMyb4 transgenic mice (black bars) versus wt littermates (white bars) from thymocyte subsets sorted as indicated on the x-axis. Data are the mean of ⩾3 experiments performed in triplicate. Error bars: s.d.
Gata3 induction following TCR signalling requires c-Myb
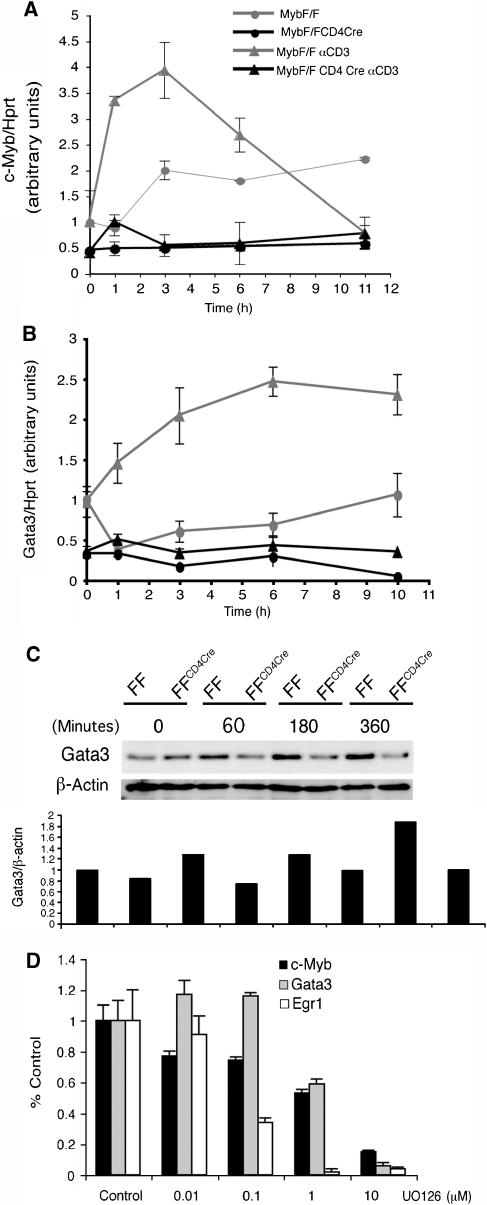
Gata3 expression following positive selection is necessary but not sufficient for CD4 lineage commitment and the phenotypes displayed in loss- or gain-of-function Gata3 mutant thymocytes are remarkably similar to those seen in the corresponding c-Myb mutant cells (Hernandez-Hoyos et al, 2003; Pai et al, 2003). To explore whether Gata3 might be a downstream target of c-Myb, we looked at the kinetics of induction of the two genes following TCR signalling in DP cells. Purified naïve CD69− DP thymocytes isolated from knockout MybF/F:CD4Cre or control MybF/F mice were activated by crosslinking of the TCR complex with αCD3 antibody. In this system, CD69 surface protein expression, an early marker of TCR signalling, was first detected 2 h after crosslinking (data not shown). c-Myb expression was quantitated by qRT–PCR relative to hprt. In MybF/F cells, c-Myb mRNA was rapidly induced, being 3.5-fold induced at 1 h and peaking at 3 h (Figure 4A, grey triangles). Levels of c-Myb mRNA in unstimulated cells also increased slightly over time, perhaps as a stress response to in vitro culture (Figure 4A, grey circles). Unsurprisingly, very low levels of full-length c-Myb mRNA were seen in Myb-knockout thymocytes (Figure 4A, black lines).
Figure 4.
c-Myb and Gata3 expression following TCR signalling. (A) qRT–PCR quantitating c-Myb expression relative to hprt. mRNA was harvested from naïve DP thymocytes stimulated for the times shown on the x-axis with nothing (grey lines), or αCD3 (black lines). Data are the mean of three experiments performed in triplicate. Error bars: s.d. (B) qRT–PCR quantitating Gata3 mRNA relative to hprt mRNA from the same experiments. (C) Gata3 expression assessed by Western blot relative to a β-actin control. Cell lysates were prepared from purified DP CD69− thymocytes of the genotypes shown, stimulated in vitro with PMA and ionomycin for the indicated times. (D) c-Myb and Gata3 expression following αCD3 stimulation of MHCI/IIo/o DP thymocytes in the presence or absence of UO126. Data are the mean of three experiments performed in triplicate. Error bars: s.d.
In MybF/F control thymocytes, αCD3 crosslinking also resulted in a rise in Gata3 mRNA by 1 h, with levels plateauing at 6 h (Figure 4B, grey triangles; compare with unstimulated, grey circles). This rise showed similar but slightly delayed kinetics to that observed for c-Myb mRNA. Notably, when knockout MybF/F:CD4Cre naïve DP were similarly activated, induction of Gata3 was completely inhibited (Figure 4B, black triangles); Gata3 mRNA remained at baseline uninduced levels (Figure 4B, black circles), which were themselves lower than in control thymocytes (Figure 4B, compare grey and black circles). Gata3 protein (Figure 4C) was also 1.3-fold induced in control DP thymocytes 1 h after activation, and had doubled its expression by 6 h (here shown using PMA/ionomycin as activation stimuli). However, as for its mRNA, it remained at baseline levels in the absence of c-Myb. These data show that the expression patterns of c-Myb and Gata3 are consistent with Gata3 being downstream of c-Myb, and also that induction of Gata3 following TCR ligation and positive selection is dependent on c-Myb.
To generate different cell fate outcomes in response to variations in the strength of TCR signalling, we hypothesised that both c-Myb and Gata3 expression should be modulated in response to modulation of the TCR signal. To test this, naïve DP thymocytes were purified from MHC-deficient mice (β2M° and H2-Ab1°) and their TCRs activated with αCD3 in the presence of increasing concentrations of the MEK inhibitor UO126, leading to a graded inhibition of MAPK activation (Supplementary Figure 3). Expression of c-Myb, Gata3 and the well-documented MAPK-induced gene Egr1 after treatment with UO126 was analysed relative to hprt at the normal peak expression time for each gene. Figure 4D shows that, in relation to their expression in the absence of UO126, there was a gradual diminution of mRNA levels for all three genes, with expression of both c-Myb and Gata3 halved after treatment with 1 μM UO126. Therefore, both c-Myb and Gata3 can be induced differentially following TCR ligation, depending on the strength of the TCR signal through the MAPK pathway.
Gata3 is a c-Myb target gene
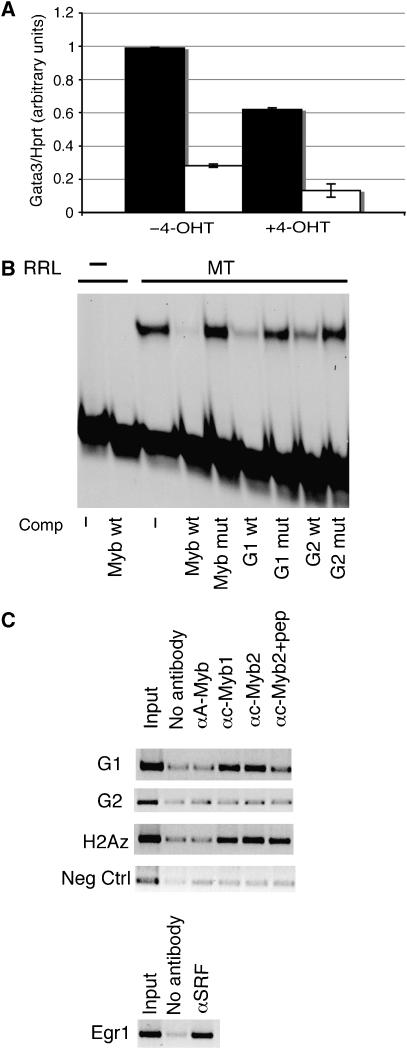
The artificially inducible Myb inhibitor MERT acts as a specific repressor of Myb activity in the presence of 4-hydroxytamoxifen (4-OHT). We previously made an EL4-derived cell line, E16C, in which MERT can be regulated by 4-OHT in an inducible fashion (Taylor et al, 1996). To determine whether Gata3 could be repressed by loss of Myb activity, we cultured E16C cells in the presence or absence of 4-OHT, and assessed endogenous levels of Gata3 mRNA by qRT–PCR relative to hprt. Figure 5A shows that after 24 h of 4-OHT treatment, Gata3 mRNA levels decreased by 40% (compare lanes 1 and 3). For comparison, we also measured repression of the previously characterised c-Myb target bcl2 (Frampton et al, 1996; Taylor et al, 1996), which showed a 50% decrease in this assay (Figure 5A, compare lanes 2 and 4). Therefore, Gata3 expression is repressed to a similar degree as a well-established c-Myb target gene.
Figure 5.
c-Myb regulates Gata3. (A) Downregulation of Gata3 upon inhibition of Myb. E16C cells were treated with 4-OHT, to induce MERT and hence repress Myb activity, or vehicle control for 24 h. qRT–PCR was performed to quantitate Gata3 (black bars) or Bcl2 (white bars) expression relative to hprt. Data are the mean of five experiments. Error bars: s.d. (B) EMSA using as probe a strong Myb-binding site (MybMBS). Lanes 1 and 2; blank reticulocyte lysate controls. Lanes 3–9; reticulocyte lysate programmed with the Myb DNA-binding domain (MT). Lane 3; no competitor. Lane 4; 10-fold excess unlabelled wt MybMBS. Lanes 5–9: 100-fold excess unlabelled mutant MybMBS (lane 5); wt MBS G1 (lane 6); mutant MBS G1 (lane 7); wt MBS G2 (lane 8), or mutant MBSG2 (lane 9). (C) Chromatin immunoprecipitation. PCR reactions from immunoprecipitated DNA specific for the indicated regions are shown. Data were identical in two separate experiments.
Examination of the Gata3 upstream and 5′untranslated regions revealed there were two Myb consensus-binding sites (MBS), at positions −545 (MBS G1) and +864 (MBS G2) relative to the previously published mRNA start (George et al, 1994). EMSA showed that, whereas neither site was able to bind strongly enough to an in vitro translated c-Myb DNA-binding domain (MT) to generate a bandshift, both sites could compete with a probe comprising the strong MBS from the mim-1 promoter (Ness et al, 1989), albeit at 100-fold molar excess (Figure 5B, compare lane 3 with lanes 4, 6 and 8). In contrast, a 100-fold excess of mutant canonical MBS, or of mutated Gata3 MBS G1 or G2 did not compete (Figure 5B, lanes 5, 7 and 9). Therefore, both sites are able to interact weakly in vitro with the c-Myb DNA-binding domain.
To establish whether c-Myb binds the Gata3 promoter in vivo, chromatin immunoprecipitations were performed using extracts from total thymocytes. As a positive control for the integrity of the chromatin, we showed (Figure 5C, lower panel) that an antibody against SRF was able to immunoprecipitate the Egr1 promoter (Christy and Nathans, 1989). When the prepared chromatin was immunoprecipitated with either of two antibodies against c-Myb, a specific band was detected by PCR in the positive control, the Myb target gene H2AZ (Figure 5C, third panel, lanes 4 and 5; JH, DM and KW, submitted) and importantly, for MBS G1 (Figure 5C, top panel, lanes 4 and 5). This band was substantially reduced when the peptide immunogen used to generate one of the antibodies was added to the immunoprecipitation reaction (Figure 5C, top panel, lane 6). No increase above background was seen without antibody, or with an antibody against A-Myb, a family member which is not expressed in thymocytes (Figure 5C, top panel, lanes 2 and 3). There was also no increase above background in PCR reactions specific for MBS G2, or for the negative control, a region of the Gata3 promoter lacking an MBS (Figure 5C, second and fourth panels). Therefore, in vivo, c-Myb is occupying the G1-binding site at −545 on the Gata3 promoter, whereas the G2 site is not recognised.
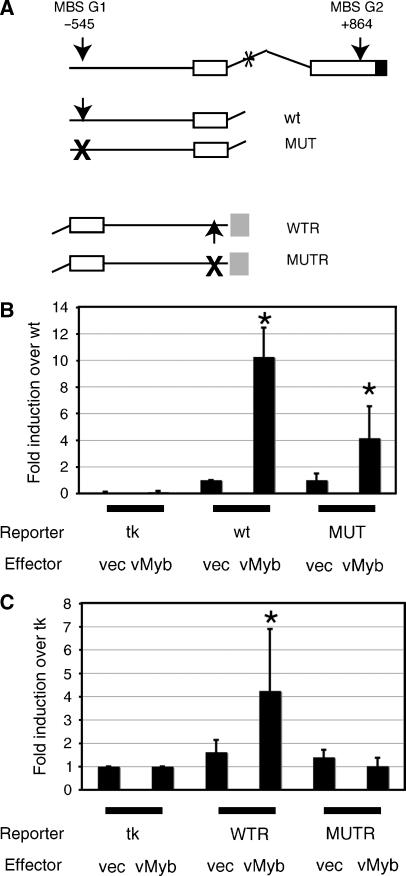
As a further test of c-Myb's ability to interact with the Gata3 promoter, we transiently transfected plasmids carrying the region of the Gata3 promoter containing MBS G1 linked to a firefly luciferase reporter gene into NIH3T3 fibroblasts, in the presence or absence of a vMyb effector plasmid and with a renilla luciferase reporter as internal control. The promoter was either in its normal orientation relative to the luciferase gene, or in reverse orientation; in the latter case the minimal thymidine kinase promoter provided a TATA box. For both orientations, MBS G1 was either wt or mutated. The four constructs are shown in Figure 6A, together with a schematic of the Gata3 promoter. Figure 6B shows that in its normal orientation, the wt promoter was stimulated 10-fold when v-Myb was present (P=0.007), whereas the mutant promoter was only stimulated 4-fold (P=0.1), presumably due to an MBS-independent mechanism. In the reverse orientation, relative to the basal tk promoter, the mutant was not stimulated at all, and the wt increased fourfold in the presence of v-Myb (Figure 6C; P=0.02). Taken together, all these data provide good evidence that Gata3 is a direct c-Myb target gene, whose promoter contains an MBS at position −545 which can be bound and activated by c-Myb.
Figure 6.
v-Myb activates the Gata3 promoter. (A) Schematic of the Gata3 promoter and constructs. Arrows, locations of wt MBSG1 and G2, crosses, mutated sites; open boxes, 5′ UTR, exons 1 and 2; black box, start of cdr. *Location of Gata3 double site in intron1; grey box, location of tk promoter in reversed constructs. (B) Transient transfection of WT and MUT constructs into NIH3T3 cells, with pGL3tkbasic as baseline control, in the presence of either empty vector or vector expressing vMyb. Data are from seven independent experiments carried out in triplicate, normalised with a renilla luciferase internal control and shown as mean and s.d. relative to the uninduced WT construct. *Indicates a P-value of <0.1 (see text). (C) Transient transfection of WTR and MUTR constructs into NIH3T3 cells, in the presence of either empty vector or vector expressing vMyb. Data are from three independent experiments carried out in triplicate, normalised with a renilla luciferase internal control and shown as mean and s.d. relative to pGL3tkbasic. *Indicates a P-value of 0.02 (see text).
Discussion
We have explored the relationship between the c-Myb transcription factor, positive selection and lineage commitment during thymopoiesis. We make three principal observations: (1) c-Myb mRNA is expressed at high levels in DP thymocytes during selection and is strongly associated with commitment to the CD4 lineage; (2) overexpression of v-Myb causes a complete loss of CD8 cells, leads to a build up of CD4+CD8lo intermediate cells, and can enhance CD4SP development, and most importantly, (3) c-Myb mediates its effects at least partly via the CD4 lineage regulator Gata3, which is a direct target.
c-Myb's effect on the ratio of CD4 SP to CD8 SP cells has been previously reported (Badiani et al, 1994; Bender et al, 2004; Lieu et al, 2004), but has not been examined in detail. Although it is often the case that such an effect may be the result of other peripheral events, rather than a direct regulation of the positive selection and/or lineage commitment decisions, we were able to exclude two such possibilities. Firstly, although loss of c-Myb has been shown to inhibit TCRβ rearrangement (Bender et al, 2004), this cannot be involved, as crossing MybF/F:LckCre or MybF/F:CD4Cre animals onto an OTIIRAG2−/− background, thus providing a ready-made CD4-specific TCR, does not result in CD4 SP cells developing, and indeed severely inhibits OTII-specific CD4 SP cells (Figure 1A; Bender et al, 2004). Secondly, it has been proposed that c-Myb can act as both an activator (Siu et al, 1992) or repressor (Allen et al, 2001) of the Cd4 gene itself, which would make CD4 an unreliable measure of CD4:CD8 lineage choice. We have seen no evidence of c-Myb being essential for CD4 activation, as CD4 expression is at normal levels on both DP and SP thymocytes in MybF/F:CD4Cre animals (Figure 1A). CD4 repression by c-Myb is unlikely as the MBS in the Cd4 silencer can be mutated with no effect on silencing (Taniuchi et al, 2002b). Therefore, we feel confident that the effects mediated by c-Myb on positive selection and lineage choice are likely caused by c-Myb being directly involved in one or both of these processes.
The pattern of c-Myb expression in DP thymocytes is consistent with its having a role both pre- and post-selection. In CD69− DP, c-Myb mRNA is increased relative to its level in DN cells, perhaps reflecting its involvement in TCR rearrangement (Bender et al, 2004). However, the increase in c-Myb expression we observed following TCR ligation in vitro, coupled with its marked downregulation in CD8 lineage-committed cells in vivo, suggest that c-Myb transcription is further controlled during selection and lineage choice. In support of this, experiments with an inhibitor of the MAPK-signalling pathway, UO126, showed that it was able to repress the initial TCR-mediated induction of c-Myb mRNA.
The main finding of this paper is that c-Myb can directly regulate a gene important for CD4 lineage commitment, namely Gata3. There are consensus-binding sites for c-Myb in both the thymus-specific Gata3 promoter, which we have analysed here and shown to be directly regulated, but also in the alternative murine upstream promoter directing expression outside the thymus, and the human promoter (Labastie et al, 1994; Asnagli et al, 2002), suggesting that c-Myb may regulate Gata3 in other contexts. In the thymus, like c-Myb, Gata3 is induced in response to TCR signalling, and is downregulated in DP thymocytes committed to the CD8 lineage (Hernandez-Hoyos et al, 2003). In retrovirally infected fetal thymic organ culture, its overexpression causes a severe reduction in CD8 SP and an enhancement of CD4 SP cells, but it cannot force CD4 differentiation in an MHC Class I-restricted transgenic model (Hernandez-Hoyos et al, 2003). Loss of Gata3 activity results in severe losses in CD4 SP cell numbers, but little or no change in the CD8 SP lineage (Hernandez-Hoyos et al, 2003; Pai et al, 2003). These data are remarkably similar to those presented here, strongly suggesting that c-Myb and Gata3 lie on the same pathway and supporting c-Myb's proposed role as a Gata3 regulator. Importantly, following αCD3 or PMA/ionomycin treatment of MybF/F:Cd4Cre conditional knockout thymocytes, induction of Gata3 mRNA and protein is completely abolished, showing that c-Myb must be required for transduction of the post-selection TCR signal. The loss of Gata3 expression in sorted thymocyte populations from MybF/F:CD4Cre mice (Figure 3A) suggests that c-Myb continues to be a crucial regulator from this stage onwards. Furthermore, downregulation of c-Myb upon CD8 lineage commitment may contribute to Gata3 being repressed in CD8 SPs.
In our experiments, the master regulator of CD4 lineage commitment, ThPOK, was initially repressed twofold in MybF/FCD4Cre CD69+ DP, but its expression was induced to normal levels as cells reached the CD4+CD8lo stage, and remained at its normal high level in CD4 SP thymocytes. ThPOK upregulation requires a TCR signal and positive selection to have occurred, and this is also required for its function; ThPOK overexpression cannot force naïve unsignalled DP thymocytes to differentiate (He et al, 2005). Three possibilities are suggested by this: (1) ThPOK induction may be independent of c-Myb and Gata3, lying on a separate post-TCR signalling pathway; (2) its expression may be triggered at a threshold level of Gata3 which is achieved in the MybF/FCD4Cre thymocytes; or (3) there is variegation in the expression of c-Myb in selected DPs due to the incomplete deletion we observe (Supplementary Figure 1), with only those cells still expressing c-Myb able to induce ThPOK and progress down the CD4 lineage.
Our data strongly implicate c-Myb as being involved in fate determination during the DP stage, although it is clearly not a master regulator of differentiation akin to ThPOK. As it is upregulated immediately upon TCR ligation, and its expression declines thereafter, it seems more likely that it is acting as a transducer of the positive selection signal; indeed c-Myb seems to enhance this process when overexpressed to generate excess CD4+CD8lo transitional cells even in the absence of a TCR signal (Figure 2A). However, like its target gene Gata3, c-Myb is also able to repress CD8 differentiation when overexpressed and CD4 differentiation when deleted. This suggests a model where variation in the level of c-Myb is an important transcriptional readout of the TCR signal received upon positive selection, acting as a sensor for TCR signal strength. In cells passing positive selection whose TCRs recognise peptide bound to MHC Class II, the CD4 co-receptor-TCR signal is strong and induces the high levels of c-Myb and hence Gata3, which we observe in sorted CD4-committed subsets (Figure 1C, black bars and Figure 2B, bottom panel; OTII). However, when the TCR sees peptide presented by MHC Class I, the CD8 co-receptor-TCR signal is weaker, and we observe lower levels of c-Myb and Gata3 (Figure 1C, white bars; and Figure 2B, bottom panel; F5 and HYRAG2−/−). Therefore, levels of c-Myb are maintained in CD4-committed cells, but drop in CD8-committed cells, perhaps enforcing the commitment decision, and allowing differentiation down the appropriate lineage. Our data (Figure 4D) showing that partial inhibition of MAPK signalling in vitro results in a reduction in both c-Myb and Gata3 expression provide further preliminary evidence that both genes exhibit an early and modulatable response to different levels of TCR signalling. By this model, when c-Myb is inappropriately overexpressed, as in the vMyb4 transgenic line, CD8-committed cells would be unable to downregulate Myb activity, and hence TCR signalling and Gata3 expression would be maintained, resulting in a differentiation block. However, CD4-committed cells would be able to continue on to maturity, although only in the presence of other TCR-dependent differentiation signals. When c-Myb is absent, CD8-committed cells would be relatively unaffected, but CD4-committed cells would not be receiving the full differentiation signal, and would therefore be unable to mature.
Materials and methods
Animals
Mice were maintained in-house according to UK Home Office regulations. All strains used have been previously published as follows: vMyb4 (Badiani et al, 1996); MybF/F mice (Emambokus et al, 2003); CD4Cre (Lee et al, 2001); OTII:RAG2−/− (Teh et al, 1988); HYRAG2−/− (B10.D2) and HYRAG2 (B10.D2-H2d) (Kisielow et al, 1988); F5 (Mamalaki et al, 1993); MHC-deficient mice (β2M° and H2-Ab1°) (Koller et al, 1990; Cosgrove et al, 1991). All mice analysed were 4–8 weeks old.
Antibodies
The following monoclonal antibodies from BD Pharmingen were used for staining: αCD4 (L3T4, RM4-5); αCD8α (Ly2, 53-6.7); αTCRβ (H57-597); αCD69 (H1.2F3); αCD44 (IM7); αCD25 (7D4); αHSA (CD24, M1/69); αHY-TCR (T3.70, Bioscience). The following antibodies were used for immunoprecipitation or immunoblotting: αc-Myb (E1105, C2X, Santa-Cruz Biotechnology Inc. (SCB)); αA-Myb (sc-9957X, SCB); αSRF (sc-13029, SCB, and α-βactin (ab 25139-100, abcam). αMyb2 is a rabbit polyclonal antibody raised against a peptide from the C terminus of c-Myb (kind gift of R Roux).
Cell preparation, purification and staining
Thymocytes were prepared by gentle disaggregation of tissue through a 70 μM nylon filter with a syringe plunger. For flow cytometry, cells were stained with antibody conjugated to FITC, PE, allophycocyanin or biotin followed by streptavidin–PE-Cy7 (BD Pharmingen). Cells were analysed on a FACS Calibur (Becton Dickinson) with CellQuest software. Events were collected and stored ungated in list mode. Live cells were gated according to their forward-scatter and side-scatter profiles. Data were analysed with FlowJo software (Treestar). Purified thymocyte populations were prepared using anti-CD69 biotinylated antibody and streptavidin-conjugated magnetic beads (Miltenyi Biotec) as follows: both positively and negatively selected CD69 fractions were stained with anti-CD4, anti-CD8 and anti-CD69 antibodies and sorted to isolate DPCD69+, CD4+CD8lo, CD4SP and CD8SP subpopulations from the CD69+ fraction. DN and DPCD69− cells were sorted from the CD69− fraction to ⩾95% purity. Purified thymocyte populations were also obtained by sorting based on the expression of TCRβ and CD69 as described previously (Hernandez-Hoyos et al, 2003). For in vitro stimulation, DP CD69− thymocytes were isolated on columns (Miltenyi Biotech) by negative selection using a cocktail of biotinylated anti-CD69, anti-CD44 and anti-CD25 and streptavidin-conjugated magnetic beads. The DP CD69− population was ⩾94% pure.
Cell stimulation
Purified DP CD69− cells were cultured in RPMI1640 medium+L-glutamine (Gibco), supplemented with 10% FCS, 10 μM 2-mercaptoethanol. Cells were cultured in the absence or presence of plate-bound αCD3 clone 2C11 or 7.5 ng/ml PMA (Sigma) and 180 ng/ml Ionomycin (Sigma). For MAPK inhibition, UO126 was added 15 min before cells were seeded on αCD3-coated plates. E16C cells were treated as described previously (Taylor et al, 1996).
Gene expression, immunoblotting and chromatin immunoprecipitation
Total RNA was extracted using Trizol (Invitrogen) and reversed-transcribed by oligo-dT priming using the Thermoscript RT–PCR kit (Invitrogen). For q-RT–PCR, pre-designed kits from Applied Biosystems were as follows: c-Myb (Mm00501741); Gata3 (Mm00484683); Tox (Mm00455231); Runx3 (Mm00490666); Perforin (Mm00812512); Th-Pok (Mm00784709). Reactions were run in triplicate according to the manufacturer's instructions on a 7700 Sequence Detector (Applied Biosystems) and normalised to Hprt (Mm00446968m1). Experiments were performed at least three times. Chromatin immunoprecipitation was as carried out as described previously (Miralles et al, 2003), using 10 μg of αA-myb, αc-Myb1 or αMyb2, and 20 μg αMyb2 blocking peptide of sequence CSEDEDNVLKAFTVPKN. PCR primers were: G1 5′ggcgtccgaatcaaagcccag3′ and 5′cagtttatatcagcttaggggc3′; G2 5′gcagagaccataacaataacg3′ and 5′aaattaggattcaagccagaacg3′; neg control 5′caatctgaccgggcaggtcac3′ and 5′cctccaaaaggagaaaagctgag3′, H2AZ 5′atagacttgtacacacggtac3′ and 5′atgcgaaattcgcaagactc3′, and egr1 5′tgcgccgacccggaaacgccatata3′ and 5′atcgcgagcgctcaggctcctggaa3′.
EMSA
EMSA was performed as described previously (Weston, 1992), using as probe 0.1 pmol of an IRD700-labelled double-stranded DNA fragment of sequence 5′ctaggacattataacggttttttagtctag3′ and as a source of Myb DNA-binding activity, 2 μl of rabbit reticulocyte lysate (Promega) either unprogrammed or programmed with plasmid pT7βMT(Badiani et al, 1994). Unlabelled oligonucleotide competitors were: wt Myb as above; mutant Myb (ctaggacattatcacggttttttagtctag); G1WT (ggcaaatcttcagttacttcgccatg); G1MUT (ggcaaatcttcaggtacttcgccatg); G2WT (tctatacccttaactgcaaacaaaccatta); G2MUT (tctatacccttacctgcaaacaaaccatta). Bands were visualised on a Li-Cor Odyssey infrared imager.
Transient transfections
DNA was transfected into NIH3T3 cells using Lipofectamine2000 (Invitrogen). All transfections were in triplicate using the renilla expression plasmid pGL4.75 (Promega) as an internal control. Gata3 promoter fragments were cloned into pGL3Basic (Promega) and contained the sequences: WT; NT_039202 nt6797094-6796212, and WTR; NT_039202 nt 6796212-6797094. WTR was cloned upstream of the tk promoter. Mutations in the Myb-binding sites were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene), and in all cases the core AAC of the MBS was replaced with ACC. Cells were harvested 30–40 h post-transfection using the Dual-Glo Luciferase Assay System (Promega), and luciferase activity quantitated on a Wallac 1420 Victor2 luminometer.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Acknowledgments
We thank D Bird, V Lawson, I Titley, D Davies and C Atkins for technical assistance, J Dyson, J Frampton, C Reis e Sousa and R Zamoyska for reagents, and D Cantrell, P Costello, R Treisman for much helpful discussion. This work was supported by Cancer Research UK.
References
- Alberola-Ila J, Hernandez-Hoyos G (2003) The Ras/MAPK cascade and the control of positive selection. Immunol Rev 191: 79–96 [DOI] [PubMed] [Google Scholar]
- Aliahmad P, O'Flaherty E, Han P, Goularte OD, Wilkinson B, Satake M, Molkentin JD, Kaye J (2004) TOX provides a link between calcineurin activation and CD8 lineage commitment. J Exp Med 199: 1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RD III, Bender TP, Siu G (1999) c-Myb is essential for early T cell development. Genes Dev 13: 1073–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RD III, Kim HK, Sarafova SD, Siu G (2001) Negative regulation of CD4 gene expression by a HES-1-c-Myb complex. Mol Cell Biol 21: 3071–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnagli H, Afkarian M, Murphy KM (2002) Cutting edge: identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J Immunol 168: 4268–4271 [DOI] [PubMed] [Google Scholar]
- Badiani P, Corbella P, Kioussis D, Marvel J, Weston K (1994) Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev 8: 770–782 [DOI] [PubMed] [Google Scholar]
- Badiani PA, Kioussis D, Swirsky DM, Lampert IA, Weston K (1996) T-cell lymphomas in v-Myb transgenic mice. Oncogene 13: 2205–2212 [PubMed] [Google Scholar]
- Barnden MJ, Allison J, Heath WR, Carbone FR (1998) Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol 76: 34–40 [DOI] [PubMed] [Google Scholar]
- Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K (2004) Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol 5: 721–729 [DOI] [PubMed] [Google Scholar]
- Bosselut R (2004) CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat Rev Immunol 4: 529–540 [DOI] [PubMed] [Google Scholar]
- Christy B, Nathans D (1989) Functional serum response elements upstream of the growth factor-inducible gene zif268. Mol Cell Biol 9: 4889–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D, Gray D, Dierich A, Kaufman J, Lemeur M, Benoist C, Mathis D (1991) Mice lacking MHC class II molecules. Cell 66: 1051–1066 [DOI] [PubMed] [Google Scholar]
- Ehlers M, Laule-Kilian K, Petter M, Aldrian CJ, Grueter B, Wurch A, Yoshida N, Watanabe T, Satake M, Steimle V (2003) Morpholino antisense oligonucleotide-mediated gene knockdown during thymocyte development reveals role for Runx3 transcription factor in CD4 silencing during development of CD4−/CD8+ thymocytes. J Immunol 171: 3594–3604 [DOI] [PubMed] [Google Scholar]
- Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J (2003) Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J 22: 4478–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton J, Ramqvist T, Graf T (1996) v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev 10: 2720–2731 [DOI] [PubMed] [Google Scholar]
- George KM, Leonard MW, Roth ME, Lieuw KH, Kioussis D, Grosveld F, Engel JD (1994) Embryonic expression and cloning of the murine GATA-3 gene. Development 120: 2673–2686 [DOI] [PubMed] [Google Scholar]
- Germain RN (2002) T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol 2: 309–322 [DOI] [PubMed] [Google Scholar]
- He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ (2005) The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433: 826–833 [DOI] [PubMed] [Google Scholar]
- Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J (2003) GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity 19: 83–94 [DOI] [PubMed] [Google Scholar]
- Hernandez-Munain C, Krangel MS (2002) Distinct roles for c-Myb and core binding factor/polyoma enhancer-binding protein 2 in the assembly and function of a multiprotein complex on the TCR delta enhancer in vivo. J Immunol 169: 4362–4369 [DOI] [PubMed] [Google Scholar]
- Hernandez-Munain C, Lauzurica P, Krangel MS (1996) Regulation of T cell receptor delta gene rearrangement by c-Myb. J Exp Med 183: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes DJ, He X, He X (2005) CD4-CD8 lineage commitment: an inside view. Nat Immunol 6: 761–766 [DOI] [PubMed] [Google Scholar]
- Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H (1988) Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature 333: 742–746 [DOI] [PubMed] [Google Scholar]
- Koller BH, Marrack P, Kappler JW, Smithies O (1990) Normal development of mice deficient in beta 2 M, MHC class I proteins, and CD8+ T cells. Science 248: 1227–1230 [DOI] [PubMed] [Google Scholar]
- Labastie MC, Bories D, Chabret C, Gregoire JM, Chretien S, Romeo PH (1994) Structure and expression of the human GATA3 gene. Genomics 21: 1–6 [DOI] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB (2001) A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15: 763–774 [DOI] [PubMed] [Google Scholar]
- Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP (2004) Requirement of c-myb in T cell development and in mature T cell function. Proc Natl Acad Sci USA 101: 14853–14858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Taylor BJ, Sun G, Bosselut R (2005) Analyzing expression of perforin, Runx3, and Thpok genes during positive selection reveals activation of CD8-differentiation programs by MHC II-signaled thymocytes. J Immunol 175: 4465–4474 [DOI] [PubMed] [Google Scholar]
- Mamalaki C, Elliott J, Norton T, Yannoutsos N, Townsend AR, Chandler P, Simpson E, Kioussis D (1993) Positive and negative selection in transgenic mice expressing a T-cell receptor specific for influenza nucleoprotein and endogenous superantigen. Dev Immunol 3: 159–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou A, Treisman R (2003) Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113: 329–342 [DOI] [PubMed] [Google Scholar]
- Nawijn MC, Ferreira R, Dingjan GM, Kahre O, Drabek D, Karis A, Grosveld F, Hendriks RW (2001) Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J Immunol 167: 715–723 [DOI] [PubMed] [Google Scholar]
- Neilson JR, Winslow MM, Hur EM, Crabtree GR (2004) Calcineurin B1 is essential for positive but not negative selection during thymocyte development. Immunity 20: 255–266 [DOI] [PubMed] [Google Scholar]
- Ness SA, Marknell A, Graf T (1989) The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 59: 1115–1125 [DOI] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC (2003) Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19: 863–875 [DOI] [PubMed] [Google Scholar]
- Palmer E (2003) Negative selection—clearing out the bad apples from the T-cell repertoire. Nat Rev Immunol 3: 383–391 [DOI] [PubMed] [Google Scholar]
- Pearson R, Weston K (2000) c-Myb regulates the proliferation of immature thymocytes following beta-selection. EMBO J 19: 6112–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Taghon T (2005) Molecular genetics of T cell development. Annu Rev Immunol 23: 601–649 [DOI] [PubMed] [Google Scholar]
- Sato T, Ohno S, Hayashi T, Sato C, Kohu K, Satake M, Habu S (2005) Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity 22: 317–328 [DOI] [PubMed] [Google Scholar]
- Singer A, Bosselut R (2004) CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv Immunol 83: 91–131 [DOI] [PubMed] [Google Scholar]
- Siu G, Wurster AL, Lipsick JS, Hedrick SM (1992) Expression of the CD4 gene requires a Myb transcription factor. Mol Cell Biol 12: 1592–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Jameson SC, Hogquist KA (2003) Positive and negative selection of T cells. Annu Rev Immunol 21: 139–176 [DOI] [PubMed] [Google Scholar]
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R (2005) The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol 6: 373–381 [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR (2002a) Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111: 621–633 [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Sunshine MJ, Festenstein R, Littman DR (2002b) Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol Cell 10: 1083–1096 [DOI] [PubMed] [Google Scholar]
- Taylor D, Badiani P, Weston K (1996) A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev 10: 2732–2744 [DOI] [PubMed] [Google Scholar]
- Teh HS, Kisielow P, Scott B, Kishi H, Uematsu Y, Bluthmann H, von Boehmer H (1988) Thymic major histocompatibility complex antigens and the alpha beta T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature 335: 229–233 [DOI] [PubMed] [Google Scholar]
- Weston K (1992) Extension of the DNA binding consensus of the chicken c-Myb and v-Myb proteins. Nucleic Acids Res 20: 3043–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B, Chen JY, Han P, Rufner KM, Goularte OD, Kaye J (2002) TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol 3: 272–280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3