Abstract
Fumonisins are a group of mycotoxins that contaminate maize and cause leukoencephalomalacia in equine, pulmonary edema in swine, and promote cancer in mice. Fumonisin biosynthesis in Fusarium verticillioides is repressed by nitrogen and alkaline pH. We cloned a PACC-like gene (PAC1) from F. verticillioides. PACC genes encode the major transcriptional regulators of several pH-responsive pathways in other filamentous fungi. In Northern blot analyses, a PAC1 probe hybridized to a 2.2-kb transcript present in F. verticillioides grown at alkaline pH. A mutant of F. verticillioides with a disrupted PAC1 gene had severely impaired growth at alkaline pH. The mutant produced more fumonisin than the wild type when grown on maize kernels and in a synthetic medium buffered at an acidic pH, 4.5. The mutant, but not the wild type, also produced fumonisin B1 when mycelia were resuspended in medium buffered at an alkaline pH, 8.4. Transcription of FUM1, a gene involved in fumonisin biosynthesis, was correlated with fumonisin production. We conclude that PAC1 is required for growth at alkaline pH and that Pac1 may have a role as a repressor of fumonisin biosynthesis under alkaline conditions.
Fusarium verticillioides (Sacc.) Nirenberg (teleomorph: Gibberella moniliformis Wineland) produces fumonisins, a group of mycotoxins structurally similar to sphinganine that can disrupt sphingolipid metabolism by inhibiting ceramide synthase (36). Fumonisins are produced in maize kernels infected with F. verticillioides prior to harvest and are stable through grain processing (2). Of the many structural forms of fumonisins, the most abundant in nature and the most studied is fumonisin B1 (FB1) (21). Concerns about the toxicity of fumonisins have led to the issuance by the Food and Drug Administration of recommendations for allowable levels of fumonisins in food and feed (22).
Important areas of current research include determining the biochemical pathway for fumonisin biosynthesis and understanding how fumonisin biosynthesis is regulated. Traditional biochemical studies found that fumonisins are polyketide secondary metabolites (10). FUM1 (previously designated FUM5), the gene encoding a polyketide synthase involved at an early step in the formation of fumonisins, and a cluster of 14 genes adjacent to FUM1 are coordinately expressed during fumonisin biosynthesis (25). In silico analysis of the nucleotide sequences has allowed predictions of enzymatic function in the fumonisin pathway (25). The disruption of FUM1, FUM6, or FUM8 blocked fumonisin production (24, 29).
None of the genes in the fumonisin biosynthetic gene cluster has a clear regulatory function. Keller and Sullivan (12) found that glucose and phosphate do not repress FB1 biosynthesis, while nitrogen limitation and acidic pH (3.0 to 4.0) enhance FB1 biosynthesis. Nitrogen metabolite repression also affects FB1 biosynthesis (30).
FCC1 also affects the regulation of fumonisin biosynthesis (31). The deduced product of FCC1 is similar to C-type cyclins, a class of proteins involved in the transcriptional activation or repression of genes associated with stress responses and development (4, 14, 15). F. verticillioides strains with a disrupted FCC1 (Δfcc1) do not produce FB1 on maize kernels (31). The inability to produce FB1 could be reversed in the Δfcc1 strain by growing the mutant on a defined medium with low pH. The mechanism by which acidic pH restores the ability of the mutant to produce FB1 is unknown.
The pH-regulatory system in filamentous fungi is best described in Aspergillus nidulans, in which PacC is a transcriptional activator of alkaline-expressed genes and a repressor of acid-expressed genes. PACC homologues also have been described in several other filamentous fungi, including Aspergillus niger, Penicillium chrysogenum, Acremonium chrysogenum, and Sclerotinia sclerotiorum (16, 27, 28, 32). There are three phenotypic classes of PACC mutants. PACC mutants that constitutively express an active PacC protein have the same phenotype as the wild type grown under alkaline conditions regardless of the external pH by activating alkaline-expressed genes. Mutants in which the PacC protein is not processed to its active form have the same phenotype as the wild type grown under acidic conditions (23). PACC-null mutants have acidic-growth phenotypes that resemble those of mutants in the PAL genes, a group of six genes that relay the signal to process the PacC protein in response to alkaline conditions (6, 7, 34). Acid-expressed genes are repressed in response to alkaline conditions when PacC binds and blocks the recognition site of a transcriptional activator, thereby preventing transcription (6).
The objective of this study was to isolate and characterize PAC1 from F. verticillioides. Based on our previous observation that ambient pH has a dominant effect on the Δfcc1 mutant in F. verticillioides, we hypothesized that PAC1 has a regulatory role in FB1 biosynthesis. Here we show that PAC1 has high homology to PACC-like genes in other filamentous fungi, that the gene is required for growth at alkaline pH, and that the gene product may repress fumonisin biosynthesis under alkaline conditions.
MATERIALS AND METHODS
Strains and media.
F. verticillioides strain 7600 (Fungal Genetics Stock Center, University of Kansas Medical School, Kansas City) was stored in 20% glycerol at −80°C. For the inoculum, the fungus was grown on potato dextrose agar (Difco Laboratories, Detroit, Mich.) at 28°C. For isolation of genomic DNA, the fungus was grown in stationary YEPD medium (0.5% yeast extract, 1% peptone, 2% glucose) at 28°C. For comparative growth and conidiation measurements, 5-mm-diameter mycelial plugs were placed at the center of plastic petri dishes (100 by 15 mm) containing solid medium consisting of 30 g of sucrose per liter, 1 g of KH2PO4 per liter, 14.0 g of NaPO4 per liter (pH 4.5, 7.0, and 8.4), 10 mg of FeSO4 per liter, 15 g of agar per liter with nitrogen sources of 8.5 g of NaNO3 per liter, 10.3 g of γ-amino-n-butyric acid (GABA) per liter, or 7.5 g of glycine per liter. After 7 days of growth, the colony diameter was recorded, and then each plate was flooded with 5 ml of 0.01% Triton X-100 to harvest conidia. Conidial production was quantified with a hemacytometer. Fungi were grown on cracked maize kernels and BSAL medium (40 g of sucrose per liter, 3.0 g of KH2PO4 per liter, 0.5 g of MgSO4 per liter, 5 g of NaCl per liter, 1.0 g of bovine serum albumin per liter) for FB1 analysis and RNA isolation (31). Resuspension experiments were conducted by inoculating 105 conidia of a fungal strain into plastic petri dishes (100 by 15 mm) containing 12 ml of YEPD medium. After 3 days of incubation at 28°C, mycelia were collected and washed, and 1 g was placed in 250-ml flasks containing 50 ml of DL medium (30) buffered at pH 4.5 or 8.4 with 0.1 M NaPO4 and with 2 mM NH4PO4 as the nitrogen source. Resuspended cultures were shaken at 150 rpm at 25°C, and samples were collected at 0, 24, 36, and 48 h post-resuspension. From each sample, 1 ml of the culture medium was analyzed for FB1, and the mycelium was stored at −80°C for RNA isolation. All experiments were repeated three times with the same results.
Nucleic acid isolation and analysis.
Bacterial plasmids were isolated by an alkaline lysis method with the Qiagen Miniprep DNA purification system (Qiagen, Valencia, Calif.). Fungal genomic DNA was isolated as previously described (37). For Southern analysis, F. verticillioides genomic DNA was digested with HindIII, size fractionated on a 0.7% agarose gel, and transferred to a nylon membrane (Nytran; Schleicher and Schuell, Keene, N.H.) by standard procedures (17). For Northern analysis, total RNA was extracted by an acid-phenol extraction procedure (5). Northern blot analysis was performed on total RNA extracted from the wild type grown in DL medium at pH 3.0 or 8.0 for 7 days. Ten micrograms of RNA was separated by electrophoresis in a formaldehyde-denaturing gel and transferred to a nylon membrane (9). The F. verticilliodes β-tubulin gene TUB2 (GenBank accession no. U27303) (38) was labeled and hybridized as a loading control. The probes used in all hybridization experiments were radiolabeled with 32P by using the Prime-It II random prime labeling kit (Amersham Biosciences, Arlington Heights, Ill.). High-stringency hybridization was performed in a mixture containing 7% sodium dodecyl sulfate (SDS), 0.5 M sodium phosphate (pH 7.5), and 10 mM EDTA (3) followed by two 30-min washes, the first at room temperature in a solution of 2× SSC (0.3 M NaCl, 30 mM sodium citrate) and 0.5% SDS, and the second at 65°C in 0.2× SSC and 0.5% SDS (17). Blots were exposed to a phosphorimaging screen and scanned with a Typhoon 9200 high-performance gel blot reader (Molecular Dynamics, Inc., Amersham Biosciences).
Total RNA for real-time PCR analysis was extracted with Trizol reagent (Invitrogen, Carlsbad, Calif.) from mycelia collected during the resuspension experiment. RNA was treated with DNase (Promega, Madison, Wis.) for 30 min at 37°C to remove contaminating genomic DNA. One microgram of total RNA was used in the first-strand cDNA synthesis reaction with Superscript II RNase H− reverse transcriptase (Invitrogen, Carlsbad, Calif.) in a 30-μl reaction volume incubated at 42°C for 2 h. Control reactions were performed without the addition of reverse transcriptase.
Real-time PCR primers and fluorogenic probes were designed from the FUM1 (FUM5 GenBank accession no. AF155773) and TUB2 genes of F. verticillioides. Probes were labeled at the 3′ end with the quencher dye TAMRA (5-carboxytetramethylrhodamine) and at the 5′ end with either TET (6-carboxy-2′,4,7,7′-tetrachlorfluorescein) or FAM (6-carboxyfluorescein) as the reporter dye. The primer pairs and probe for FUM1 were 5′-ACACCAAAGCCTCTACAGGTGA-3′ as the forward primer, 5′-AGGTATCGGGCACCGCT-3′ as the reverse primer, and 5′-[TET]TCGCAGCAGCACGGCCGAGTCCACC[TAMRA]-3′ as the probe. The primer pairs and probe for TUB2 were 5′-TGCTCATTTCCAAGATCCGCG-3′ as the forward primer, 5′-GTAGTTGAGGTCACCGTAGGAGG-3′ as the reverse primer, and 5′-[6-FAM]CCACCCTCTCCGTCCACCAGCTGGTC[TAMRA]-3′ as the probe. All PCR primers and the FUM1 probe were obtained from IDT (Coralville, Iowa); the TUB2 probe was obtained from Qiagen Operon (Valencia, Calif.).
Real-time PCRs consisted of 10 μl of Qiagen Quantitect PCR master mix, 5 μl of single-stranded cDNA template, 500 nM forward primer, 500 nM reverse primer, and 100 nM of fluorogenic probe in a final volume of 20 μl. Additionally, control reactions were performed with no template and RNA template without reverse transcriptase. The cycling conditions were 2 min at 50°C (1 cycle), 10 min at 95°C (1 cycle), and 15 s at 95°C followed by 1 min at 60°C (40 cycles). Reactions were performed in 96-well optical reaction plates (Applied Biosystems, Foster City, Calif.) sealed with optical adhesive covers (Applied Biosystems). Real-time PCR and data analysis were performed with the ABI 7700 sequence detection system with Sequence Detection software version 1.7 (Applied Biosystems).
The expression level of FUM1 was normalized to the expression of TUB2 by the comparative cycle threshold (Ct) method (separate tubes) (ABI User Bulletin 2; Applied Biosystems). TUB2 was chosen as an expression standard because microarray analyses and Northern data indicate that it is constitutively expressed under a variety of culture conditions (38) (see Fig. 1). Before data analysis, the relative efficiencies of the FUM1 and TUB2 primers were assessed as described in the ABI User Bulletin 2 and were found to be approximately equal. For each sample, average Ct values were calculated from three replicate PCRs for both TUB2 and FUM1. ΔCt values were calculated by subtracting the average Ct values of TUB2 from those of FUM1. ΔΔCt values were obtained by subtracting the value for ΔCtwild-type pH 8.4 from all other ΔCt values. Levels of FUM1 expression were calculated as relative expression = 2−ΔΔCt with the range determined by 2−ΔΔCt + s and 2−ΔΔCt − s, where s = the standard deviation of the ΔΔCt value.
FIG. 1.
Northern blot analysis of the wild type cultured at pHs 3.0 and 8.0. Total RNA (10 μg) isolated from the wild-type strain cultured for 7 days at pHs 3.0 (lane 1) and 8.0 (lane 2) was separated by electrophoresis in 1.2% agarose-formaldehyde gels, transferred to a nylon membrane, and hybridized with a 32P-labeled PAC1-specific probe (A) and 32P-labeled β-tubulin-specific probe (B).
Isolation of PAC1.
Degenerate PCR primers were designed based on highly conserved regions of amino acids within the coding regions of PacC homologues from five filamentous fungi. Included were PacC from A. chrysogenum (GenBank accession no. AJ251521), PacC from A. nidulans (GenBank accession no. Z47081), Pac1 from S. sclerotiorum (GenBank accession no. AY005467), PacC from P. chrysogenum (GenBank accession no. U44726), and PacC from A. niger (GenBank accession no. X98417). A 960-bp product was obtained from F. verticillioides genomic DNA (100 ng) by PCR amplification with 20 pmol each of the forward primer 5′-TTCAAGCGYCCHCARGAYYTSAAGAARCATG-3′ and reverse primer 5′-GMRCTGGAGGGHGGBGT-3′ and the following reaction conditions: 2 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 52°C, and 1 min at 72°C. The PCR product was subcloned into a pGEM-T easy vector (Promega, Valencia, Calif.) to yield a plasmid designated as pPAC-TA. Subsequently, a 45-kb cosmid clone, designated pPACcos1, was identified by screening a genomic library of F. verticillioides (24). Standard protocols (17) were used to plate and screen the library with 32P-labeled insert from pPAC-TA.
Nucleotide sequence analysis.
Nucleic acid sequence was obtained from plasmids pPAC-TA and pPACsub, a 2.9-kb PstI fragment subcloned from the cosmid pPACcos1. The remainder of PAC1 sequence was obtained directly from cosmid pPACcos1. Bidirectional sequencing was performed by the Plant-Microbe Genomics Facility, Ohio State University, Columbus.
To obtain cDNA clones of PAC1, total RNA was extracted from strain 7600 grown in DL medium at pH 8.0 for 7 days and used to synthesize cDNAs. Regions of the PAC1 cDNAs were amplified with PCR primers pac5c1 (5′-CTCCTTCGGCTCCAGAG-3′), pac5c2 (5′-CAACAACAACAACAGCAGG-3′), and pac3c1 (5′-ACTGCGTTAGTTCCATCTG-3′) that yielded two products (1.0 and 1.2 kb) spanning segments of the open reading frame that were interrupted by introns. The reaction conditions were the same as those described for the isolation of PAC1. The PCR products were subcloned into pGEM T-easy vector and sequenced by the Purdue Genomics Core Facility (West Lafayette, Ind.). All DNA sequences were analyzed, and predicted amino acid sequences were deduced with MacDNASIS software (Hitachi Software Engineering America, Ltd., San Bruno, Calif.). Homology searches were conducted by using the BLAST algorithm (1). The PSORTII program was used to analyze the deduced Pac1 peptide sequence for subcellular localization predictions (20). Multiple alignments were conducted by using ClustalW software (http://www.ch.embnet.org/software/ClustalW.html) (33).
Disruption of PAC1.
The PAC1-disruption vector (pWE70) was constructed by inserting a 1.4-kb HpaI fragment containing a hygromycin resistance gene cassette from pCB1003 (Fungal Genetics Stock Center) into the Eco47III site of the PAC1 DNA fragment in pPAC-TA. The pWE70 insert was amplified by PCR with primers pacV5 (5′-ATCCCAGGATCCTCAAG-3′) and pacV3 (5′-TGTTGCGGTAACCATTG-3′). The PCR conditions were as described for the isolation of PAC1. The 2.3-kb product was gel purified and used for fungal transformation. Protoplasts of F. verticillioides strain 7600 were produced and transformed as described by Proctor et al. (24). Transformants were selected on regeneration medium containing 60 μg of hygromycin B per ml (Roche Molecular Biochemicals, Indianapolis, Ind.) (24). Transformants were screened by PCR with primers that distinguished homologous crossover events. The primer set pacD5 (5′-GGACTCTGTACTTGTTCG-3′) and h3P (5′-CGATAGTGGAAACCGACG-3′) produced a 450-bp DNA product from the homologous crossover at the 5′ end of the insertion DNA, and primer set pacD3 (5′-GACCTGTGAGAGGTAG-3′) and h5P (5′-GATCAGAAACTTCTCGACAG-3′) produced a 950-bp product from the 3′ end. The conditions for PCR amplification were the same as those described for the isolation of PAC1, with the exception of the annealing temperature (54°C).
One transformant (PAC2A) out of 24 analyzed contained a disrupted PAC1 gene and was complemented with the cosmid pPACcos1 and pPAC2.7, a subclone containing a 2.7-kb DNA fragment that spans PAC1 from 500 bp upstream of the ATG to 200 bp downstream of the translational stop codon. The DNA fragment was amplified from pPACcos1 in a PCR containing a high-fidelity polymerase (Pwo; Roche Molecular Biochemicals) with forward primer pacA (5′-GTCAGTACCTCTGATTCTTG-3′) and reverse primer pacB (5′-AATCCGACTCAGGTCCATG-3′). The amplification conditions were as follows: 2 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 54°C, and 2 min at 72°C. The amplified DNA fragment was subcloned into pGEM T-easy vector. Two micrograms of pPAC2.7 or pPACcos1 was coincubated with 1 μg of plasmid pSM334, which contains the Geneticin resistance gene (neomycin phosphotransferase) (18) from E. coli under the transcriptional control of a GPDA promoter from Cochliobolus heterostrophus (35). Geneticin-resistant transformants were selected on regeneration medium containing 75 μg of Geneticin per ml (Sigma, St. Louis, Mo.). Analysis of the transformants with primer sets pacA and pacB and pacD3 and pacD5 identified 1 of 25 transformants with pPACcos1 and 2 of 20 transformants with pPAC2.7.
Fumonisin analysis.
Fumonisins were extracted from cultures, and concentrations of FB1 were determined by high-pressure liquid chromatography as previously described (30). Analysis of variance (ANOVA) was conducted with SuperANOVA software v.1.11 (SAS, Inc., Cary, N.C.), and means were separated by Fisher's protected least significant difference.
Nucleotide sequence accession number.
The nucleic acid sequence and predicted amino acid sequences of PAC1 were submitted to GenBank (accession no. AY216461).
RESULTS
Isolation and characterization of a PacC homolog.
Alignment of the amino acid sequences of available PacC homologues indicated highly conserved areas in the N-terminal zinc finger region and less-conserved areas across the rest of the peptides. Degenerate PCR primers designed from amino acids near the zinc finger region and in a serine-threonine-rich region were used to amplify a single band of 960-bp from genomic DNA of F. verticillioides strain 7600. Nucleic acid sequence analysis of the PCR fragment indicated a single open reading frame with similarity to other PacC homologues ranging from 38 to 69%. The DNA fragment was used to screen a cosmid genomic library from which a single 45-kb cosmid, pPACcos1, was isolated.
PAC1 was identified in a 3.4-kb region of pPACcos1. The genomic sequence was compared with sequences from three cDNA clones, and three introns were identified. The third intron sequence in one cDNA clone remained unprocessed. Both transcripts were detectable in the fungus grown in medium at pH 8.4 by conventional reverse transcription-PCR (RT-PCR) techniques (data not shown). Removal of the three introns results in an uninterrupted open reading frame of 1,836 nucleotides that encodes a predicted polypeptide of 612 amino acids. If the third intron is not removed, the reading frame is not shifted, and the resulting protein contains an additional 25 amino acid residues. Three PacC consensus binding sequences (5′-GCCARG-3′) were identified within 500 bp upstream of the predicted PAC1 translational start codon. This motif is found in promoters of genes regulated by PacC, including PACC (34).
The deduced translation product of PAC1 has 38 and 69% amino acid identity to the PacC homologues in A. nidulans and A. chrysogenum, respectively. Pac1 shares the highest identity to other PacC homologues in the region comprising three Cys2His2 zinc finger domains. The most similar homologue is PacC from A. chrysogenum (96%), and the least similar is Pac1 from S. sclerotiorum (85%). Pac1 from F. verticillioides also contains the peptide sequence KRTYDMVDDFFGSAKRR beginning at amino acid position 244, which fits the pattern of a bipartite nuclear localization signal (NLS) (26). This peptide sequence aligns with a putative bipartite NLS in other PacC homologues.
Expression of PAC1.
Northern analysis of total RNA isolated from cultures grown for 7 days in defined medium, adjusted to pH 8.0, revealed a 2.2-kb band of hybridization with the PAC1 probe (Fig. 1). No hybridization signal was detected from cultures grown in defined medium at pH 3.
Disruption of PAC1.
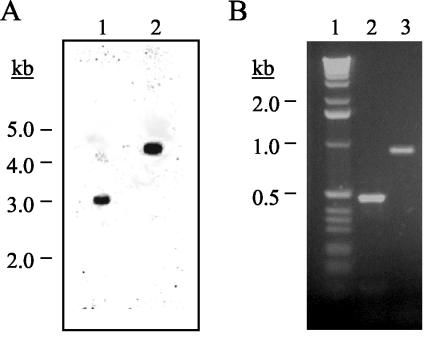
PAC1 was disrupted in F. verticillioides by homologous recombination. Of the 25 hygromycin-resistant colonies resulting from the transformation of strain 7600 with the disruption vector, one contained a disrupted PAC1. The disrupted strain was designated PAC2A. Southern blot analysis of wild-type and PAC2A genomic DNA indicated a 1.4-kb increase in the size of the single band hybridizing to the PAC1 probe (Fig. 2A). The increase in size resulted from the insertion of the hygromycin cassette (1.4 kb) into PAC1. PCR products generated from the ends of the hygromycin cassette indicated that the disruption occurred by a double homologous crossover event (Fig. 2B).
FIG. 2.
Analysis of the PAC1 disruption strain. (A) Southern analysis. Genomic DNA (2 μg) from the wild-type strain (lane 1) and strain PAC2A (PAC1 disruption strain) (lane 2) was digested with HindIII, separated by electrophoresis in a 0.7% agarose gel, transferred to a nylon membrane, and probed with a 32P-labeled DNA fragment of PAC1. The molecular size standards (kilobases) are indicated. (B) PCR Analysis. PAC2A genomic DNA was amplified with primer pairs pacD5:h3p (lane 2) and pacD3:h5p (lane 3). The molecular size standards (kilobases) are shown in lane 1. The DNA was separated by electrophoresis in a 1% agarose gel. The gel was stained with ethidium bromide and photographed under UV illumination.
Radial growth of PAC2A on agar media buffered at pHs of 4.5, 7.0, and 8.4 was compared to that of the wild-type strain, 7600, and PAC2A-C, a derivative of PAC2A complemented with the wild-type PAC1 gene. The growth rates of all three strains were similar at pHs 4.5 and 7.0 (Table 1). PAC2A did not grow at pH 8.4, but the wild-type and PAC2A-C strains grew with equal vigor. Similar results were obtained when the nitrogen source of the media was GABA or glycine (data not shown).
TABLE 1.
Effect of pH on radial growth and conidiation of the wild-type strain, PAC2A, and PAC2A-Ca
| Strain | Growth (cm) at pHb:
|
Conidiation (105 conidia/ml) at pHc:
|
||||
|---|---|---|---|---|---|---|
| 4.5 | 7.0 | 8.4 | 4.5 | 7.0 | 8.4 | |
| Wild Type | 5.4 ± 0.3 | 5.0 ± 0.1 | 3.7 ± 0.3 | 27.6 ± 1.5 | 19.8 ± 2.2 | 12.0 ± 4.0 |
| PAC2A | 5.4 ± 0.3 | 5.0 ± 0.1 | NDd | 6.0 ± 2.1 | 0.3 ± 0.1 | ND |
| PAC2A-C | 5.4 ± 0.3 | 5.0 ± 0.1 | 4.6 ± 0.2 | 29.7 ± 1.5 | 12.0 ± 3.5 | 15.3 ± 4.2 |
Mycelial plugs 5-mm diameter were placed at the center of plastic petri dishes containing solid medium containing NaNO3 as a nitrogen source and incubated for 7 days at 28°C.
Mean colony diameter (in centimeters) of three replicates ± standard error.
Mean number of conidia (105) per milliliter ± standard error for three replicates.
ND, none detected.
Disruption of PAC1 also affected conidiation (Table 1). When grown in medium buffered at pH 4.5, PAC2A produced 77% less conidia than the wild type and PAC2A-C. At pH 7, the mutant produced 95% less conidia. Similar results were observed on media containing other nitrogen sources (data not shown).
Impact of PAC1 on fumonisin biosynthesis.
FB1 production by PAC2A was compared to that by the wild type and PAC2A-C after 14 days of growth on cracked maize kernels. PAC2A consistently produced more FB1 than either strain 7600 or PAC2A-C (Table 2). The mutant also produced higher levels of FB1 than the other strains when grown on DL medium buffered at pH 4.5 (Table 2).
TABLE 2.
Comparison of FB1 production by the wild-type, PAC2A, and PAC2A-C strainsa
| Strain | FB1 production onb:
|
|
|---|---|---|
| Maize kernels (μg of FB1/g of maize)b | DL medium (μg of FB1/ml) | |
| Wild type | 480 ± 20 | 120 ± 17 |
| PAC2A | 650 ± 50* | 160 ± 20* |
| PAC2A-C | 475 ± 29 | 120 ± 20 |
FB1 was extracted from cultures grown for 14 days on maize kernels and DL medium (pH 4.5).
Values represent the mean of three replicates ± standard error. *, significantly different from other values in the same column (P < 0.04, Fisher's protected least significant difference).
When mycelia were resuspended in medium buffered at pH 4.5, FB1 was detected after 48 h in cultures of the wild type, PAC2A, and PAC2A-C (Table 3). When resuspended in medium buffered at pH 8.4, only PAC2A produced FB1. The level of expression of FUM1 was measured by real-time PCR after 0, 24, 36, and 48 h of incubation in the resuspension medium and normalized to TUB2 expression. The transcript of FUM1 at 36 h was lowest in wild-type cultures at pH 8.4 and was ∼10-fold higher at pH 4.5 (Table 3). In contrast, FUM1 transcription in PAC2A at pH 8.4 was ∼47-fold higher than the wild type at pH 8.4, and at pH 4.5, the level of transcription was ∼135-fold higher than that in the wild type at pH 8.4. FUM1 expression in the PAC1-complemented strain was similar to that in the wild-type strain at both pHs 4.5 and 8.4. These data are consistent with the hypothesis that PAC1 is sufficient to repress fumonisin biosynthesis at alkaline pH.
TABLE 3.
Comparison of FB1 production and relative FUM1 expression by wild-type, PAC2A, and PAC2A-C strains
| Strain | FB1 production (μg of FB1/ml) and FUM1 expression (relative to wild type) at:
|
|||
|---|---|---|---|---|
| pH 4.5
|
pH 8.4
|
|||
| FB1a | FUM1b | FB1a | FUM1b | |
| Wild type | 20 ± 0 | 10 (8.0-13) | NDc | 1.0 (0.9-1.1) |
| PAC2A | 25 ± 2 | 140 (120-150) | 10 ± 2 | 48 (43-52) |
| PAC2A-C | 20 ± 2 | 39 (37-43) | ND | 4.3 (3.9-4.7) |
Data are presented for results obtained 48 h after resuspension. Values are means of three replicates ± standard error.
FUM1 expression was evaluated by real-time PCR 36 h after resuspension, normalized to TUB2 expression, and quantified relative to wild-type FUM1 expression at pH 8.4. FUM1 expression data represent the results of a single experiment (repeated three times with similar results). FUM1 expression was calculated as 2−ΔΔCt, with the range (in parentheses) calculated as 2−ΔΔCt+ s− 2−ΔΔCt−s, where s = the standard deviation of the ΔΔCt value.
ND, none detected.
DISCUSSION
Prior to this study, five PACC homologues had been described for filamentous fungi (16, 27, 28, 32, 34). The PACC genes from the industrially important fungi A. niger and P. chrysogenum and from the phytopathogenic fungus S. sclerotiorum are functional homologues of the A. nidulans PACC (16, 27, 32). The PACC homologues from these filamentous fungi have the highest amino acid similarity over the protein's zinc finger DNA binding domain. At the nucleotide level, this region (∼150 bp) also has the highest nucleotide sequence identity (>80%). Sequence analysis of PAC1 from F. verticillioides indicated a predicted translation product of 612 amino acids (splice variant I, with all three introns processed) or 637 (splice variant II, with only the first two introns processed). Pac1 has total amino acid identity to the other PacC homologues ranging from 38 to 69%. The protein has three zinc finger motifs, with the order and spacing conserved relative to the other PacC homologues. The three introns in PAC1 are similar in size and position to those in PACC of A. chrysogenum and PAC1 of S. sclerotiorum (27, 28). The data also indicated that a transcript is made from PAC1 in which the third intron is unprocessed. This splice variant resembles PACC genes from A. nidulans and A. niger, in which only two introns are processed. These similarities all suggest that PAC1 encodes a PacC homologue.
The PAC1 disruptant contains an insertion at amino acid position 235 that is immediately downstream of the zinc finger motifs but upstream of the putative NLS. It is possible that a stable truncated PAC1 transcript is produced in this mutant and that a protein containing the N-terminal 235 amino acids is produced as well. Similar mutants have been studied in A. nidulans that express truncated PacC proteins (238 and 310 N-terminal amino acids). These mutants had phenotypes identical to that of the acid-mimicking null mutation (19). Based on sequence similarity between PacC and Pac1, it is unlikely that a truncated Pac1 protein produced by the PAC1-disrupted strain of F. verticillioides would give rise to a gain-of-function, alkaline-mimicking phenotype. However, in contrast to the PACC-null mutation in A. nidulans, no growth defects were exhibited by the PAC1-disrupted strain when cultured under neutral or acidic pH conditions, although the mutant did not grow at alkaline pH.
The level of conidiation by the mutant at neutral or acidic pH was lower, relative to that of the wild type, with greatest inhibition at neutral pH. This observation suggests that expression of some (or all) alkaline-expressed genes and repression of acid-expressed genes decreases at neutral to alkaline pH leading to growth inhibition. In A. nidulans, PacC participates in the regulation of the acid-expressed GABA gene, encoding GABA permease, suggesting that the PacC gain-of-function mutant fails to utilize GABA in the growth medium (6). We compared radial growth of wild type F. verticillioides, PAC2A, and PAC2A-C on media containing GABA, glycine or nitrate as the sole nitrogen source. Growth and conidiation were the same in all three strains, indicating that the PAC1-disrupted strain could use these nitrogen sources. Additional experiments are necessary to determine if Pac1 is involved in GABA gene regulation in F. verticillioides. Furthermore, growth of the PAC1-disrupted strain was the same as that of the wild type when cultured on media containing various amounts of glycerol or salt (data not shown), indicating that osmoregulation in the PAC1-disrupted strain was not impaired.
PacC impacts the production of secondary metabolites by filamentous fungi. For example, in the β-lactam antibiotic-producing fungi, PacC is a positive regulator of genes involved in penicillin and cephalosporin production (8, 28). The PacC recognition sequence, 5′-GCCARG-3′, is found multiple times in the promoters of the structural genes ACVA and IPNA of A. nidulans, P. chrysogenum, and A. chrysogenum and CEFEF and CEFG of A. chrysogenum. Recombinant PacC from A. chrysogenum can competitively bind to these promoters in vitro (28). Six FUM genes in the fumonisin gene cluster (25) contain at least one copy of the PacC recognition sequence within 600 bp upstream of the translational start codon.
Alkaline growth conditions and PacC gain-of-function mutations (PACCc) increase penicillin production in A. nidulans and P. chrysogenum (8, 32) and repress the accumulation of sterigmatocystin, an aflatoxin precursor in A. nidulans (11). Aflatoxin production by Aspergillus flavus and Aspergillus parasiticus is inversely correlated with pH of the growth medium. Fumonisins also are produced preferentially at acidic pH (12, 13, 31). In this study, growth of the PAC1-disruption mutant was completely inhibited at pH 8.4; however, FB1 production and expression of FUM1 were detectable when the mutant was resuspended in liquid medium buffered at pH 8.4. In contrast, the wild-type and complemented (PAC2A-C) strains did not produce FB1 at pH 8.4, and levels of expression of FUM1 in these strains were ∼45- and ∼10-fold less, respectively, than that in PAC2A. These results suggest that PAC1 is a negative regulator of fumonisin biosynthesis.
In this study, we have shown that PAC1 in F. verticillioides is required for normal growth at alkaline pH but not for growth and fumonisin biosynthesis at acidic pH. Our data are consistent with the hypothesis that Pac1 represses fumonisin biosynthesis by the fungus under alkaline conditions. Based on elevated expression of a fumonisin pathway gene and increased production of FB1 by the PAC1-disrupted strain on cracked maize and in synthetic liquid medium, expression of the gene may reduce the amount of fumonisin produced by the wild-type strain.
Previous studies indicated that the Δfcc1 mutant grows normally under alkaline conditions (31). In comparison, the PAC1-disruption mutant could not grow under these conditions, suggesting that Fcc1 does not operate upstream of Pac1 in the regulation of fumonisin biosynthesis. Furthermore, the Δfcc1 mutant does not produce FB1 on maize kernels (31). The PAC1-disruption mutant produces FB1 on maize kernels, suggesting that Pac1 does not operate upstream of Fcc1. These observations suggest that Fcc1 and Pac1 act independently of each other with respect to fumonisin biosynthesis.
Acknowledgments
We thank Su-Joung Shim for technical assistance, J.-R. Xu for providing the transformation vector pSM334, and R. Proctor for providing the F. verticillioides genomic DNA library.
Financial support was provided by the USDA NRI Competitive Grants Program, award no. 02-35201-11542.
Footnotes
This report constitutes Journal Publication 16997 of the Purdue University Agricultural Research Program.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullerman, L. B., D. Ryu, and L. S. Jackson. 2002. Stability of fumonisins in food processing. Adv. Exp. Med. Biol. 504:195-204. [DOI] [PubMed] [Google Scholar]
- 3.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper, K. F., M. J. Mallory, J. B. Smith, and R. Strich. 1997. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p). EMBO J. 16:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries, S. C., J. Springer, and J. H. Wessels. 1982. Diversity of abundant mRNA sequences and patterns of protein synthesis in etiolated and greened pea seedlings. Planta 156:129-135. [DOI] [PubMed] [Google Scholar]
- 6.Espeso, E. A., and H. N. Arst, Jr. 2000. On the mechanism by which alkaline pH prevents expression of an acid-expressed gene. Mol. Cell. Biol. 20:3355-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espeso, E. A., and M. A. Penalva. 1996. Three binding sites for the Aspergillus nidulans PacC zinc-finger transcription factor are necessary and sufficient for regulation by ambient pH of the isopenicillin N synthase gene promoter. J. Biol. Chem. 271:28825-28830. [DOI] [PubMed] [Google Scholar]
- 8.Espeso, E. A., J. Tilburn, H. N. Arst, Jr., and M. A. Penalva. 1993. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 12:3947-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaherty, J. E., and G. A. Payne. 1997. Overexpression of aflR leads to upregulation of pathway gene transcription and increased aflatoxin production in Aspergillus flavus. Appl. Environ. Microbiol. 63:3995-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelderblom, W. C., K. Jaskiewicz, W. F. Marasas, P. G. Thiel, R. M. Horak, R. Vleggaar, and N. P. Kriek. 1988. Fumonisins—novel mycotoxins with cancer-promoting activity produced by Fusarium moniliforme. Appl. Environ. Microbiol. 54:1806-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller, N. P., C. Nesbitt, B. Sarr, T. D. Phillips, and G. B. Burow. 1997. pH regulation of sterigmatocystin and aflatoxin biosynthesis in Aspergillus spp. Phytopathology 87:643-648. [DOI] [PubMed] [Google Scholar]
- 12.Keller, S. E., and T. M. Sullivan. 1996. Liquid culture methods for the production of fumonisin. Adv. Exp. Med. Biol. 392:205-212. [DOI] [PubMed] [Google Scholar]
- 13.Keller, S. E., T. M. Sullivan, and S. Chirtel. 1997. Factors affecting the growth of Fusarium proliferatum and the production of fumonisin B1: oxygen and pH. J. Ind. Microbiol. Biotechnol. 19:305-309. [DOI] [PubMed] [Google Scholar]
- 14.Kuchin, S., P. Yeghiayan, and M. Carlson. 1995. Cyclin-dependent protein-kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc. Natl. Acad. Sci. USA 92:4006-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao, S. M., J. H. Zhang, D. A. Jeffrey, A. J. Koleske, C. M. Thompson, D. M. Chao, M. Viljoen, H. J. J. Vanvuuren, and R. A. Young. 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374:193-196. [DOI] [PubMed] [Google Scholar]
- 16.MacCabe, A. P., J. P. T. W. van den Hombergh, J. Tilburn, H. N. Arst, Jr., and J. Visser. 1996. Identification, cloning and analysis of the Aspergillus niger gene pacC, a wide domain regulatory gene responsive to ambient pH. Mol. Gen. Genet. 250:367-374. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Marek, E. T., C. L. Schardl, and D. A. Smith. 1989. Molecular transformation of Fusarium solani with an antibiotic resistance marker having no fungal DNA homology. Curr. Genet. 15:421-428. [DOI] [PubMed] [Google Scholar]
- 19.Mingot, J.-M., J. Tilburn, E. Diez, E. Bignell, M. Orejas, D. A. Widdick, S. Sarkar, C. V. Brown, M. X. Caddick, E. A. Espeso, H. N. Arst, Jr., and M. A. Peñalva. 1999. Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signaling is bypassed. Mol. Cell. Biol. 19:1390-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakai, K., and M. Kanehisa. 1992. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson, P. E., A. E. Desjardins, and R. D. Plattner. 1993. Fumonisins, mycotoxins produced by Furarium species: biology, chemistry, and significance. Annu. Rev. Phytopathol. 31:233-252. [DOI] [PubMed] [Google Scholar]
- 22.Park, D. L., and T. C. Troxell. 2002. U.S. perspective on mycotoxin regulatory issues. Adv. Exp. Med. Biol. 504:277-285. [DOI] [PubMed] [Google Scholar]
- 23.Peñalva, M. A., and H. N. Arst, Jr. 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66:426-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proctor, R. H., A. E. Desjardins, R. D. Plattner, and T. M. Hohn. 1999. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 27:100-112. [DOI] [PubMed] [Google Scholar]
- 25.Proctor, R. H., D. W. Brown, R. D. Plattner, and A. E. Desjardins. 2003. Co-expression of fifteen contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet. Biol. 38:237-249. [DOI] [PubMed] [Google Scholar]
- 26.Reinhardt, A., and T. Hubbard. 1998. Using neural networks for prediction of the subcellular location of proteins. Nucleic Acids Res. 26:2230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rollins, J. A., and M. B. Dickman. 2001. pH signaling in Sclerotinia sclerotiorum: identification of a pacC/RIM1 homolog. Appl. Environ. Microbiol. 67:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt, E. K., R. Kempken, and U. Kuck. 2001. Functional analysis of promoter sequences of cephalosporin C biosynthesis genes from Acremonium chrysogenum: specific DNA-protein interactions and characterization of the transcription factor PACC. Mol. Genet. Genomics 265:508-518. [DOI] [PubMed] [Google Scholar]
- 29.Seo, J. A., R. H. Proctor, and R. D. Plattner. 2001. Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 34:155-165. [DOI] [PubMed] [Google Scholar]
- 30.Shim, W.-B., and C. P. Woloshuk. 1999. Nitrogen repression of fumonisin B1 biosynthesis in Gibberella fujikuroi. FEMS Microbiol. Lett. 177:109-116. [DOI] [PubMed] [Google Scholar]
- 31.Shim, W.-B., and C. P. Woloshuk. 2001. Regulation of fumonisin B1 biosynthesis and conidiation in Fusarium verticillioides by a cyclin-like (C-type) gene, FCC1. Appl. Environ. Microbiol. 67:1607-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suarez, T., and M. A. Penalva. 1996. Characterization of a Penicillium chrysogenum gene encoding a PacC transcription factor and its binding sites in the divergent pcbAB-pcbC promoter of the penicillin biosynthetic cluster. Mol. Microbiol. 20:529-540. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Penalva, and H. N. Arst, Jr. 1995. The Aspergillus PACC zinc finger transcription factor mediates regulation of both acid-expressed and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Wert, S. L., and O. C. Yoder. 1992. Structure of the Cochliobolus heterostrophus glyceraldehydes-3-phosphate dehydrogenase gene. Curr. Genet. 22:29-35. [DOI] [PubMed] [Google Scholar]
- 36.Wang, E., W. P. Norred, C. W. Bacon, R. T. Riley, and A. H. Merrill, Jr. 1991. Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 266:14486-14490. [PubMed] [Google Scholar]
- 37.Woloshuk, C. P., and G. A. Payne. 1994. The alcohol dehydrogenase gene adh1 is induced in Aspergillus flavus grown on medium conducive to aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:670-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan, K., and M. B. Dickman. 1996. Isolation of a β-tubulin gene from Fusarium moniliforme that confers cold-sensitive benomyl resistance. Appl. Environ. Microbiol. 62:3053-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]