Abstract
In the unfolded protein response (UPR) signaling pathway, accumulation of unfolded proteins in the endoplasmic reticulum (ER) activates a transmembrane kinase/ribonuclease Ire1, which causes the transcriptional induction of ER-resident chaperones, including BiP/Kar2. It was previously hypothesized that BiP/Kar2 plays a direct role in the signaling mechanism. In this model, association of BiP/Kar2 with Ire1 represses the UPR pathway while under conditions of ER stress, BiP/Kar2 dissociation leads to activation. To test this model, we analyzed five temperature-sensitive alleles of the yeast KAR2 gene. When cells carrying a mutation in the Kar2 substrate-binding domain were incubated at the restrictive temperature, association of Kar2 to Ire1 was disrupted, and the UPR pathway was activated even in the absence of extrinsic ER stress. Conversely, cells carrying a mutation in the Kar2 ATPase domain, in which Kar2 poorly dissociated from Ire1 even in the presence of tunicamycin, a potent inducer of ER stress, were unable to activate the pathway. Our findings provide strong evidence in support of BiP/Kar2-dependent Ire1 regulation model and suggest that Ire1 associates with Kar2 as a chaperone substrate. We speculate that recognition of unfolded proteins is based on their competition with Ire1 for binding with BiP/Kar2.
INTRODUCTION
Accumulation of unfolded proteins in the endoplasmic reticulum (ER) results in the transcriptional induction of various genes including those encoding ER-resident chaperones and folding enzymes. This cellular mechanism is called the unfolded protein response (UPR), and several important features of the UPR signaling pathway have been revealed initially through studies in the budding yeast Saccharomyces cerevisiae. Yeast Ire1 is a 1115-amino acid transmembrane protein that transmits the unfolded protein signal across the ER membrane (Cox et al., 1993; Mori et al., 1993). The cytoplasmic domain (C-terminal half) of Ire1 possesses both serine/threonine kinase and site-specific endoribonuclease activities (Mori et al., 1993; Sidrauski and Walter, 1997). Ire1 dimerizes in response to the accumulation of unfolded proteins, resulting in its trans-autophosphorylation and activation (Shamu and Walter, 1996; Welihinda and Kaufman, 1996). Activated Ire1 in turn promotes splicing of HAC1 precursor mRNA to produce the mature form (Cox and Walter, 1996; Sidrauski and Walter, 1997), which is effectively translated into a functional transcription factor (Mori et al., 2000; Ruegsegger et al., 2001). Mature form of Hac1 efficiently induces transcription of UPR target genes containing the UPR element (UPRE) in their promoter region (Mori et al., 1992; Kohno et al., 1993).
In mammalian cells, accumulation of unfolded proteins initiates signaling from the ER via more complicated pathways. Two homologues of Ire1, Ire1α and Ire1β, have been identified (Tirasophon et al., 1998; Wang et al., 1998; Iwawaki et al., 2001). According to recent reports (Yoshida et al., 2001; Calfon et al., 2002), IRE1 functions to promote splicing of an mRNA encoding the transcription factor XBP1. This Ire1-XBP1 signaling pathway is reported to act synergistically with another signaling pathway involving an ER-located transmembrane protein ATF6 to promote the mammalian UPR (Yoshida et al., 2001; Shen et al., 2001; Lee et al., 2002). Moreover, accumulation of unfolded proteins in the mammalian ER attenuates bulk protein synthesis. This occurs by phosphorylation of the eukaryotic translation initiation factor 2 subunit by PERK (PKR-like ER kinase), a transmembrane kinase containing an Ire1-like lumenal domain (Harding et al., 1999) and by cleavage of the 28S rRNA by Ire1β (Iwawaki et al., 2001).
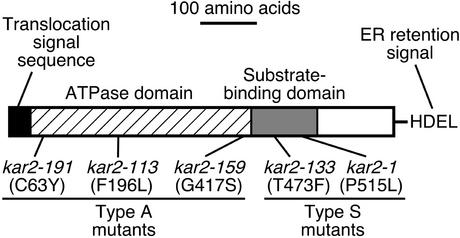
Kar2 is the yeast orthologue of mammalian BiP, an ER-resident member of the Hsp70 family (Normington et al., 1989; Rose et al., 1989), and the expression of KAR2 gene is upregulated by the UPR. Like other members of the family, Kar2 has an amino-terminal ATPase domain adjacent to a substrate-binding domain (Figure 1; Bukau and Horwich, 1998). It is believed that the ATPase domain is involved in regulating the interaction between the substrate-binding domain and unfolded protein substrates. Because KAR2 is essential for viability, studies of its physiological functions have been performed primarily using conditional lethal kar2 alleles. When cultured at the restrictive temperature, some of the temperature-sensitive kar2 mutant strains exhibit a block in the translocation of secretory proteins into the ER (Vogel et al., 1990; Sanders et al., 1992; Brodsky et al., 1995). Several molecular mechanisms have been proposed to explain the function of Kar2 in the protein translocation machinery (Hamman et al., 1998; Matlack et al., 1999). Kar2 has also been shown to serve as a molecular chaperone during protein folding in the ER. For instance, refolding of carboxypeptidase Y (CPY), after denaturation by dithiothreitol (DTT), was impaired in some kar2 mutant strains (Simons et al., 1995). In addition, other reports implicate Kar2 involvement in ER-associated protein degradation (ERAD; Plemper et al., 1997; Brodsky et al., 1999; Nishikawa et al., 2001).
Figure 1.
Schematic representation of the Kar2 protein and kar2 mutant alleles used in this study. Based on the sites of mutation, kar2 mutant alleles were categorized either as ATPase domain mutants (type A) or substrate-binding domain mutants (type S).
We have proposed that KAR2 is not only a UPR target, but is itself a critical negative regulator of the UPR pathway (Okamura et al., 2000). In the current model, Kar2 associates with Ire1 to repress the activation of Ire1 in nonstressed cells, and in response to accumulation of unfolded proteins in the ER, Kar2 dissociates from Ire1, resulting in dimerization and activation of Ire1. This proposed mechanism, hereafter called the BiP/Kar2-dependent Ire1 regulation, is supported by several findings in our previous studies in yeast (Kohno et al., 1993; Okamura et al., 2000). First, overproduction of Kar2 in yeast cells was shown to attenuate the UPR. Furthermore, Kar2 coimmunoprecipitated with Ire1 from lysates of nonstressed cells. Notably, the amount of Kar2 that coimmunoprecipitated with Ire1 extremely decreased when cells were cultured in the presence of reagents that activate the UPR pathway. Similar observations were reported by A. Bertolotti et al. (2000), suggesting that BiP acts to regulate Ire1 and PERK in mammalian cells. Although these studies provided correlative support for the model, firm confirmation awaits direct evidence that the binding state of Ire1 by BiP/Kar2 regulates its activity.
To obtain the further evidence of the BiP/Kar2-dependent Ire1 regulation model, we used a different approach in which the phenotypes of kar2 mutations were analyzed. Mutations stabilizing Kar2 association with Ire1 impaired UPR signaling even under conditions of ER stress. Conversely, mutations disrupting this association constitutively activate the pathway. Our results suggest a molecular mechanism by which unfolded proteins are detected by the sensor protein in the UPR pathway.
MATERIALS AND METHODS
Yeast Strains and Culturing Conditions
Yeast strains used in this study are listed in Table 1. The kar2 mutant genes were cloned from these strains and sequenced for confirmation of their mutation points that are indicated in Figure 1. Culturing and genetic manipulation of yeast strains were by standard techniques described in Kaiser et al. (1994). Basically, cells were cultured in YPD-rich medium, synthetic complete (SC) medium lacking uracil, or minimal synthetic medium plus dextrose (SD) supplemented with appropriate nutrients. For induction of the GAL1 promoter, cells were cultured in synthetic complete galactose (SCG) medium, which is identical to SC medium except that 2% dextrose was replaced with 2% galactose. Cells were grown at 23°C except where noted.
Table 1.
Yeast strains
| Strain | Background | Genotype | Source |
|---|---|---|---|
| MS10 | MS10 | MATa ura3-52 ade2-101 leu2-3,112 | M. Rose |
| MS137 | MS10 | MATa ura3-52 ade2-101 leu2-3,112 kar2-159 | M. Rose |
| MS192 | MS10 | MATa ura3-52 ade2-101 leu2-3,112 kar2-113 | M. Rose |
| MS958 | MS10 | MATa ura3-52 ade2-101 leu2-3,112 kar2-191 | M. Rose |
| MS193 | MS10 | MATa ura3-52 ade2-101 leu2-3,112 kar2-133 | M. Rose |
| W303 | W303 | MATa ura3-1 ade2-1 trp1-1 leu2-3,112 his3-11,15 can1-100 | |
| KMY81 | W303 | MATa ura3-1 ade2-1 leu2-3,112 his4-Δ15 leu1 his3-11,15 can1-100 kar2-1 | This studya |
A kar2-1 strain KNH1 (Normington et al., 1989) was backcrossed four times with W303 to obtain KMY81
Plasmids
The XbaI site in the HIS3 centromeric vector pRS313 (Sikorski and Hieter, 1989) was converted to SphI by insertion of the appropriate oligonucleotide. The vector was then digested with BamHI and SphI, and a 5.6-kbp BamHI-SphI DNA fragment containing the IRE1 gene from plasmid pERN1-EM (a generous gift of Dr. K. Mori, Graduate School of Biostudies, Kyoto University, Kyoto, Japan; Mori et al., 1993) was inserted to generate plasmid pRS313-IRE1. An SphI recognition sequence was inserted into the site 5′-adjacent to the hemagglutinin (HA) epitope coding sequence on plasmid phIRE1-HA2 (a generous gift of A. Hosoda, Nara Institute of Science and Technology, Japan), and a 0.17-kbp SphI-NotI DNA fragment encoding three tandem copies of the HA epitope (YPYDVPDYAGS) followed by a termination codon was isolated. The termination codon (TAA) of the IRE1 gene in pRS313-IRE1 was converted to an SphI site by PCR mediated mutagenesis, the resulting plasmid was digested with SphI and NotI, and the 0.17-kbp DNA fragment was ligated into these sites. From the resulting plasmid, a 4.9-kbp DNA fragment encoding Ire1 tagged with a C-terminal triple HA epitope tag was excised by digesting with BamHI and NotI and cloned into an URA3 2-μm vector pRS426 (Christianson et al., 1992) to obtain plasmid pRS426-IRE1HA.
An URA3 2-μm plasmid pCZY1 (Mori et al., 1992) containing a lacZ reporter gene driven by the CYC1 core promoter fused with the UPRE was generously provided by Dr. K. Mori and used to monitor cellular UPR activity. A 2-μm plasmid pYPR3841U (a generous gift of Dr. M. Takagi, Department of Biotechnology, The University of Tokyo, Tokyo, Japan) was derived from pYPR3841 (Umebayashi et al., 1997) by replacement of the TRP1 selectable maker with URA3 and used for cellular expression of the Δpro from the GAL1 promoter. pKAR2HSE-lacZ (Oka et al., 1997) is an URA3 2-μm plasmid containing a lacZ reporter gene driven by the CYC1 core promoter fused with the KAR2 heat shock element (HSE).
Preparation of Cell Lysates, Immunoprecipitation, and Western Blotting
Yeast cells (10 OD600 equivalent) carrying pRS426-IRE1HA were harvested, washed with PBS, and suspended in 200 μl of buffer A (50 mM Tris-Cl, pH 7.9, 5 mM EDTA, and 1% Triton X-100) supplemented by protease inhibitors (2 mM phenylmethanesulfonyl fluoride, 10 μg/ml each of pepstatin, leupeptin, and aprotinin). Glass beads (0.5 mm) were added up to the meniscus. The cells were broken by vortexing six times for 30 s each at maximum speed. After removal of the glass beads, the cell lysates were clarified by centrifugation (15,000 × g for 10 min).
The cell lysates were then diluted with 800 μl buffer B (the same composition as buffer A except containing 180 mM NaCl and 6% skim milk) and incubated for 1 h at 4°C with 2 μg of the anti-HA mAb 12CA5 (Roche Diagnostics, Basel, Switzerland), followed by the addition of 15 μl of protein A–conjugated Sepharose beads (Protein A Sepharose 4 FF; Amersham Biosciences, Uppsala, Sweden). After further incubation at 4°C for 1 h, the Sepharose beads were collected by centrifugation, washed five times with buffer C (the same composition as buffer B except for containing no skim milk), and used as immunoprecipitates.
For cells carrying pYPR3841U or the control empty vector pRS426, lysates were prepared as described above except that buffer C supplemented by the protease inhibitors was used instead of buffer A. Next, lysates (190 μl) were ultracentrifuged (Beckman TLA-100.3 rotor, 100,000 × g, 15 min, 4°C), and both supernatant and pellet fractions were collected. The pellet fractions were washed twice with buffer C, and resuspended in 190 μl of buffer C.
The samples described above were added to an equal volume of gel loading buffer containing 125 mM Tris-HCl (pH 6.8), 20% glycerol, 4% SDS, 20 mM DTT, and 0.02% bromophenol blue. Then they were incubated at 95°C for 2 min and fractionated on 8% SDS-PAGE gels. Proteins were then transferred to nitrocellulose membranes (PROTORAN; Schleicher & Schuell, Dassel, Germany) by electrophoresis at 1 mA/cm2 for 1.5 h in transfer buffer containing 48 mM Tris, 38.6 mM glycine, 0.037% SDS, and 20% methanol. The blots were blocked overnight at 4°C in PBS containing 0.1% Tween 20 and 5% skim milk (blocking solution). The following day, the blots were incubated with the same blocking solution, but containing the primary antibody (12CA5 anti-HA antibody at 0.4 μg/ml, rabbit anti-Kar2 antiserum [Tokunaga et al., 1992] at a 1:1000 dilution, or rabbit anti-RNAP-I antiserum [a generous gift of Dr. M. Takagi; Fukuda et al., 1994]) at a 1:5000 dilution. Incubation was for 1 h at room temperature. The blots were subsequently washed five times with PBS containing 0.1% Tween 20 (washing solution) at room temperature for 3 min each. The blots were next incubated with a 1:1000 dilution of horseradish peroxidase–coupled, goat anti-rabbit or anti-mouse IgG (Amersham Biosciences) in blocking solution for 1 h at room temperature. The blots were then washed five times with the washing solution at room temperature for 3 min each, and specific protein bands were detected using enhanced chemiluminescence (ECL; Amersham Biosciences) and x-ray films (RX-U; Fuji Photo Film, Ashigara, Japan).
In case of chemical cross-linking experiments, cells (25 OD600 equivalent) were suspended in 200 μl of PBS containing 1% Triton X-100 and the protease inhibitors and were broken by the glass beads. After clarification by centrifugation (3000 × g for 30 s), 90 μl of the lysates was subjected to chemical cross-linking reaction with 2 mM dithiobissuccinimidyl propionate (DSP; 1.8 μl of 100 mM stock [freshly prepared in DMSO] was added) at 27°C for 30 min. The reaction was quenched by the addition of 90 μl of 1 M Tris-Cl, pH 7.5, and the samples were then ultracentrifuged (Beckman TLA-100.3 rotor, 100,000 × g, 15 min, 4°C). The pellet fractions were dissolved in 100 μl of resuspend buffer (50 mM Tris-Cl, pH 7.5, 1 mM EDTA, 1% SDS) by incubation at 95°C for 3 min, diluted by 900 μl of buffer C, and incubated with the anti-Kar2 antiserum (1:120 dilution) or a control preimmune serum for 1 h at 4°C. The immunocomplexes were collected by protein A–conjugated Sepharose beads and analyzed by Western blotting as described above.
RNA Analysis
DNA probes for Northern blot analysis were prepared by PCR amplification of yeast genomic DNA. The HAC1, KAR2, and SSA probes, respectively, correspond to nucleotide -11–654 of HAC1, nucleotide 9–2047 of KAR2, and nucleotide 1016–1756 of SSA1.
Total RNA was prepared using the hot phenol method (Collart and Oliviero, 1993). For Northern blot analysis, 5 μg of total RNA was separated on a 1% agarose, 1.8% formaldehyde gel and transferred to a nylon membrane (Hybond-N; Amersham Biosciences). The membrane was prehybridized in 500 mM sodium phosphate, pH 7.0, 1 mM EDTA, and 7% SDS. The membrane was then incubated with the random primed 32P-labeled DNA probe. After washing, the membrane was exposed to an imaging screen (BAS-MS2040; Fuji), and signal intensity was quantified using a Fuji BAS2500 image analyzer. The percentage of HAC1 mRNA cleavage was calculated using the equation (It - I u)/It × 100%, where It is the intensity of total HAC1 mRNA species and Iu is the intensity of uncleaved HAC1 (HAC1u) mRNA.
Assay of Cellular β-Galactosidase Activity
β-Galactosidase activity of cells was determined using the protocol of Kaiser et al. (1994). Cells (0.5 OD600 equivalent) were suspended in 800 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.27% 2-mercaptoethanol, pH 7.0), and 20 μl of 0.1% SDS and 50 μl of chloroform was added. The mixture was vortexed vigorously for 20 s. After equilibration at 28°C for 5 min, o-nitrophenyl-β-d-galactoside was added to a final concentration of 0.8 mg/ml. The reaction was stopped at various times by adding 0.5 ml of 1 M Na2CO3, and the concentration of the product, o-nitrophenol (ONP), was measured by optical density at 420 nm. One unit of β-galactosidase activity is defined as 1 nmol of ONP/min of reaction for 1 ml of culture at 1 OD600 unit.
RESULTS
Regulation of the UPR Is Disrupted in kar2 Mutant Strains
In this study, we used yeast strains carrying various temperature-sensitive kar2 mutant alleles originally generated by Polaina and Conde (1982) and by us. As shown in Figure 1, all of these are single-point mutants. Based on the location of the mutations, we categorized each as an ATPase domain mutant (kar2-159, kar-113, kar2-191; hereafter called type A) or a substrate-binding domain mutant (kar2–1, kar2–133; hereafter called type S). As shown in Table 1, two different strain backgrounds were used. The kar2-159, -113, -191 and -133 strains are of the MS10 background, and the kar2-1 strain is of the W303 background. Thus, the parental MS10 and W303 strains, respectively, were used as KAR2 wild-type controls. All of the kar2 mutant strains grew well at 23°C, but exhibited various degrees of growth defects when cultured at 37°C. Hence, 23°C was used as the permissive temperature and 37°C as the restrictive temperature, unless otherwise described.
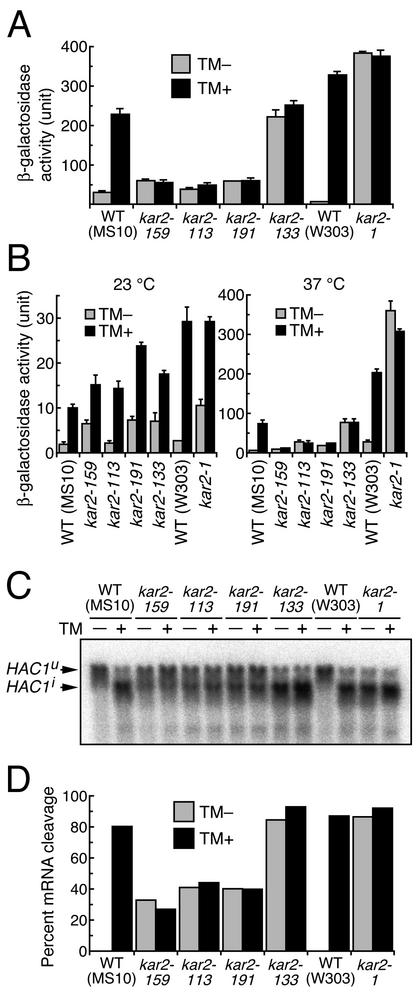
We first wished to determine whether any of the mutations altered the cell's ability to regulate the UPR pathway. To monitor cellular activity of this pathway, we used a lacZ reporter plasmid for expression of β-galactosidase under the control of the UPRE. At first, cells carrying this plasmid were cultured in SC medium at 23°C and shifted to 37°C for 200 min. Extrinsic ER stress was imposed by 2 μg/ml tunicamycin, an N-glycosylation inhibitor (Takatsuki et al., 1975), which was added immediately after the temperature shift. In the absence of tunicamycin, β-galactosidase activity was slightly higher in kar2 type A mutant cells than in wild-type cells (Figure 2A). However, in the presence of tunicamycin, a strong induction was observed in the wild-type, but was completely lacking in the kar2 type A mutant cells. In contrast, constitutively high β-galactosidase activity was observed in the kar2 type S mutant cells, regardless of the presence of tunicamycin. This activity was similar to that seen in tunicamycin-treated wild-type cells.
Figure 2.
Altered UPR in kar2 mutant strains. (A) Cells transformed with the UPRE-lacZ reporter plasmid pCZY1 were exponentially grown to an OD600 of 0.3 at 23°C in SC medium, and at time (t = 0), the cultures were shifted to 37°C. Tunicamycin was added to a final concentration of 0 μg/ml (TM -)or2 μg/ml (TM +) at 3 min, and the cultures were subjected to a β-galactosidase assay at 200 min. Data presented are the averages of three independent transformants with SD as indicated by error bars. (B) Cells transformed with pCZY1 were exponentially grown to an OD600 of 0.2 at 23°C in SD medium, and at time (t = 0), the cultures were shifted to 37°C (indicated as 37°C) or further cultured at 23°C (indicated as 23°C). Tunicamycin was added to a final concentration of 0 μg/ml (TM -)or 2 μg/ml (TM +) at 3 min, and the cultures were subjected to β-galactosidase assay at 200 min. Data are presented as described in the legend to the A. (C) Cells were exponentially grown to an OD600 of 0.4 at 23°C in YPD medium, and at time (t = 0), the cultures were shifted to 37°C. Tunicamycin was added to a final concentration of 0 μg/ml (TM -)or 2 μg/ml (TM +) at 3 min, and the cells were collected at 60 min to extract total cellular RNA for Northern blot analysis using the HAC1 DNA probe. (D) Percentages of HAC1 mRNA cleavage in C were calculated as described in MATERIAL AND METHODS.
As a control, we also monitored UPR activity at the permissive temperature. For reasons unknown, we were not able to detect any β-galactosidase activity in any of the seven strains tested in this study when cultured at 23°C in SC medium (unpublished data), and we suspected that the UPRE-lacZ reporter does not work in this culturing condition. Therefore, the cells were cultured in SD medium for monitoring the UPR at the permissive temperature. As shown in the left panel of Figure 2B, all of the kar2 mutant strains as well as the wild-type strains exhibited almost normal induction of UPR by tunicamycin at 23°C. However, when cells were cultured at 37°C, the UPR was not induced by tunicamycin in the type A mutant cells, but was constitutively active in the type S mutant cells (Figure 2B, right panel). It should be noted that expression of β-galactosidase from this UPRE-lacZ reporter somehow depends on the strain background. The W303 background strains exhibited higher β-galactosidase activity than the MS10 background strains.
Because the HAC1 mRNA precursor (uninduced form; HAC1u; Cox and Walter (1996)) is the substrate of Ire1, a more direct measure of Ire1 activity is HAC1u splicing. For this, we monitored HAC1u cleavage by Northern blot analysis (Figure 2, C and D). We cultured cells in rich YPD medium at 23°C, shifted to 37°C for 60 min, and then extracted total cellular RNA. ER stress was imposed by the addition of 2 μg/ml tunicamycin immediately after the temperature shift. As expected, in the wild-type cells, HAC1u was effectively cleaved and converted to the mature HAC1 mRNA (induced form: HAC1i; Cox and Walter, 1996) only in the presence of tunicamycin. In the type A mutant cells, partial cleavage of HAC1u was observed even in the absence of tunicamycin, and addition of tunicamycin did not enhance the extent of cleavage. In contrast, the type S mutant cells exhibited nearly complete cleavage of HAC1u both in the absence and presence of tunicamycin. Importantly, these results are consistent with the observations obtained by the UPRE-lacZ reporter assay described above. We therefore concluded that at the restrictive temperature, Ire1 is constitutively and highly activated in the type S mutant strains and constitutively repressed in the type A mutant strains.
Impaired Association and Dissociation of Kar2 and Ire1 in kar2 Mutant Strains
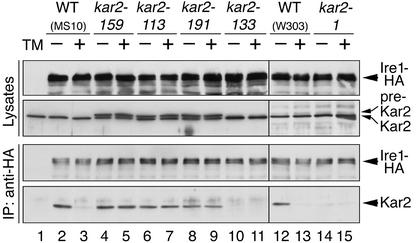
We next analyzed the in vivo physical interaction between Kar2 and Ire1 by employing a coimmunoprecipitation assay (Figure 3). Ire1 bearing a C-terminal triple influenza virus HA epitope tag (Ire1-HA) was expressed in the kar2 mutant and control wild-type strains. Our previous study had indicated that Ire1-HA functions similar to wild-type Ire1 to transmit the unfolded protein signal across the ER membrane (Okamura et al., 2000). In this study, cells carrying an Ire1-HA expression plasmid were cultured in SC medium at 23°C and shifted to 37°C for 60 min, and lysates were prepared. ER stress was induced by the addition of 2 μg/ml tunicamycin immediately after the temperature shift. Anti-HA Western blot analysis of the cell lysates indicated that Ire1-HA was expressed at similar levels in all of the kar2 mutant and control wild-type strains (Figure 3, uppermost panel). The precursor form of Kar2 (pre-Kar2) was detected in lysates from the type A mutant cells by anti-Kar2 Western blotting (Figure 3, second panel, lanes 4–9), suggesting that protein translocation is impaired in these cells (Vogel et al., 1990).
Figure 3.
Altered association or dissociation of Kar2 with Ire1 in kar2 mutant strains. The indicated strains transformed with the Ire1-HA expression plasmid pRS426-Ire1HA (lanes 2–15), or MS10 cells containing an empty vector pRS426 (lane 1) were exponentially grown to an OD600 of 0.4 at 23°C in SC medium. At time (t = 0), the cultures were shifted to 37°C. Tunicamycin was added to a final concentration of 0 μg/ml (TM -) or 2 μg/ml (TM +) at 3 min, and cells were collected at 60 min to prepare their lysates (In lane 1, no tunicamycin was added). Anti-HA immunoprecipitates equivalent to 1 × 108 cells for the lowest panel, and lysates or anti-HA immunoprecipitates equivalent to 1 × 107 cells for the other panels were fractionated by 8% SDS-PAGE. Proteins were detected by subsequent Western blotting with the indicated antibodies. After treatment with ECL chemiluminescence reagents, the blots were exposed to x-ray films for the following times: 10 s for the lowest panel and 2 s for the other panels.
The cell lysates were next subjected to anti-HA immunoprecipitation. Anti-HA Western blot analysis of the immunoprecipitates showed that nearly equal amounts of Ire1-HA were immunoprecipitated from each lysate (Figure 3, third panel). Kar2 in the immunocomplexes was then detected by Western blot analysis. From lysates prepared from wild-type cells cultured under nonstressed conditions, Kar2 was found to be complexed with Ire1-HA (Figure 3, lowest panel, lanes 2 and 12). Consistent with our previous observation (Okamura et al., 2000), the amount of coimmunoprecipitated Kar2 was sharply reduced in these cells after treatment by tunicamycin (Figure 3, lowest panel, lanes 3 and 13). Kar2 was undetectable in a control strain not expressing Ire1-HA, showing that the signal is specific (Figure 3, lowest panel, lane 1). When the assay was performed using lysates from the mutant strains, Kar2 displayed distinctly different binding characteristics to Ire1, depending on the strain. Treatment of the type A mutant cells with tunicamycin did not cause dissociation of Kar2 from Ire1 (Figure 3, lowest panel, compare lanes 5-4, 7-6, and 9-8). In type S mutants, little association between the two proteins was observed even when cultured in the absence of tunicamycin (Figure 3, lowest panel, lanes 10 and 14).
Type S Mutant Strains Exhibit Impaired Association between Kar2 and a Model Chaperone Substrate
We next examined in vivo association of Kar2 with a model unfolded protein. Aspartic proteinase-I (RNAP-I) of the filamentous fungus Rhizopus niveus is a secretory protein synthesized as a precursor form containing a presequence (translocation signal; 21 amino acid residues) and prosequence (45 amino acid residues) in the N-terminus of the mature portion (323 amino acid residues). When the fulllength form of the RNAP-I precursor is expressed heterologously in yeast cells, the protein is secreted extracellularly as the mature form (Horiuchi et al., 1990). The prosequence of RNAP-I, similar to that of other secretory proteins, acts to support proper folding of the mature protein (Fukuda et al., 1994). Therefore, a truncated mutant RNAP-I lacking the prosequence (Δpro) remains incompletely folded and aggregates in the ER when expressed in yeast cells (Umebayashi et al., 1997). Kar2 has been shown to coaggregate with Δpro (Umebayashi et al., 1997), and we have used this heterologous protein as a model chaperone substrate to monitor in vivo association with Kar2.
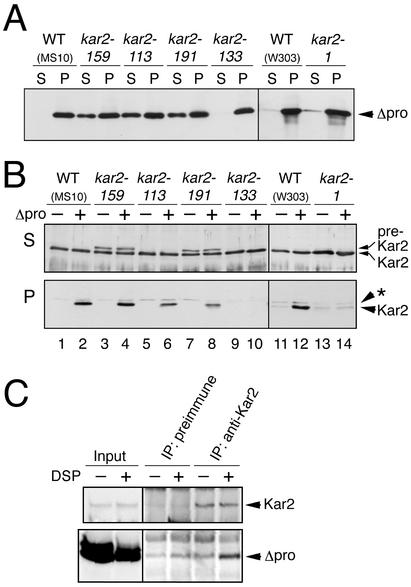
As shown in Figure 4, Δpro was expressed from the GAL1 promoter in the kar2 mutant and wild-type strains. As a control, yeast strains were transformed with an empty vector. Cells were cultured in galactose-based medium (SCG) at 23°C and shifted to 37°C for 60 min, except for the kar2-133 mutant, which was shifted to 34°C in order to avoid Δpro independent aggregation of the Kar2-133 protein at 37°C. Lysates of the cells were then subjected to ultracentrifugation at 100,000 × g, and both supernatant and pellet fractions were collected. Note that to dissolve membraneous components, the cell lysates were prepared in the presence of the nonionic detergent Triton X-100. Anti–RNAP-I Western blot analysis of the supernatant and pellet fractions showed that Δpro was almost perfectly aggregated in the wild-type and type S mutant cells (Figure 4A). In the type A mutant cells, Δpro was also detected in the supernatant fractions. Although we speculated that the accumulation of this soluble version of Δpro might be caused by translocation block in these mutant cells, a protease protection assay showed that this soluble Δpro resides in a closed membranous compartment (unpublished data). Thus the soluble Δpro may locate in the ER lumen in the type A mutant cells under the restricted temperature by unknown reasons. On the other hand, anti-Kar2 Western blotting showed little or no accumulation of pre-Kar2 even in the type A mutant cells (Figure 4B, lanes 3–8). This may be because cells grow slower in galactose medium than in glucose medium. Western blotting of the pellet fractions with anti-Kar2 showed that Kar2 coaggregates with Δpro in both wild-type cells (Figure 4B, lower panel, compare lanes 2-1 and 12-11) and type A mutant cells (Figure 4B, lower panel, compare lanes 4-3, 6-5, and 8-7). As shown in Figure 4C, Δpro was cross-linked with Kar2 by DSP, which indicates that Kar2 directly contacts Δpro. Furthermore, Δpro cofractionated with Kar2 in a sucrose gradient analysis of the microsomal fraction, which suggests that Δpro and Kar2 are present in the same complex (Umebayashi et al., 1997). In contrast, Δpro-dependent aggregation of Kar2 was not observed in the type S mutant cells (Figure 4B, lanes 9, 10, 13, and 14).
Figure 4.
Impaired coaggregation of Kar2 with a model unfolded protein in kar2 type S mutant strains. Cells transformed with the Δpro expression plasmid pYPR3841U or an empty vector pRS426 were exponentially grown to an OD600 of 0.2 at 23°C in SCG medium, and the cultures were shifted to 37°C (or 34°C for the kar2-133 strain) for 60 min. Cell lysates were prepared from the resulting cultures and ultracentrifuged at 100,000 × g for 15 min, and the supernatant and pellet fractions were collected (A and B). (A) Supernatant (S) and pellet (P) fractions (equivalent to 1 × 107 cells) prepared from cells containing pYPR3841U were analyzed by anti-RNAP1 Western blotting. (B) Supernatant (S; equivalent to 1 × 107 cells) and pellet (P; equivalent to 5 × 107 cells) fractions prepared from cells containing pYPR3841U (Δpro +) or pRS426 (Δpro -) were analyzed by anti-Kar2 Western blotting. The asterisk indicates a nonspecific band. In both panels A and B, the blots treated with ECL reagent were exposed to x-ray films for 2 s. (C) Lysates from W303 cells carrying pYPR3841U were treated with control DMSO or 2 mM DSP and ultracentrifuged. The resulting pellet fraction (input) were dissolved in 1% SDS, and subjected to immunoprecipitation with anti-Kar2 antiserum or control preimmune serum. The inputs (equivalent to 1 × 107 cells for the upper panel or 3 × 107 cells for the lower panel) or immunoprecipitates (equivalent to 3 × 107 cells for the upper panel or 9 × 107 cells for the lower panel) were analyzed by reducing SDS-PAGE followed by anti-Kar2 and anti-RNAP Western blotting.
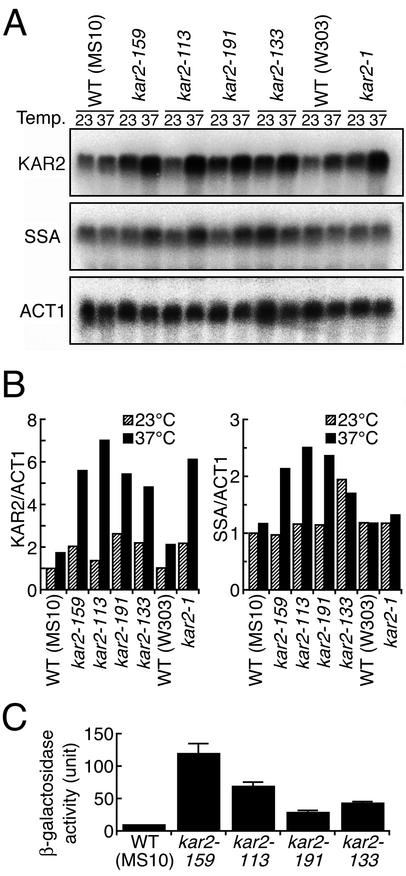
Upregulation of KAR2 Transcription in kar2 Mutants Is Due to Both the UPR and the Heat Shock Response
It has been previously reported that mutation of KAR2 results in its own transcriptional induction (Scidmore et al.,1993 for kar2-159 and Kohno et al., 1993 for kar2–1). Therefore we monitored KAR2 mRNA abundance in all of the kar2 mutant strains used in this study. As shown in Figure 5A, the kar2 mutant and wild-type strains were cultured at 23°C or shifted to 37°C, and gene expression was examined by Northern blotting. The mRNAs were then quantitated and normalized to ACT1 mRNA (Figure 5B, left). Culturing at 37°C for 60 min caused only weak induction of the KAR2 gene in the wild-type strains. In contrast, all of the kar2 mutant strains exhibited about twofold higher expression of the KAR2 gene than the wild-type strains when cultured at 23°C and exhibited greatly enhanced expression of the KAR2 gene when shifted to 37°C. However, as described above, the type A and type S mutant strains have different unfolded protein responses, implying that this transcriptional upregulation of the KAR2 gene in kar2 mutant strains cannot be explained simply by activation of the UPR pathway. Regulation of the KAR2 gene involves not only the UPRE but also the heat shock element (HSE), which is present in its promoter region (Mori et al., 1992; Kohno et al., 1993). We therefore examined whether the heat shock response pathway is activated in kar2 mutant strains. SSA1, 2, 3, and 4 are highly similar genes encoding cytosolic Hsp70, and all except SSA2 are induced by the heat shock response (Ziegelhoffer and Craig, 1997). Northern blotting using a probe for SSA1 revealed enhanced SSA expression in type A mutant cells cultured at 37°C for 60 min, but not in the wild-type or kar2-1 cells (Figure 5, A and B). In addition, the SSA genes were highly expressed both at 23 and 37°C in the kar2-133 cells (Figure 5, A and B). Activation of the heat shock response pathway in the type A and kar2-133 mutant strains was confirmed by an HSE-lacZ reporter assay, in which expression of lacZ is under the control of the KAR2 HSE. As shown in Figure 5C, both the type A and kar2-133 mutant strains exhibited higher lacZ expression than the wild-type strain, when they were cultured at 37°C. These observations indicate that KAR2 mRNA induction in these mutant strains is at least partially caused by the heat shock response.
Figure 5.
Induction of the KAR2 gene in kar2 mutants results partially from activation of the heat shock response pathway. (A) Cells were exponentially grown to an OD600 of 0.4 at 23°C in YPD medium, and shifted to 37°C (37°C) or further cultured at 23°C (23°C) for 60 min. The resulting cells were used to extract total cellular RNA for Northern blot analysis using the KAR2, SSA1, and ACT1 DNA probes. (B) The RNAs shown in A were quantitated and normalized to ACT1 mRNA. Normalized mRNA values are presented relative to their levels of expression in MS10 cells cultured at 23°C. (C) Cells transformed with the HSE-lacZ reporter plasmid pKAR2HSE-lacZ were exponentially grown to an OD600 of 0.3 at 23°C in SC medium and shifted to 37°C for 200 min, followed by β-galactosidase assay. For an unknown reason, this HSE-lacZ reporter did not work in the W303 background strains.
DISCUSSION
Previously, two studies reported findings supporting a BiP/Kar2-dependent mode of Ire1 regulation (Bertolotti et al., 2000; Okamura et al. 2000). Of most significance was the observation that inactivation and activation of the UPR correlated with the binding state of BiP/Kar2 with Ire1. However, the causality between regulation of Ire1 (or PERK) and its physical interaction with BiP/Kar2 was not firmly established. The assertion that BiP/Kar2 plays a negative regulatory role upstream of Ire1 (or PERK) is derived from the observation that activation can be attenuated by overproduction of BiP/Kar2. However, an alternative explanation must be considered. The overproduction of BiP/Kar2, in some circumstances, is sufficient to alleviate ER stress (Umebayashi et al., 1999). Therefore, the observed attenuation of the response might be a secondary effect.
Hence, we sought to provide more definitive evidence for the BiP/Kar2-dependent Ire1 regulation model. In the present study, we analyzed a pool of KAR2 mutants that display allele-specific phenotypes with respect to Ire1 association and activation. As shown in Figure 2, examination of five kar2 mutant alleles revealed two different types of UPR abnormalities, as monitored by the UPRE-lacZ reporter assay and Northern blot analysis for HAC1u mRNA cleavage. When cultured at the restrictive temperature of 37°C, the kar2 type A mutant strains (Figure 1) exhibited weak activation of the UPR pathway that was not enhanced by treatment with tunicamycin. In contrast, the type S mutant strains (Figure 1) showed high activation of the UPR pathway both in the absence and presence of tunicamycin. We then investigated the in vivo interaction between Kar2 and Ire1 by monitoring coimmunoprecipitation of Kar2 with HA-tagged Ire1 from cell lysates (Figure 3). Consistent with our previous report (Okamura et al., 2000), dissociation of Kar2 from Ire1 in response to tunicamycin was clearly observed in the wild-type cells. In contrast, this dissociation was not observed in the type A mutant cells, whereas in the type S mutant cells, association between Kar2 and Ire1 was almost undetectable even in the absence of tunicamycin. One attractive interpretation of these results, which we believe most likely, is that the abnormalities in the UPR pathway observed in kar2 mutants are the results of altered association and dissociation between Kar2 and Ire1. This mechanism would be consistent with the BiP/Kar2-dependent Ire1 regulation model.
We should note that Kar2 has multiple biological roles in the cell, as described in the Introduction, and thus other interpretations for the UPR abnormalities in the kar2 mutant strains are possible. Because Kar2 acts to promote both proper folding and ERAD of unfolded proteins (Simons et al., 1995; Plemper et al., 1997; Brodsky et al., 1999; Nishikawa et al., 2001), it is likely that mutations in the KAR2 gene cause activation of the UPR pathway through increased amount of unfolded proteins in the ER. Indeed, all of the kar2 mutant strains displayed higher activation of the UPR pathway than the wild-type strains when cultured at 37°C in the absence of tunicamycin (Figure 2). However, this idea alone does not explain the different levels of UPR activity between the type A and S mutant strains. In addition, Kar2 plays important roles in the protein translocation machinery (Hamman et al., 1998; Matlack et al., 1999). As shown in the second panel of Figure 3, pre-Kar2 accumulated in the type A mutant cells but not in the wild-type or type S mutant cells when cultured at 37°C. We also observed accumulation of an unglycosylated form of CPY only in the type A mutant cells (unpublished data). These findings are consistent with previous reports (Vogel et al., 1990; Sanders et al., 1992) and suggest that type A and type S mutants correspond to the class I and class II kar2 mutants, respectively, defined by Brodsky and Rose (1997), based on their phenotypes. These observations strongly suggest that protein translocation into the ER is blocked only in the type A mutant strains. Thus, it is possible that in Type A mutants, protein influx into the ER is reduced to the extent that even under ER-stressed conditions unfolded proteins do not accumulate to levels sufficient to activate the UPR pathway. Therefore, as we discussed below, we think that experimental approaches not involving kar2 mutant strains are required for further support of the BiP/Kar2-dependent Ire1 regulation model.
We next investigated the in vivo association of Kar2 mutant proteins with chaperone substrate using Δpro as a model unfolded protein. As shown in Figure 4, coaggregation of Kar2 with Δpro was observed in the wild-type and type A mutant cells, but not in the type S mutant cells. Impaired association of Kar2 with its chaperone substrate in the type S mutant strains is reasonable because these kar2 mutant alleles carry single-point mutations corresponding to the Kar2 substrate-binding domain (Figure 1; Bukau and Horwich, 1998). On the other hand, te Heesen and Aebi (1994) expressed a nonglycosylatable mutant version of CPY (CPY*) in several kar2 mutant strains and detected a complex of CPY* and Kar2 in kar2-159 cells but not in wild-type or kar2-1 cells. Furthermore, Simons et al. (1995) reported that in vivo association of Kar2 with DTT-denatured CPY was enhanced in kar2-159 and kar2-113 mutants. Thus it is likely that dissociation between Kar2 and its chaperone substrate is impaired in the type A mutant strains. This idea is further supported by the low affinity of purified Kar2–159 protein to ATP (Brodsky et al., 1995), because in Hsp70 chaperones, binding of ATP to the ATPase domain promotes release of substrate from the substrate-binding domain (Bukau and Horwich, 1998). In the case of kar2-113, Brodsky et al. (1995) reported no defect in ATP binding or ATPase activity with purified Kar2-113 protein. Thus the abovementioned observation in kar2-113 strain may be caused by impaired interaction of the Kar2–113 ATPase domain to cochaperone(s).
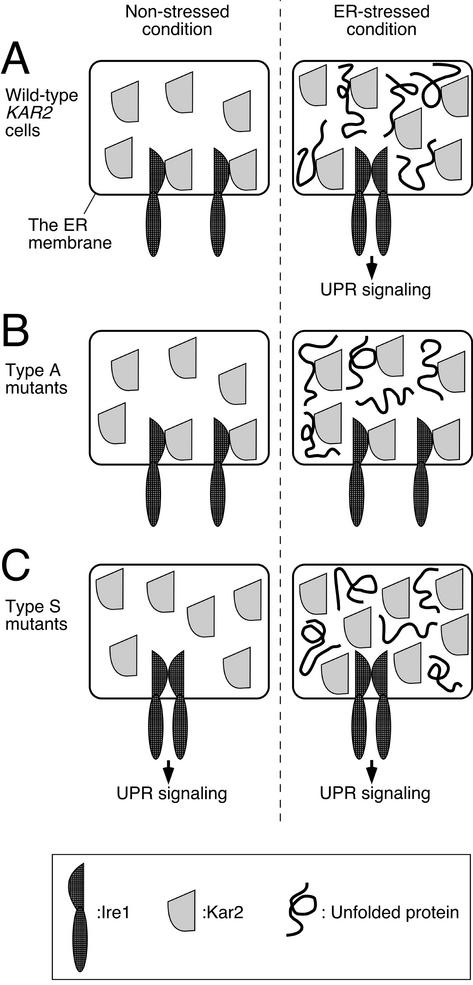
It is notable that the type S Kar2 mutant proteins showed defective association with both Ire1 and a model unfolded protein. In addition, the type A Kar2 mutant proteins, of which dissociation from Ire1 was impaired, are reported to exhibit insufficient release of unfolded proteins. Thus it is likely that Ire1 association with Kar2 is analogous or identical to that of a chaperone substrate. On the basis of this idea, we envision that Ire1 competes with unfolded proteins for binding to the same site on Kar2. This is consistent with the BiP/Kar2-dependent Ire1 regulation model and explains the various results reported in this study. The explanation for the results in this study is presented in Figure 6. In wild-type cells under nonstressed conditions, Kar2 binds with Ire1 and represses Ire1 activity. In ER-stressed conditions, unfolded proteins compete with Ire1 for binding to Kar2, Ire1 is released and activated, and activated Ire1 in turn induces the UPR signaling pathway (Figure 6A). In contrast, type A mutant Kar2 binds constitutively to its chaperone substrates, including Ire1, and does not release them even under ER-stressed conditions. This results in the constitutive repression of Ire1 activity (Figure 6B). In the third case, Type S mutant Kar2 binds poorly with its chaperone substrates, and this results in free Ire1 molecules and constitutive induction of the UPR signaling pathway even in the absence of ER stress (Figure 6C).
Figure 6.
A schematic model to interpret our present observations. See text for explanation.
This interpretation implies that the major role of the Ire1 lumenal domain may merely be homodimerization, which is inhibited by Kar2. Generally speaking, the association of proteins involves interactions between their hydrophobic segments, which are often masked by Hsp70 chaperones. Thus it may be possible to sustain the function of Ire1 by a replacement of its lumenal domain with another dimerforming polypeptide. Indeed, Liu et al. (2000) reported that a chimeric protein in which the lumenal domain of Ire1 was replaced with a functional leucine zipper motif from the transcription factor Maf or Jun was able to moderately activate the UPR pathway in response to ER stress. However, in view of the strong response of native Ire1 to unfolded proteins, we believe that its lumenal domain has additional properties that further facilitate its function as a sensor. For instance, the balance between the rates of Ire1 homodimerization and Kar2-Ire1 association could be an important factor in determining Ire1 activation in response to accumulated unfolded proteins.
Both our present and previously published observations indicate that any mutation of KAR2 results in induction of the KAR2 gene, especially at the restrictive temperature (Figure 5; Kohno et al., 1993; Scidmore et al., 1993). We also observed that the heat shock response pathway is highly activated in the type A and kar2-133 strains but not in the wild-type or kar2-1 strains, as monitored by expression of the SSA genes and by an HSE-lacZ reporter assay (Figure 5). This indicates that both the UPR pathway and the heat shock response pathway contribute to the upregulation of the KAR2 gene expression in kar2 mutant strains, albeit this contribution varies with the particular mutant. It should be noted that generally speaking, heat shock induced by shifting cells to high temperature is quickly attenuated even if culturing at this temperature is continued. Indeed, we could not detect induction of the SSA genes by culturing the wild-type cells at 37°C for 60 min (see Figure 5, A and B). One possible explanation for the high state of activation of the heat shock response pathway in the type A mutant strains cultured at 37°C is that untranslocated secretory and membrane proteins accumulate and act as unfolded cytosolic proteins to induce this pathway (Morimoto, 1997; Oka et al., 1997). However, we are currently unable to explain the reason why the heat shock response pathway is upregulated by the kar2-133 mutation but not by the kar2–1 mutation both at 23 and 37°C.
In conclusion, we believe our findings in the present study are strong evidence to support the BiP/Kar2-dependent Ire1 regulation model and suggest that recognition of unfolded proteins by the UPR pathway is based on competition between Ire1 and unfolded proteins for binding with Kar2. Interestingly, Shen et al. (2002) have recently proposed that BiP negatively regulates ATF6 activity. Thus, it appears that negative regulation by BiP/Kar2 is a common feature of transmembrane signal transducers for the ER stress response.
To clarify the detailed mechanism by which Ire1 is regulated by BiP/Kar2, we think that additional experimental approaches are needed. Characterization of the purified Ire1 lumenal domain, such as undertaken by Liu et al. (2002), may be an important experimental approach. Furthermore, mutational analysis of the Ire1 lumenal domain, an effort we are currently pursuing, may also provide new insights into the molecular mechanism regulating the Ire1 activity.
Acknowledgments
We thank Jeff Brodsky (University of Pittsburgh, Pittsburgh, PA) for the information about kar2 mutants; Kazutoshi Mori (Kyoto University, Kyoto, Japan) for plasmids; Masamichi Takagi and Akinori Ohta (The University of Tokyo, Tokyo, Japan) for plasmids and an antibody; Miki Matsumura for excellent technical assistance; Davis Ng (Pennsylvania State University, State College, PA) for useful comments on the manuscript; and Marc Lamphier for critical reading of the manuscript. This work was supported by Grants-in-Aid for Scientific Research (13480239 to K.K.) and Scientific Research on Priority Areas (14035239 to Y.K., 14037240 to K.K.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by grants from Novartis Foundation (K.K.), Senri Life Science Foundation (Y.K.), and Yamada Science Foundation (K.K.).
Abbreviations used: BiP, immunoglobulin heavy-chain binding protein; CPY, carboxypeptidase Y; DSP, dithiobissuccinimidyl propionate; DTT, dithiothreitol; ECL, enhanced chemiluminescence; ER, endoplasmic reticulum; ERAD, ER-associated protein degradation; HA, hemagglutinin; HSE, heat shock element; ONP, o-nitrophenol; PERK, PKR-like ER kinase; RNAP-I, aspartic proteinase-I; SC, synthetic complete; SCG, synthetic complete galactose; SD, synthetic medium plus dextrose; UPR, unfolded protein response; UPRE, UPR element; Δpro, truncated mutant RNAP-I lacking the prosequence.
References
- Bertolotti, A., Zhang, Y., Hendershot, L.M., Harding, H.P., and Ron, D. (2000). Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2, 326-332. [DOI] [PubMed] [Google Scholar]
- Brodsky, J.L., Goeckeler, J., and Schekman, R. (1995). BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92, 9643-9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J.L., and Rose, M.D. (1997). Saccharomyces cerevisiae Kar2p. In: Molecular Chaperones and Protein-Folding Catalysts, ed. M.J. Gething, New York: Oxford University Press, 33-36.
- Brodsky, J.L., Werner, E.D., Dubas, M.E., Goeckeler, J.L., Kruse, K.B., and McCracken, A.A. (1999). The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 274, 3453-3460. [DOI] [PubMed] [Google Scholar]
- Bukau, B., and Horwich, A.L. (1998). The Hsp70 and Hsp60 chaperone machines. Cell 92, 351-366. [DOI] [PubMed] [Google Scholar]
- Calfon, M., Zeng, H., Urano, F., Till, J.H., Hubbard, S.R., Harding, H.P., Clark, S.G., and Ron, D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92-96. [DOI] [PubMed] [Google Scholar]
- Christianson, T.W., Sikorski, R.S., Dante, M., Shero, J.H., and Hieter, P. (1992). Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119-122. [DOI] [PubMed] [Google Scholar]
- Collart, M.A., and Oliviero, S. (1993). In: Current Protocols in Molecular Biology, ed. F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smoth, and K. Struhl, New York: Greene Publishing Associates and John Wiley & Sons, 13.12.1-13.12.2.
- Cox, J.S., Shamu, C.E., and Walter, P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197-1206. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., and Walter, P. (1996). A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391-404. [DOI] [PubMed] [Google Scholar]
- Fukuda, R., Horiuchi, H., Ohta, A., and Takagi, M. (1994). The prosequence of Rhizopus niveus aspartic proteinase-I supports correct folding and secretion of its mature part in Saccharomyces cerevisiae. J. Biol. Chem. 269, 9556-9561. [PubMed] [Google Scholar]
- Hamman, B.D., Hendershot, L.M., and Johnson, A.E. (1998). BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell 92, 747-758. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Zhang, Y., and Ron, D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271-274. [DOI] [PubMed] [Google Scholar]
- Horiuchi, H., Ashikari, T., Amachi, T., Yoshizumi, H., Takagi, M., and Yano, K. (1990). High-level secretion of a Rhizopus niveus aspartic proteinase in Saccharomyces cerevisiae. Agric. Biol. Chem. 54, 1771-1779. [PubMed] [Google Scholar]
- Iwawaki, T., Hosoda, A., Okuda, T., Kamigori, Y., Nomura-Furuwatari, C., Kimata, Y., Tsuru, A., and Kohno, K. (2001). Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat. Cell Biol. 3, 158-164. [DOI] [PubMed] [Google Scholar]
- Kaiser, C., Michaelis, S., and Mitchell, A. (1994). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Kohno, K., Normington, K., Sambrook, J., Gething, M.J., and Mori, K. (1993). The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 13, 877-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Tirasophon, W., Shen, X., Michalak, M., Prywes, R., Okada, T., Yoshida, H., Mori, K., and Kaufman, R.J. (2002). IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16, 452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.Y., Schroder, M., and Kaufman, R.J. (2000). Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 275, 24881-24885. [DOI] [PubMed] [Google Scholar]
- Liu, C.Y., Wong, H.N., Schauerte, J.A., and Kaufman, R.J. (2002). The Protein kinase/endoribonuclease IRE1alpha that signals the unfolded protein response has a luminal N-terminal ligand-independent dimerization domain. J. Biol. Chem. 277, 18346-18356. [DOI] [PubMed] [Google Scholar]
- Matlack, K.E., Misselwitz, B., Plath, K., and Rapoport, T.A. (1999). BiP acts as a molecular ratchet during posttranslational transport of pre-pro-alpha factor across the ER membrane. Cell 97, 553-564. [DOI] [PubMed] [Google Scholar]
- Mori, K., Ma, W., Gething, M.J., and Sambrook, J. (1993). A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74, 743-756. [DOI] [PubMed] [Google Scholar]
- Mori, K., Ogawa, N., Kawahara, T., Yanagi, H., and Yura, T. (2000). mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 97, 4660-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, K., Sant, A., Kohno, K., Normington, K., Gething, M.J., and Sambrook, J.F. (1992). A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 11, 2583-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, R.I. (1997). Transcriptional regulation of eukaryotic heat shock genes. In: Molecular Chaperones and Protein-Folding Catalysts, ed. M.J. Gething, New York: Oxford University Press, 534-541.
- Nishikawa, S.I., Fewell, S.W., Kato, Y., Brodsky, J.L., and Endo, T. (2001). Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153, 1061-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normington, K., Kohno, K., Kozutsumi, Y., Gething, M.J., and Sambrook, J. (1989). S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell 57, 1223-1236. [DOI] [PubMed] [Google Scholar]
- Oka, M., Kimata, Y., Mori, K., and Kohno, K. (1997). Saccharomyces cerevisiae KAR2 (BiP) gene expression is induced by loss of cytosolic HSP70/Ssa1p through a heat shock element-mediated pathway. J. Biochem. (Tokyo) 121, 578-584. [DOI] [PubMed] [Google Scholar]
- Okamura, K., Kimata, Y., Higashio, H., Tsuru, A., and Kohno, K. (2000). Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 279, 445-450. [DOI] [PubMed] [Google Scholar]
- Plemper, R.K., Bohmler, S., Bordallo, J., Sommer, T., and Wolf, D.H. (1997). Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388, 891-895. [DOI] [PubMed] [Google Scholar]
- Polaina, J., and Conde, J. (1982). Genes involved in the control of nuclear fusion during the sexual cycle of Saccharomyces cerevisiae. Mol. Gen. Genet. 186, 253-258. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., Misra, L.M., and Vogel, J.P. (1989). KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57, 1211-1221. [DOI] [PubMed] [Google Scholar]
- Ruegsegger, U., Leber, J.H., and Walter, P. (2001). Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107, 103-114. [DOI] [PubMed] [Google Scholar]
- Sanders, S.L., Whitfield, K.M., Vogel, J.P., Rose, M.D., and Schekman, R.W. (1992). Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell 69, 353-365. [DOI] [PubMed] [Google Scholar]
- Scidmore, M.A., Okamura, H.H., and Rose, M.D. (1993). Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol. Biol. Cell 4, 1145-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu, C.E., and Walter, P. (1996). Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 15, 3028-3039. [PMC free article] [PubMed] [Google Scholar]
- Shen, J., Chen, X., Hendershot, L., and Prywes, R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99-111. [DOI] [PubMed] [Google Scholar]
- Shen, X. et al. (2001). Complementary signalling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107, 893-903. [DOI] [PubMed] [Google Scholar]
- Sidrauski, C., and Walter, P. (1997). The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90, 1031-1039. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, J.F., Ferro-Novick, S., Rose, M.D., and Helenius, A. (1995). BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J. Cell Biol. 130, 41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuki, A., Kohno, K., and Tamura, G. (1975). Inhibition of biosynthesis of polyisoprenol sugars in chick embryo microsomes by tunicamycin. Agric. Biol. Chem. 39, 2089-2091. [Google Scholar]
- te Heesen, S., and Aebi, M. (1994). The genetic interaction of kar2 and wbp1 mutations. Distinct functions of binding protein BiP and N-linked glycosylation in the processing pathway of secreted proteins in Saccharomyces cerevisiae. Eur. J. Biochem. 222, 631-637. [DOI] [PubMed] [Google Scholar]
- Tirasophon, W., Welihinda, A.A., and Kaufman, R.J. (1998). A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12, 1812-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga, M., Kawamura, A., and Kohno, K. (1992). Purification and characterization of BiP/Kar2 protein from Saccharomyces cerevisiae. J. Biol. Chem. 267, 17553-17559. [PubMed] [Google Scholar]
- Umebayashi, K., Hirata, A., Fukuda, R., Horiuchi, H., Ohta, A., and Takagi, M. (1997). Accumulation of misfolded protein aggregates leads to the formation of russell body-like dilated endoplasmic reticulum in yeast. Yeast 13, 1009-1020. [DOI] [PubMed] [Google Scholar]
- Umebayashi, K., Hirata, A., Horiuchi, H., Ohta, A., and Takagi, M. (1999). Unfolded protein response-induced BiP/Kar2p production protects cell growth against accumulation of misfolded protein aggregates in the yeast endoplasmic reticulum. Eur. J. Cell Biol. 78, 726-738. [DOI] [PubMed] [Google Scholar]
- Vogel, J.P., Misra, L.M., and Rose, M.D. (1990). Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J. Cell Biol. 110, 1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.Z., Harding, H.P., Zhang, Y., Jolicoeur, E.M., Kuroda, M., and Ron, D. (1998). Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 17, 5708-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda, A.A., and Kaufman, R.J. (1996). The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J. Biol. Chem. 271, 18181-18187. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., and Mori, K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881-891. [DOI] [PubMed] [Google Scholar]
- Ziegelhoffer, T., and Craig, E.A. (1997). Saccharomyces cerevisiae Ssa proteins. in Molecular Chaperones and Protein-Folding Catalysts, ed. M.J. Gething, New York: Oxford University Press, 26-28.