Abstract
C/EBPα is a key transcription factor indispensable for the onset of gluconeogenesis in perinatal liver. However, C/EBPα was already expressed in fetal liver, suggesting that the expression of C/EBPα alone does not account for the dramatic increase of the expression of metabolic genes, and hence an additional factor(s) is expected to function cooperatively with C/EBPα in perinatal liver. We show here that expression of Foxo1 was sharply increased in the perinatal liver and augmented C/EBPα-dependent transcription. Foxo1 bound C/EBPα via its forkhead domain, and Foxo1 bound to the promoter of a gluconeogenic gene, phosphoenolpyruvate carboxykinase (PEPCK), in a C/EBPα-dependent manner in vivo. Insulin inhibited the expression of PEPCK in a culture of fetal liver cells, and also the C/EBPα-dependent transcription enhanced by Foxo1. These results indicate that Foxo1 regulates gluconeogenesis cooperatively with C/EBPα, and also links insulin signaling to C/EBPα during liver development.
Keywords: C/EBPα, Foxo1, gluconeogenesis, hepatocyte maturation
Introduction
During mammalian development, characteristics of the liver change drastically from a major hematopoietic tissue in fetus toward a central metabolic tissue in adult (Girard, 1990). While nutrients are provided from mother via the placenta in prenatal stage, nutritional supplies from mother stop abruptly after birth. Thus, newborns have to survive by their own metabolic functions immediately after birth, and the liver changes gene expression dramatically during the perinatal stage for the adaptation, which requires the changes in transcription factors (Schrem et al, 2004). It is well established that C/EBPα is an essential factor for glucose metabolism during perinatal stage. In C/EBPα-deficient mice, expression of gluconeogenic genes is blunted and the blood glucose level is significantly low, leading to neonatal death (Darlington et al, 1995; Wang et al, 1995). The blood glucose level is important for energy homeostasis and is strictly regulated by insulin. In fetus at a late gestational stage, the plasma insulin level is high and gluconeogenic genes are suppressed, whereas it decreases rapidly after birth, resulting in upregulation of gluconeogenic enzymes (Blazquez et al, 1970; Croniger et al, 1997). Although a variety of extracellular signals, including insulin, have been suggested to regulate C/EBPα activity, the precise mechanism how insulin regulates C/EBPα activity is not fully elucidated.
Foxo1 is a member of the forkhead family transcription factors and regulates glucose metabolism in the adult liver (Puigserver et al, 2003; Accili and Arden, 2004). Foxo1 regulates the expression of phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) genes by direct binding to its target DNA sequence and also through the interaction with nuclear receptors. Foxo1 activity is regulated by phosphorylation and acetylation. Insulin induces phosphorylation of Foxo1 through the PI3K-Akt signaling pathway. Phosphorylated Foxo1 is excluded from the nucleus, and thereby its transcriptional activity is attenuated (Brunet et al, 1999; Kops et al, 1999).
Dramatic expression of metabolic enzymes just before birth requires C/EBPα. However, the underlying mechanism for the drastic enhancement of gene expression is not well understood. We found that C/EBPα was already expressed in fetal liver, suggesting that there is a mechanism that regulates the C/EBPα function. We show here that Foxo1 enhances C/EBPα-dependent transcription in the developing liver cells, through the direct interaction between these molecules.
Results
Expression of C/EBPα during liver development
As C/EBPα plays a key role for expression of various metabolic enzymes at a perinatal stage (Wang et al, 1995), we first examined the expression of C/EBPα during liver development, and revealed that it was already expressed in E14.5 liver and was not changed drastically in the E18.5 and neonatal liver (Figure 1A). Consistently with the in vivo result, the expression of C/EBPα mRNA was not changed in fetal hepatocytes culture, in which they differentiated (Figure 1A; Kamiya et al, 1999). As C/EBPα is regulated translationally, we also examined the isoforms (p42 and p30) of C/EBPα protein (Ossipow et al, 1993; Calkhoven et al, 2000). The levels of isoforms were not changed during liver development in vivo and also in fetal hepatocyte culture, in which hepatocyte differentiation was induced (Figure 1B). These results suggest that there is a post-translational regulatory mechanism that enhances C/EBPα activity in perinatal liver.
Figure 1.
Expression of C/EBPα during hepatocyte maturation and effects of insulin on perinatal hepatocytes. (A) Expression of C/EBPα mRNA during hepatocyte maturation. RNA was prepared from livers at various developmental stages and the C/EBPα mRNA level was examined by Northern blot and also from E14.5 fetal liver cells cultured in the undifferentiation condition (Dex−, OSM−), or differentiation condition (Dex+, OSM+). Note: although images of fetal liver culture are separated by a space, the results are obtained from the same original blot. (B) Protein levels of C/EBPα isoforms (p42 and p30) during development. Nuclear extract was prepared from livers at various developmental stages of and cultured fetal liver cells and the levels of C/EBPα isoforms were examined by Western blot. Note: although images of fetal liver culture are separated by a line, the results are obtained from the same original blot. (C) Repression of gluconeogenic gene expression by insulin during liver maturation. E14.5 fetal liver cells were cultured in the presence or absence of insulin and harvested for qPCR. Results were normalized to the expression level of GAPDH. The data shown are the mean±error of triplicate experiments and at least two independent RNA preparations.
Insulin regulation of metabolic enzymes during liver maturation
Insulin is a critical regulator for glucose metabolism in the liver (Saltiel and Kahn, 2001). Insulin secreted from the pancreas in response to the blood glucose level represses gluconeogenesis and enhances glycolysis in the adult liver and muscle (Saltiel and Kahn, 2001). We examined the effect of insulin on the expression of gluconeogenic genes in a primary culture of fetal liver cells, by quantitative PCR (qPCR). Expression of C/EBPα and Foxo1 was not affected by insulin. While PEPCK and G6Pase were expressed in differentiated fetal liver cells in the presence of insulin, depletion of insulin from the culture at day 3 enhanced expression of PEPCK and G6Pase compared with the standard culture condition containing insulin throughout the culture, indicating that insulin negatively regulates gluconeogenesis as is known in adult liver (Figure 1C). In contrast to wild-type cells, PEPCK was not expressed in C/EBPα KO liver culture, and insulin depletion did not affect the expression (Supplementary Figure S1). Interestingly, the expression of another C/EBPα target, glycogen synthase (Gys2), was expressed in the liver from E14 to adult and was not affected by insulin (Figure 1C).
Expression of Foxo1 during the liver development
On the basis of the observation that insulin affects gluconeogenesis, we assumed that Foxo1, a target of insulin signaling, might be involved in gluconeogenesis in fetal as well as in adult liver. To investigate whether Foxo1 plays a role for liver development, we first examined the expression of Foxo1 during liver development. Foxo1 mRNA expression in liver was barely detectable at E14.5 and dramatically increased at E18.5 (Figure 2A). Furthermore, we examined Foxo1 protein in nuclear extracts. Foxo1 was abundantly present in the nuclear extract of neonate liver, but only small amount was detected in the nuclear extracts from E14.5 and adult liver (Figure 2B). In spite of the abundant presence of Foxo1 protein in neonatal liver, phosphorylated Foxo1 protein in neonatal liver was comparable to that in E18 liver and Foxo1 protein was mostly in the nucleus (Figure 2B).
Figure 2.
Foxo1 expression during liver development and effects of Foxo1 on C/EBPα-dependent PEPCK expression. (A) Expression of Foxo1 during liver development. Expression of the Foxo1 mRNA in liver was examined by Northern blot. (B) Foxo1 protein level was examined by Western blot with an anti-Foxo1 antibody (upper panel) or with an anti-phospho-Foxo1 antibody (lower panel). (C) Dex enhanced Foxo1 expression. E14.5 fetal liver cells were cultured with or without Dex (1 μM) and cells harvested at various time points were used for Northern blot analysis. (D) Localization of Foxo1 protein in the neonate liver. Neonatal liver was sectioned and stained with an anti-Foxo1 antibody and an Alexa Fluor 488-conjugated anti-rabbit Ig antibody, and nucleus was stained with PI. (E) Effects of Foxo1 mutants on the PEPCK and G6Pase mRNA expression. E14.5 fetal liver cells were infected with control retrovirus or retrovirus expressing dominant-negative form (Δ256), or constitutive active form (3A) of Foxo1. Cells were cultured under the differentiation condition and the expression level was determined by qPCR.
Glucocorticoid is also required for liver maturation, especially for the expression of gluconeogenic genes in vivo and also in vitro (Cole et al, 1995). As glucocorticoid has been shown to induce Foxo1 expression in the muscle (Furuyama et al, 2003), we evaluated the effect of glucocorticoid on Foxo1 expression in the liver. E14.5 liver cells were cultured with or without a glucocorticoid agonist, dexamethasone (Dex). As shown in Figure 2C, Dex augmented the expression of Foxo1 in the primary hepatocytes. On the other hand, insulin did not affect expression of Foxo1 in the hepatocyte culture (Figure 1C). Foxo1 is transcriptionally active in the nucleus and is inactivated by the exclusion from the nucleus by phosphorylation (Brunet et al, 1999; Kops et al, 1999). We therefore examined the localization of Foxo1 protein by immunohistochemistry. In the neonate liver, Foxo1 protein was located in the nucleus as well as cytoplasm (Figure 2D), as confirmed by the Western blot (Supplementary Figure S2), suggesting that Foxo1 was actively involved in gene expression. We then evaluated the effect of Foxo1 on the expression of PEPCK and G6Pase during maturation. Fetal liver cells were infected with retrovirus expressing a dominant-negative form (Δ256) or a constitutively active form (3A) of Foxo1, and their effect was examined. The expression of each mutant was examined by Western blotting using antibody against Foxo1 N-terminal domain (Supplementary Figure S3) and infection efficiency was estimated to be ∼50%, by FACS analysis (Supplementary Figure S3). The expression of PEPCK and G6Pase was repressed by Δ256 Foxo1 and enhanced by 3A Foxo1 (Figure 2E). Next, we employed shRNA to test the involvement of C/EBPα and Foxo1 in gluconeogenic gene expression. While suppression of C/EBPα and Foxo1 by shRNAs was incomplete, expression of both PEPCK and G6Pase was significantly reduced (data not shown). These results from in vivo and in vitro studies implicate Foxo1 to the expression of the gluconeogenic genes in the perinatal liver.
Enhancement of C/EBPα-dependent transcriptional activation by Foxo1
Based on these results, we reasoned that Foxo1 would be involved in perinatal gluconeogenesis, and it might cooperatively function with C/EBPα. To test if Foxo1 is involved in C/EBPα function, we first examined the effects of Foxo1 and C/EBPα on the PEPCK promoter by employing luciferase reporter assays. Foxo1 and C/EBPα activated the PEPCK promoter as reported previously, and coexpression of these two proteins further enhanced promoter activity (Figure 3A). To analyze the cooperative function of C/EBPα and Foxo1 in detail, we used a synthetic C/EBPα response element (αRE). While Foxo1 alone failed to activate transcription from αRE, Foxo1 augmented C/EBPα-dependent transcription in a dose-dependent manner, whereas the Δ256 mutant of Foxo1 that lacks transcriptional activity on its cognate response element failed to enhance C/EBPα activity (Figure 3B). PGC-1α, a coactivator of Foxo1, enhanced transcription from the αRE in the presence of Foxo1 and C/EBPα, and insulin suppressed the transcriptional enhancement by Foxo1 (Figure 3C and D). These results indicate that Foxo1 regulates transcriptional activity of C/EBPα during liver development, which is regulated by insulin. The results also suggest that Foxo1 and PGC-1α not only cooperate on the Foxo1 binding element but also on the C/EBPα binding element.
Figure 3.
Effects of Foxo1 on C/EBPα-dependent transcription. (A) E14.5 fetal liver cells were transfected with PEPCK promoter-Luc, together with expression plasmids, as indicated. Cells were collected for luciferase assays after incubation with DMEM supplemented with 10% FCS for 16 h and medium was replaced with DMEM and 1% FCS, with or without insulin, and incubated for further 24 h. Data shown are normalized by Renilla luciferase activity and are the means±s.d. of a representative experiment performed in triplicate and at least three independent cell preparations. (B, C) E14.5 fetal liver cells were transfected with αRE-Luc along with expression plasmids, as indicated. Cells were collected for luciferase assays as in panel A, 48 h after transfection. (D) E14.5 fetal liver cells were transfected with indicated plasmids and after incubation with DMEM supplemented with 10% FCS for 16 h, and medium was replaced with DMEM and 1% FCS, with or without insulin, and incubated for further 24 h. Cells were collected for luciferase assay as in panel A. (E) Effects of acetylation mutants were confirmed on Foxo1-dependent 3xIRS element. Assays were performed as described above. (F) Effects of acetylation mutants were on C/EBPα-dependent transcription. Assays were performed as described above.
Foxo1 directly interacts with C/EBPα via the forkhead domain
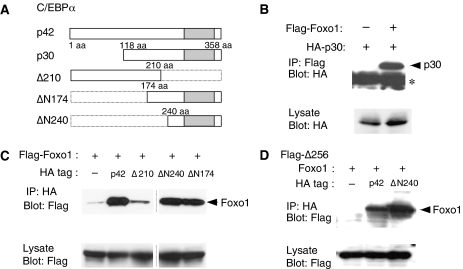
We considered the possibility that cooperation between Foxo1 and C/EBPα is through their physical interaction. To test this idea, co-immunoprecipitation experiments were performed using neonate liver. Foxo1 was co-immunoprecipitated with C/EBPα (Figure 4A). However, such an interaction was not detected in the E14.5 liver (data not shown). To demonstrate direct binding between the two proteins, we expressed GST-Foxo1 fusion protein in Escherichia coli. Fusion protein was purified and mixed with HA-C/EBPα produced by in vitro translation. C/EBPα was pulled down by GST-Foxo1, indicating that these two protein physically interact (Figure 4B). We then delineated the domains responsible for the interaction between these two proteins (Figure 4C and D). Full-length Foxo1 (WT) protein efficiently co-immunoprecipitated C/EBPα and the mutant containing the N-terminal portion, that is, Δ256, also co-immunoprecipitated C/EBPα. However, mutant lacking the N-terminal domain, ΔN, failed to immunoprecipitate C/EBPα. Thus, N-terminal portion of Foxo1 is necessary and sufficient for the interaction with C/EBPα. We further examined the interaction between the two proteins using point mutants of Foxo1 proteins. 3A Foxo1 is a constitutively active mutant, in which three amino-acid residues responsible for phosphorylation are replaced with alanine. The WH mutant has amino-acid substitutions, that is, substitution of the tryptophan residue at 206 to glycine (W206G) and substitution of the histidine residue at 212 to proline (H212P), in the helix 3 in the DNA binding region, and is devoid of DNA binding to and transactivation from the Foxo1 responsive DNA element (Schmoll et al, 2000). The 3A mutant bound C/EBPα as efficiently as WT Foxo1. Interestingly, WH mutant did not bind C/EBPα and single substitution mutants, W206G Foxo1 and H212P Foxo1, also failed to bind C/EBPα (Figure 4E). These results indicate that the helix 3 in the DNA binding region of Foxo1 is required for the interaction between the two proteins (Lai et al, 1993; Schmoll et al, 2000).
Figure 4.
Interaction between C/EBPα and Foxo1 proteins. (A) Nuclear extracts from neonate liver cells were prepared for immunoprecipitation with an anti-C/EBPα antibody or control IgG, followed by Western blotting using an anti-Foxo1 antibody. (B) Direct biding between C/EBPα and Foxo1 was examined by GST pull-down assay. GST-Foxo1 fusion protein was prepared (lower panel) and incubated with in vitro translated C/EBPα protein and examined by Western blot. (C) A schematic representation of Foxo1 mutants. WH has both W206G and H212P substitutions. 3A has T24A, S253A and S316A substitutions. Δ256 is the C-terminal truncated form at 256 aa. ΔN lacks N-terminal 256 amino-acid residues. WG and HP have a single amino-acid residue substitution, W206G and H212P, respectively. Acetylation mutants, 3KR and 3KA, have substitutions of three lysine residues (K242, K245 and K262) with arginine and alanine, respectively. Approximate positions of mutated amino-acid residues are shown as asterisk, A or m. (D–F) 293T cells were transfected with various Flag-tagged Foxo1 mutants and HA-tagged C/EBPα. (D) Binding of 3A, WH and deletion mutants of Foxo1 protein with C/EBPα. (E) Binding of Helix3 mutants in forkhead domain of Foxo1 protein with C/EBPα. (F) Binding of acetylation mutants of Foxo1 protein with C/EBPα. Flag-tagged Foxo1 mutants were precipitated with an anti-Flag antibody and subjected to Western blot analysis using an anti-HA antibody. * represents the IgG heavy chain of antibody used for immunoprecipitation.
We then examined acetylation mutants of Foxo1 because it was reported that CBP-dependent acetylation and Sirt1-dependent deacetylation modulate transcriptional activity of Foxo1 by regulating its DNA binding (Matsuzaki et al, 2005). To test this possibility, we utilized 3KA and 3KR mutants, in which three lysine residues (K242, K245 and K262) were replaced with alanine and arginine, respectively. They are considered to mimic an acetylated and non-acetylated form of Foxo1, respectively. Both 3KA and 3KR bound C/EBPα (Figure 4F). These mutant enhanced C/EBPα dependent transcription comparably (Figure 3E and F). These results suggest that the helix 3 in the forkhead domain is required for the binding to C/EBPα, but acetylation of the three lysine residues of Foxo1 and the DNA binding activity of Foxo1 did not have a major impact on the interaction between Foxo1 and C/EBPα, though the helix 3 is necessary for DNA binding (Schmoll et al, 2000).
Next we mapped the domain of C/EBPα responsible for binding to Foxo1 and found that that the C-terminal domain of C/EBPα has a strong binding ability to the N-terminal portion of Foxo1 (Figure 5).
Figure 5.
Binding domain of C/EBPα. (A) A schematic representation of various C/EBPα mutants. (B) Flag-tagged Foxo1 and HA-tagged p30 isoform of C/EBPα were coexpressed in 293T and binding was examined by immunoprecipitation with anti-Flag antibody and Western blot with an anti-HA antibody. * represents the IgG light chain of antibody used for immunoprecipitation. (C) Foxo1 strongly binds to the C-terminal region of C/EBPα. Flag-tagged Foxo1 and HA-tagged C/EBPα mutants were coexpressed, and their binding was examined by immunoprecipitation as in panel B. Note: images are separated by a line, the results are obtained from the same original blot. (D) N-terminal domain of Foxo1 binds to C/EBPα. Flag-tagged Foxo1 deletion Δ256 was expressed with C/EBPα mutants, as indicated, and their binding was evaluated as in panel B.
C/EBPα–Foxo1 complex on target DNA
To test whether the interaction between C/EBPα and Foxo1 occurs on DNA, we employed avidin–biotin-conjugated DNA binding (ABCD) assay. C/EBPα alone efficiently bound to αRE and the binding of C/EBPα to the αRE was inhibited by coexpression of a dominant-negative form of C/EBPα, showing specific binding of C/EBPα to the beads (data not shown). Whereas Foxo1 alone did not bind this DNA, Foxo1 was efficiently precipitated together with C/EBPα by the αRE-conjugated magnetic beads when Foxo1 was coexpressed with C/EBPα (Figure 6A). However, unrelated DNA sequence failed to precipitate these proteins. These results indicate that binding of Foxo1 to αRE is achieved through the interaction with C/EBPα.
Figure 6.
Recruitment of C/EBPα and Foxo1 to target DNA. (A) Complex formation of C/EBPα and Foxo1 on target DNA element was assessed by avidin–biotin-conjugated DNA binding assay. 293T cells transfected with vectors expressing HA-C/EBP and/or Flag-Foxo1 as indicated were lysed and precipitated with DNA-conjugated magnetic beads. Beads were collected and proteins bound to the beads were analyzed by Western blotting with the respective antibody against tags. (B) ChIP assays with neonatal liver. Association of C/EBPα, Foxo1 and acetylated histone H3 with the endogenous PEPCK gene promoter (−210∼+67) in neonate liver cells was examined by ChIP assay, as described in Materials and methods. The β-actin gene promoter (−75∼+252) failed to recruit C/EBPα and Foxo1, indicating specific recruitment of these proteins to the PEPCK promoter. Note that acetylation of histone H3 was observed on the β-actin promoter. (C) ChIP assays with C/EBPα knockdown hepatocytes. Assays were performed as in panel B.
To verify the recruitment of C/EBPα and Foxo1 to the promoter in vivo, we utilized the chromatin immunoprecipitation (ChIP) assay. Cell lysates prepared from neonate liver were used for ChIP assays to test if endogenous proteins were recruited to the PEPCK promoter in vivo. We immunoprecipitated the cell lysate with antibodies against Foxo1, C/EBPα and acetylated histone H3, and immunoprecipitates were subjected to PCR using primers to amplify the DNA fragment (−210 to +67 nucleotide) of the PEPCK promoter. These antibodies immunoprecipitated the promoter fragment from neonatal liver (Figure 6B), but not from E14.5 liver (data not shown), indicating that both Foxo1 and C/EBPα were recruited to the promoter.
To address whether the recruitment of Foxo1 to the promoter depends on the presence of C/EBPα, we performed ChIP assays using C/EBPα-deficient liver cells. Neither C/EBPα nor Foxo1 was recruited to the PEPCK promoter in C/EBPα-deficient cells (Figure 6B). To exclude effects of C/EBPα on hepatocyte development, we knocked down C/EBPα expression in wild-type cells by shRNA and examined occupancy of Foxo1 on the PEPCK promoter (Figure 6C). The results were consistent with the C/EBPα knockout cells. These results indicate that C/EBPα is necessary for the recruitment of Foxo1 to the PEPCK promoter.
Discussion
As the C/EBPα expression alone cannot account for the expression of metabolic genes in the perinatal liver and in primary cultured hepatocytes, an additional factor(s) is necessary for the dramatic increase of metabolic genes in the perinatal liver (Figure 1). We show that Foxo1 expression in the liver was abruptly upregulated before birth, and that Foxo1 augmented C/EBPα-dependent transcription through direct interaction. As the expression of gluconeogenic genes is regulated cooperatively by multiple transcription factors, including C/EBPs, CREB, HNFs, GR, PPARs and AP-1, it is rather difficult to dissect their complex interactions in vivo (Desvergne et al, 2006). Nevertheless we provide several lines of evidence that Foxo1 participates in the C/EBPα-dependent transcription of gluconeogenic genes in the perinatal liver, and PGC-1α also participates in this coregulation. Immunoprecipitation demonstrated the direct interaction between C/EBPα and Foxo1. ABCD assays revealed that Foxo1 binds to the αRE only when it is coexpressed with C/EBPα, and ChIP assays also showed that Foxo1 is recruited to the PEPCK promoter only in the presence of C/EBPα. Although somewhat controversial results were reported on the function of C/EBPα for glucose metabolism in adult liver (Lee et al, 1997; Inoue et al, 2004; Qiao et al, 2006), it is considered that C/EBPα also plays an important role for glucose metabolism in adult liver. The C/EBPα–Foxo1 complex may exhibit different functions, depending on cellular context, for example, fetal versus adult liver. Although both C/EBPα and Foxo1 were shown to have a role in growth arrest, it is unlikely that they cooperate in this case, because growth arrest by C/EBPα does not require the Foxo1 interaction domain.
As the DNA binding domain of Foxo1 was required for the interaction with C/EBPα (Figure 4C and D), we examined acetylation mutants of Foxo1 that modulate DNA binding (Matsuzaki et al, 2005). Mutants considered to mimic an acetylated and non-acetylated form of Foxo1, bound to C/EBPα, there was no significant difference in C/EBPα-dependent transcriptional activity between these mutants compared with wild type Foxo1 (Figures 3E, F and 4F). However, as these results were obtained by overexpression experiments, it remains possible that in the physiological condition, acetylation of Foxo1 may affect the cooperation with C/EBPα. Because the third helix of Foxo1 was required for C/EBPα binding and is conserved among the Forkhead family members (Lai et al, 1993), the family members other than Foxo1 may also interact with C/EBPα. In fact, we found that Foxo3 was also expressed in E18.5 liver and interacted with C/EBPα (data not shown). The C-terminal region of C/EBPα was shown to interact with Foxo1, and this region is relatively conserved among the C/EBP family members (Figure 5). C/EBPβ also bound to Foxo1 (data not shown), suggesting that C/EBPβ may also function together with Foxo1 to regulate gene expression. In fact, C/EBPβ was reported to play a role for glucose metabolism in the perinatal liver; although some of C/EBPβ KO mice were viable, all C/EBPβ KO mice in the context of C/EBPα+/− background died immediately after birth, because of impaired glucose homeostasis, similar to C/EBPα KO mice (Begay et al, 2004).
Among various liver enzymes, those involved in glucose metabolism are subject to insulin regulation in adult liver. Insulin regulates the blood glucose level by repressing gluconeogenesis and stimulating glycolysis (Saltiel and Kahn, 2001). While the plasma insulin level is high and gluconeogenic genes are suppressed in fetus, it rapidly decreases and gluconeogenic genes are upregulated after birth (Blazquez et al, 1970; Girard et al, 1992). Foxo1 is phosphorylated by insulin through the PI3 kinase/Akt pathway, and the phosphorylated Foxo1 is excluded from the nucleus, resulting in the attenuation of its transcription activity (Brunet et al, 1999; Kops et al, 1999). Consistently, we show that insulin suppressed expression of PEPCK and G6Pase in primary culture of fetal liver cells and that insulin also inhibited C/EBPα-dependent transcription enhanced by Foxo1. In contrast to PEPCK and G6Pase that are sharply upregulated at the perinatal stage and subject to insulin regulation, expression of the other C/EBPα target genes, Gys2 and glucokinase (Gck), which are involved in glycogen synthesis and glycolysis, were not altered by insulin in the fetal liver. Interestingly, Gys2 is constantly expressed in the liver from E14 to adult, and Gck expression in E14 and neonate liver as well as fetal liver cells induced to differentiate in vitro was very low, that is, undetectable by Northern blot (data not shown). Thus, while there are several types of gene regulation in C/EBPα target genes, at least two key enzymes for gluconeogenesis are subject to insulin regulation via Foxo1 and C/EBPα.
Although both insulin and C/EBPα have been known to regulate gluconeogenesis, the link between insulin and C/EBPα has not been understood. Our results provide evidence for the first time that Foxo1 regulates C/EBPα function and links the insulin signaling to C/EBPα in the perinatal liver.
Materials and methods
Plasmids and antibodies
C/EBPα cDNA was cloned by PCR into pcDNA3 vector (Invitrogen), based on the reported sequence. C/EBPα response element (αRE; CGCGTATTGGCCAATATTGG CCAATCTCGA) and PEPCK promoter (−450∼−1) were inserted into pGL3 vector (Promega). The 3xIRS Foxo1 response element was described previously (Daitoku et al, 2004). Mouse Foxo1 mutants, 3A Foxo1 and WH Foxo1 were described previously (Daitoku et al, 2004), and a series of mutants were generated on the basis of these constructs. Antibodies used in this study are the following: anti-C/EBPα (14AA, SantaCruz), anti-Foxo1 (H-128, N-18, SantaCruz, and C3; Daitoku et al, 2003), anti-phospho-Foxo1 (#9461, Cell Signaling Technology), anti-HA tag (12CA5, Roche), anti-Flag tag (SIGMA), anti-acetylated histone H3 (Upstate), anti-actin (I-19, SantaCruz), anti-tubulin (T5168, SIGMA) and anti-TBP (N-12, SantaCruz). shRNA for Foxo1 and C/EBPα were of Expression Arrest Mouse retroviral shRNAmir (Open Biosystems).
Cell culture, transfection, and luciferase assays
Fetal liver cells were cultured as reported previously (Kamiya et al, 1999). Briefly, E14.5 fetal liver cells were dissected in the liver perfusion medium (Invitrogen), and a single cell suspension was obtained by collagenase digestion and seeded on the gelatin-coated dishes. Cells were maintained in the presence of OSM, Dex and insulin throughout the culture, unless otherwise indicated. 293T cells were maintained in DMEM with 10%FCS.
Transfection was performed by using the Lipofectamine and plus reagent, according to the manufacturer's instruction (Invitrogen). Luciferase assay was performed according to the manufacturer's instruction (Promega). Each experiment was performed in triplicate.
All experiments using cultured fetal liver cells were performed with at least three different cell preparations.
C/EBPα KO mice (Wang et al, 1995) were generously provided by Dr Darlington (Baylor College of Medicine). Pups obtained by crossing of heterozygotes were collected, and at least three different pups were used for analysis. Genotypes were determined by PCR using following primers: for wild-type allele, 5′-AGACCAGAAAGCTGAGTTGTGAGTT-3′ and 5′-CAAAACCAAAACAAAACAAAAGACC-3′; for mutant allele, 5′-ACGACGGGCGTTCCTTGCGCAGCTGTG-3′ and 5′-TCAGAAGAACTCGTCAAGAAGGCGATA-3.
qPCR
Total RNA was prepared from livers by Trizol (Invitrogen), and was further purified using the High Pure RNA isolation kit (Roche). Reverse transcription was performed by using the QuantiTect Reverse Transcription kit (Qiagen) and qPCR analysis was performed by using TaqMan Gene Expression Assays (Applied Biosystems), using GeneAmp 5700 Sequence Detection System (Applied Biosystems). Each experiment was performed in triplicate, and with at least two different RNA preparations.
Immunohistochemistry
Neonate liver was embedded in 4% carboxymethyl cellulose gel and sectioned. Sections were fixed with 4% paraformaldehyde and stained with anti-Foxo1 antibody. Alexa Fluor 488-conjugated anti-rabbit IgG antibody was used as the secondary antibody. Nuclei were stained with propidium iodide.
Nuclear and cytosolic extraction
Nuclear and cytosolic extracts were prepared by the standard procedure (Timchenko et al, 1999).
Immunoprecipitation and Western blotting
Neonatal liver cells or 293T cells transfected with expression vectors for a series of Flag-tagged Foxo1 mutants with HA-tagged C/EBPα were lysed with lysis buffer (10 mM Hepes–KOH (pH7.9), 100 mM KCl, 0.1% NP-40, 0.5 mM EDTA, protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, and 5 μg/ml leupeptin)) and incubated with anti-C/EBPα antibody or anti-Flag-tag antibody-linked agarose beads. C/EBPα antibody was immunoprecipitated with protein G agarose beads. The immunoprecipitates were washed with lysis buffer, and subjected to SDS–PAGE, and immunoblotted using antibodies against Foxo1 or HA-tag.
GST pull-down assay
GST-Foxo1 fusion protein was expressed in BL21(DE3) strain of E. coli by using the pGEX-5X-1 vector. HA-tagged C/EBPα protein was prepared using the TnT reticulocyte lysate system (Promega), according to the manufacturer's instruction. GST fusion protein was purified with glutathione sepharose 4B (GE healthcare). Purified glutathione sepharose -bound-GST-Foxo1 was mixed with in vitro translated HA-C/EBPα protein in lysis buffer. The glutathione sepharose beads were washed three times with lysis buffer, and subjected to SDS–PAGE, and immunoblotted using antibodies against HA tag.
ABCD assay
The DNA–avidin beads complex was prepared by adding the annealed biotinylated DNA to 30 μl of the streptavidin-conjugated magnetic beads (iMag Streptavidin Particles Plus-DM, BD), and mixed by a mixer overnight and washed with TE to eliminate unbound DNA. To prepare cell lysates, cultured cells were lysed with lysis buffer and the insoluble materials were removed by centrifugation. The supernatant was incubated with DNA–avidin beads and 50 μg of poly(dI-dC). After 20–60 min at 4°C, beads were collected by magnet and washed with lysis buffer. After washing, SDS sample buffer was added and the samples were subjected to electrophoresis followed by Western blot. Biotinylated DNA used were the following: αRE, 5′-TATTGGCCAATA-3′; control, 5′-ATATCGCGATAT-3′.
ChIP assay
ChIP assay was performed with the ChIP assay kit (Upstate), according to the protocols provided by the manufacturer. Briefly, whole livers were dissected and homogenized, and cells were fixed with 1% formaldehyde for 10 min and sonicated in the SDS-lysis buffer (50 mM Tris–HCl (pH8.1), 10 mM EDTA, 1% SDS). Fragmented soluble chromatin was immunoprecipitated with antibodies. After washing, the precipitates were reverse crosslinked for DNA isolation and PCR analysis. PCR was performed with the following primers: PEPCK sense (−210), 5′-GAGGCCTCCCAACATTCAT-3′, antisense (+67), 5′-CGCTGAGCGCCTTGCCGGA-3′; β-actin sense (−75), 5′-GTTCCGAAAGTTGCCTTTTATG-3′, antisense (+252), 5′-ATGTGGCTGCAAAGAGTCTACA-3′.
Supplementary Material
Supplementary Figures
Supplementary Figure Legends
Acknowledgments
We thank Drs A Kamiya, T Itoh and T Sato for helpful discussion and critical reading of the manuscript, Drs E Saijou and T Naiki for plasmid construction and also thank Dr H Daitoku for Foxo1 expression vectors and antibody, and Dr M Shiina for mutant C/EBP expression vectors and proteins. We are grateful to Drs G Darlington and M Takiguchi for providing us with C/EBPα mutant mice and Dr Y Ohmori for his kind support in completing this work. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and from the CREST program of Japan Science and Technology Agency.
References
- Accili D, Arden KC (2004) FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117: 421–426 [DOI] [PubMed] [Google Scholar]
- Begay V, Smink J, Leutz A (2004) Essential requirement of CCAAT/enhancer binding proteins in embryogenesis. Mol Cell Biol 24: 9744–9751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez E, Montoya E, Lopez Quijada C (1970) Relationship between insulin concentrations in plasma and pancreas of foetal and weanling rats. J Endocrinol 48: 553–561 [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868 [DOI] [PubMed] [Google Scholar]
- Calkhoven CF, Muller C, Leutz A (2000) Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev 14: 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, Aguzzi A, Fantuzzi G, Hummler E, Unsicker K, Schutz G (1995) Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev 9: 1608–1621 [DOI] [PubMed] [Google Scholar]
- Croniger C, Trus M, Lysek-Stupp K, Cohen H, Liu Y, Darlington GJ, Poli V, Hanson RW, Reshef L (1997) Role of the isoforms of CCAAT/enhancer-binding protein in the initiation of phosphoenolpyruvate carboxykinase (GTP) gene transcription at birth. J Biol Chem 272: 26306–26312 [DOI] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A (2004) Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA 101: 10042–10047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A (2003) Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes 52: 642–649 [DOI] [PubMed] [Google Scholar]
- Darlington GJ, Wang N, Hanson RW (1995) C/EBP alpha: a critical regulator of genes governing integrative metabolic processes. Curr Opin Genet Dev 5: 565–570 [DOI] [PubMed] [Google Scholar]
- Desvergne B, Michalik L, Wahli W (2006) Transcriptional regulation of metabolism. Physiol Rev 86: 465–514 [DOI] [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Yamashita H, Mori N (2003) Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J 375: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J (1990) Metabolic adaptations to change of nutrition at birth. Biol Neonate 58 (Suppl 1): 3–15 [DOI] [PubMed] [Google Scholar]
- Girard J, Ferre P, Pegorier JP, Duee PH (1992) Adaptations of glucose and fatty acid metabolism during perinatal period and suckling–weaning transition. Physiol Rev 72: 507–562 [DOI] [PubMed] [Google Scholar]
- Inoue Y, Inoue J, Lambert G, Yim SH, Gonzalez FJ (2004) Disruption of hepatic C/EBPalpha results in impaired glucose tolerance and age-dependent hepatosteatosis. J Biol Chem 279: 44740–44748 [DOI] [PubMed] [Google Scholar]
- Kamiya A, Kinoshita T, Ito Y, Matsui T, Morikawa Y, Senba E, Nakashima K, Taga T, Yoshida K, Kishimoto T, Miyajima A (1999) Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J 18: 2127–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM (1999) Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398: 630–634 [DOI] [PubMed] [Google Scholar]
- Lai E, Clark KL, Burley SK, Darnell JE Jr (1993) Hepatocyte nuclear factor 3/fork head or ‘winged helix' proteins: a family of transcription factors of diverse biologic function. Proc Natl Acad Sci USA 90: 10421–10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Sauer B, Johnson PF, Gonzalez FJ (1997) Disruption of the c/ebp alpha gene in adult mouse liver. Mol Cell Biol 17: 6014–6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A (2005) Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci USA 102: 11278–11283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipow V, Descombes P, Schibler U (1993) CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA 90: 8219–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1alpha interaction. Nature 423: 550–555 [DOI] [PubMed] [Google Scholar]
- Qiao L, MacLean PS, You H, Schaack J, Shao J (2006) Knocking down liver ccaat/enhancer-binding protein alpha by adenovirus-transduced silent interfering ribonucleic acid improves hepatic gluconeogenesis and lipid homeostasis in db/db mice. Endocrinology 147: 3060–3069 [DOI] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR (2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414: 799–806 [DOI] [PubMed] [Google Scholar]
- Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG (2000) Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem 275: 36324–36333 [DOI] [PubMed] [Google Scholar]
- Schrem H, Klempnauer J, Borlak J (2004) Liver-enriched transcription factors in liver function and development. Part II: the C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol Rev 56: 291–330 [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Wilde M, Darlington GJ (1999) C/EBPalpha regulates formation of S-phase-specific E2F–p107 complexes in livers of newborn mice. Mol Cell Biol 19: 2936–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ (1995) Impaired energy homeostasis in C/EBP alpha knockout mice. Science 269: 1108–1112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Figure Legends