Abstract
A method was developed for the rapid detection and enumeration of Aureococcus anophagefferens, the cause of harmful algal blooms called “brown tides” in estuaries of the Mid-Atlantic United States. The method employs a monoclonal antibody (MAb) and a colorimetric, enzyme-linked immunosorbent assay format. The MAb obtained exhibits high reactivity with A. anophagefferens and very low cross-reactivities with a phylogenetically diverse array of other protists and bacteria. Standard curves are constructed for each 96-well microtiter plate by using known amounts of a preserved culture of A. anophagefferens. This approach allows estimation of the abundance of the alga in natural samples. The MAb method was compared to an existing method that employs polyclonal antibodies and epifluorescence microscopy and to direct microscopic counts of A. anophagefferens in samples with high abundances of the alga. The MAb method provided increased quantitative accuracy and greatly reduced sample processing time. A spatial survey of several Long Island estuaries in May 2000 using this new approach documented a range of abundances of A. anophagefferens in these bays spanning nearly 3 orders of magnitude.
The identification and enumeration of microorganismal species in natural aquatic assemblages is an essential prerequisite for ecological studies of these populations. The ability to distinguish between closely related taxa is especially critical when these species pose health and environmental risks. Traditionally, protistan species (microalgae and protozoa) have been identified morphologically and enumerated by using light or electron microscopy. Light microscopy (bright field, phase, and differential interference contrast) has been used to identify many protists that possess distinct morphological features, whereas electron microscopy has been used effectively for many small algae and protozoa (e.g., cell sizes of ≤10 μm). Protists typically have been counted by epifluorescence microscopy (25) or by using settling techniques and inverted light microscopy (30).
Unfortunately, these approaches have significant disadvantages for ecological studies in which it is necessary to identify and count small protists in large numbers of samples in a timely manner. Morphological features that are relevant for species identification are not always easy to discern by methods that are most commonly used for enumeration. For example, transmitted and epifluorescence microscopy do not allow visualization of morphological features that are pertinent for species identifications of many small protists (e.g., striations on frustules of diatoms or body scales on chrysomonads that can be observed only by electron microscopy). In addition, microscopic analyses generally are time-consuming, and the processing of large numbers of samples that are typical in ecological surveys and experiments may require weeks or months to complete. In order to circumvent these shortcomings, new approaches based in modern immunology and genetics have emerged that are able to provide rapid and accurate identification and enumeration of microbial species.
Immunological approaches for identifying and enumerating marine microalgae have become commonplace within the last two decades. These methods and their ecological applications for the identification of phytoplankton have been summarized (19, 31). Both polyclonal antibodies (PAbs) and monoclonal antibodies (MAbs) have been developed for use by microbial ecologists. Immunological probes have proven useful for identifying species of cyanobacteria (10), raphidophytes (29), dinoflagellates (22), pelagophytes (3, 21), and other minute algal taxa (11, 24) and even for distinguishing between toxic and nontoxic strains of harmful algae (6). An added advantage of this approach is that these methods often can be converted to formats that are significantly more rapid than routine microscopical counts (32).
Aureococcus anophagefferens is a pelagophyte alga that typifies the difficulties of accurately identifying and enumerating small protistan species in natural water samples. The alga is minute (∼2 to 4 μm in diameter) and spherical, lacks flagella and body scales, and has few other features that might easily distinguish it from a variety of co-occurring algae of similar size. Unfortunately, A. anophagefferens is unique in that it has been the cause of recurring harmful algal blooms in estuaries of New York, New Jersey, Maryland, and Rhode Island in the mid-Atlantic United States. These “brown tides” have resulted in ecological damage and destruction of commercial shellfisheries (9). For this reason, considerable effort has been expended to document the abundances and distribution of A. anophagefferens (23).
The small size and nondescript morphology of A. anophagefferens has made it difficult to distinguish accurately by transmitted light microscopy from co-occurring eukaryotic algae of similar size and shape. The most popular method for accomplishing this goal for natural water samples has been immunofluorescent staining of A. anophagefferens with a PAb (3). A.anophagefferens cells stained in this manner are distinguished and counted by using epifluorescence microscopy. The development of this method has enabled studies of the geographic distribution of A. anophagefferens, as well as ecological studies of the harmful algal blooms caused by the species (2, 12, 14, 17, 23).
The polyclonal method has been a considerable improvement over previous methods for identifying and counting A. anophagefferens, and it has fostered research on this interesting species. Nevertheless, the PAb approach is time-consuming and suffers from the potential for variability in the effectiveness of staining due to differences among different batches of PAb. In addition, variability can arise from differences in the level of skill and experience among investigators both in the immunological staining procedure and in the microscopic analysis.
In order to address these issues, we devised an improved method for immunodetection of A. anophagefferens that relies on the application of a newly developed MAb that has high reactivity with the target species but very low cross-reactivity with a wide array of other species of protists and bacteria. This MAb has been adapted to a colorimetric, enzyme-linked immunosorbent assay (ELISA) performed in 96-well microtiter plates. The use of this new, indirect method allows for rapid, accurate determination of the abundance of A. anophagefferens in large numbers of natural samples.
MATERIALS AND METHODS
Generation of MAbs.
MAbs against A. anophagefferens were generated by using strain CCMP1784 isolated in 1986 by Elizabeth Cosper, Cosper Environmental Services, Inc., Bohemia, N.Y. The alga was grown at 20°C in modified f/2 medium (13, 14, 18) on a 12:12-h light-dark cycle. The culture was grown to the late exponential growth phase and preserved with 10% glutaraldehyde (prepared from a 50% aqueous solution in natural filtered seawater) to a final concentration of 1% and stored at 4°C in the dark. This preservation procedure mimics the manner in which field samples are treated. Cells were pelleted by centrifugation and resuspended three times in 0.1 M phosphate-buffered saline (PBS) (16) to remove the glutaraldehyde. Cells were resuspended in 0.1 M PBS after the last centrifugation at a final cell concentration of 2.0 × 108 ml−1 for inoculating mice.
Immunization of mice, development of hybridoma cell lines, and initial screenings against A. anophagefferens were conducted by Maine Biotechnology Services, Inc., Portland, Maine. Female BALB/c mice were immunized subcutaneously with 0.5 ml of a 1:1 emulsion of the A. anophagefferens cell suspension and Freund complete adjuvant (16). Immunizations were repeated at 3-week intervals for 12 weeks. Four days prior to fusion the mice received ca. 108 A. anophagefferens cells in 0.1 M PBS by intraperitoneal injection.
Spleen cells were harvested and fused with F/0 myeloma cells by using standard methods and then plated in 96-well culture plates. Primary screening of the emerging hybridoma lines was conducted after 9 to 12 days to determine the production of antibodies against preserved A. anophagefferens cells (strain CCMP1784). Wells showing positive reactions were subcloned and rescreened. Subcloned cell lines showing the production of antibodies against A. anophagefferens from the second screening were subjected to a second subcloning to assure monoclonality and then cryopreserved. The resulting cell lines were tested for cross-reactivity to a wide variety of protists (algae and protozoa) and bacteria.
Strains of protists and bacteria.
Three strains of A. anophagefferens were tested for reactivity to the MAb. All three are available through the Provasoli-Guillard National Center for the Culture of Marine Phytoplankton, Boothbay Harbor, Maine. Strain CCMP1784 (designated GSB in Table 1) was obtained in 1986 by Elizabeth Cosper from Great South Bay, Long Island, New York. Strain CCMP1708 (designated WNB in Table 1) was isolated in 1995 by Reyhan Mehran from West Neck Bay in the Peconic Bay Estuaries system, Long Island, N.Y. Strain CCMP1794 (designated BB in Table 1) was obtained in 1997 by John Mahoney from Barnegat Bay, New Jersey. These locations represent areas of recurring outbreaks of brown tides.
TABLE 1.
Reactivities of MAbs (as measured by the OD450) against A. anophagefferens and a diverse array of microalgae, protozoa, and bacteriaa
| Microbial taxon | OD450 | Microbial taxon | OD450 | |
|---|---|---|---|---|
| Pelagophytes | ||||
| Aureococcus anophagefferens (WNB) | 0.96 | |||
| Aureococcus anophagefferens (GSB) | 1.03 | |||
| Aureococcus anophagefferens (BB) | 0.97 | |||
| Aureoumbra lagunensis | <0.01 | |||
| Pelagococcus subviridis | <0.01 | |||
| Cyanobacteria | ||||
| Synechocystis sp. (Syn 145) | 0.03 | |||
| Synechococcus sp. (8012) | <0.01 | |||
| Synechococcus sp. (8101) | <0.01 | |||
| Prochlorophytes (Prochlorococcus sp.) | <0.01 | |||
| Cryptophytes (Rhodomonas salina [3C]) | 0.01 | |||
| Chlorophytes | ||||
| Ankistrodesmus falcatus | <0.01 | |||
| Chlamydomonas sp. | 0.01 | |||
| Chlorella stigmatophora (CL993) | <0.01 | |||
| Chlorella capsulata (Fla-E) | <0.01 | |||
| Nanochloris sp. (Nanno) | <0.01 | |||
| Prasinophytes | ||||
| Tetraselmis sp. | <0.01 | |||
| Micromonas pusilla (IB4) | <0.01 | |||
| Prymnesiophytes | ||||
| Prymnesiophyte (H3-14) | <0.01 | |||
| Isochrysis galbana (T-Iso) | <0.01 | |||
| Chrysochromulina ericina (NEPCC109A) | <0.01 | |||
| Chrysophytes | ||||
| Paraphysomonas bandaiensis (Hflag) | 0.01 | |||
| Paraphysomonas imperforata (VS1) | <0.01 | |||
| Ochromonas sp. (VT1) | 0.04 | |||
| Bacillariophytes | ||||
| Thalassiosira weissflogii (Actin) | <0.01 | |||
| Nitzschia frustulem | 0.01 | |||
| Minutocellus polymorphus | 0.01 | |||
| Pedinellids (Pteridomonas sp. [NB1]) | <0.01 | |||
| Synurophytes (Synura petersenii) | 0.01 | |||
| Bicosoecids (Cafeteria sp. [Cflag]) | 0.02 | |||
| Hamatores (Caecitellus parvulis) | 0.02 | |||
| Dictyophytes (Rhizochromulina sp.) | <0.01 | |||
| Raphidophytes (Heterosigma akashiwo) | 0.01 | |||
| Xanthophytes (Botrydiopsis intercedens) | 0.03 | |||
| Jakobids (Jakoba libera [DB9]) | <0.01 | |||
| Eustigmatophytes | ||||
| Eustigmatos magna | 0.02 | |||
| Nannochloropsis gaditana | <0.01 | |||
| Euglenids (Euglena gracilis) | <0.01 | |||
| Dinoflagellates | ||||
| Gymnodinium beii | 0.01 | |||
| Scrippsiella sp. | 0.02 | |||
| Ciliates | ||||
| Scuticociliate (BBcil) | 0.03 | |||
| Hymenostome (Hcil) | <0.01 | |||
| Bacteria (Halomonas halodurans) | 0.01 | |||
| Unidentified algae | ||||
| Long Island mixture (LI16a)* | 0.01 | |||
| Long Island mixture (LI15a)* | <0.01 | |||
| BT3 (unidentified, 2- to 3-μm alga)* | <0.01 | |||
| BT8 (unidentified, 2- to 3-μm alga)* | <0.01 |
Values of <0.01 were undetectable under the conditions employed. Cultured A. anophagefferens from three locales were tested (WNB, West Neck Bay, New York; GSB, Great South Bay, New York; BB, Barnegat Bay, New Jersey; see Materials and Methods for strain identifications). Asterisks indicate unidentified taxa (verified as not A. anophagefferens) of similar size and shape to the brown tide alga that were enriched and cultured from natural water samples with a high abundance of A. anophagefferens. OD450, optical density at 450 nm.
Thirty-nine strains and two mixed enrichment cultures of protists representing 19 major taxonomic groupings were examined for their cross-reactivity to the MAb generated against A. anophagefferens (Table 1). These species included two pelagophyte strains: Aureoumbra lagunensis (the cause of Texas brown tides) and Pelagococcus subviridis. These two species have been grouped with A. anophagefferens in this recently erected class of algae (1) and thus serve as an in-group comparison for MAbs developed against A. anophagefferens. The other protistan taxa were chosen because they represent a wide diversity of algal classes and they are common species from estuarine ecosystems. A number of these taxa are similar in size and shape to A. anophagefferens and thus difficult to distinguish morphologically by microscopical examination. Some are species that are known to be present in Long Island waters. In addition, three strains of chroococcoid cyanobacteria (Synechococcus spp.), one prochlorophyte strain, and one bacterium were tested for cross-reactivity.
The protistan strains Aureoumbra lagunensis (CCMP1681), P. subviridis (CCMP1429), Chrysochromulina ericina (CCMP387), Synura petersenii (CCMP859), Euglena gracilis (ATCC 12716), Eustigmatos magna (CCMP387), Rhizochromulina sp. (CCMP234), Heterosigma akashiwo (CCMP452), and Botrydiopsis intercedens (UTEX296) were obtained from the Provasoli-Guillard National Center for the Culture of Marine Phytoplankton, Boothbay Harbor, Maine; the American Type Culture Collection, Manassas, Va.; or The Culture Collection of Algae at the University of Texas at Austin. Ankistrodesmus falcatus, Nitzschia frustulem, and Chlamydomonas sp. were obtained from Robert W. Sanders, Temple University, Philadelphia, Pa. Rhodomonas salina and Isochrysis galbana were provided by Scott Gallager of Woods Hole Oceanographic Institution (WHOI), Woods Hole, Mass. Thalassiosira weissflogii was obtained from Donald Anderson (WHOI). Chlorella stigmatophora, Chlorella capsulata, and Nanochloris sp. were obtained from Joel C. Goldman, Institute of Marine Sciences, University of California, Santa Cruz. Jakoba libera was provided by Delma Bratvold, College of Charleston, Charleston, S.C. The prymnesiophyte strain H3-14 was obtained from Lynda Shapiro, Oregon Institute of Marine Biology, University of Oregon, Eugene. All remaining strains of protists were isolated from a variety of aquatic environments and cultured in the laboratory of D. Caron.
Strains BT3 and BT8 are unidentified algae that are spherical cells, nonflagellated, and similar in size to A. anophagefferens but not to the brown tide alga (i.e., they showed no significant reactivity to the PAb of Anderson et al. [3]). These strains were isolated from Great South Bay, New York, in 1987 during a brown tide in that bay. Cultures LI16a and LI15a were water samples obtained during the same bloom, enriched with inorganic nutrients to promote growth of microalgae, and kept in an incubator at a light intensity ∼200 μE m−2 s−1 on a 12:12-h light-dark cycle for several months. The enrichments contained a mixture of minute algae but showed no reactivity with repeated application of the PAb method of Anderson et al. (3). Cyanobacterial strains were obtained from John B. Waterbury and the prochlorophyte strain was provided by Robert Olson (both of WHOI).
Screening of hybridoma cell lines.
Hybridoma cell lines were screened for reactivity to A. anophagefferens and subsequently for cross-reactivity to other microbial species by an ELISA. Cultures of A. anophagefferens and the other species of microorganisms were harvested in mid- to late exponential growth phase, preserved with glutaraldehyde (1% final concentration), and dried onto the bottom of wells of flat-bottom, 96-well microtiter plates at normalized biomass. The wells were blocked with 1% bovine serum albumin (BSA; Fisher Scientific, BP16705-100) in 1× PBS-Tween (100 ml of 10× PBS and 100 μl of Tween 20, brought to 1 liter with distilled water and filtered [0.22-μm-pore-size filters]) prior to the addition of cells. The ELISA procedure used for these tests was the same as the method applied to natural water samples (see below) except that the method for natural samples used 96-well filter plates to retain cells instead of adhering them to the bottoms of the microtiter plates. Solution exchange for the screening and cross-reactivity tests was accomplished by decanting the liquid from the plates by inverting them, whereas solutions were removed by filtration in the method applied to natural samples. Also, cross-reactivity tests were conducted by allowing color development in the wells with A. anophagefferens to proceed to completion and then processing all samples at that time. Based on these tests, a single hybridoma cell line (LIZ 1F7-1E12) was chosen for ascites production in (BALB/c × ICR)F1 hybrid mice. Ascites were clarified by centrifugation and filtration through gauze and then purified by protein affinity chromatography.
Immunofluorescent staining with MAbs.
Immunofluorescent staining of the brown tide alga was performed to examine the labeling intensity and cellular location of the MAb. A culture of A. anophagefferens cells in the exponential growth phase was preserved in 1% glutaraldehyde for 1 h at 4°C and then dried down overnight onto coverslips. Samples were rinsed in 1× PBS for 10 min, blocked for 75 min in 1× PBS-Tween containing 1% BSA, and incubated in primary antibody solution (a 1:25,000 dilution of MAb LIZ 1F7-1E12 in block solution) for an additional 75 min. The coverslips were rinsed twice for 5 min in 1× PBS-Tween before being incubated in secondary antibody solution (a 1:400 dilution of fluorescently labeled goat anti-mouse antibody conjugated to Alexa-Fluor 350 [Molecular Probes, Inc.] in 1× PBS-Tween) for 75 min. The coverslips were then given three 10-min rinses in 1× PBS-Tween, and mounted on glass slides by using a mounting solution of 4:1 glycerol to 1× PBS at pH 8.75. The samples were examined by using transmitted light microscopy and epifluorescence microscopy with a Leica DM IRBE microscope equipped with a Hamamatsu charge-coupled device camera (C4742-95). Images were captured by using OpenLab v2.2.5. All capture times for images were kept constant to allow direct comparison of treatments examining staining intensity of positive and negative controls and cross-reactivity to nontarget cells.
Application of the MAb to natural water samples.
Natural samples were analyzed for the abundance of A. anophagefferens by using the selected MAb and an ELISA format conducted in 96-well filter plates (Millipore Multiscreen MAHV N45). The bottoms of the wells of these plates consist of 0.45-μm-pore-size Durapore PVDF membrane filters. The filters allow retention of particulate material but easy exchange of liquids in the wells by means of a vacuum manifold (Millipore MAVM 096 01) that applies vacuum to the entire plate in one step. Samples of 5 to 7 ml were collected in borosilicate glass tubes, preserved immediately with 10% glutaraldehyde to a final concentration of 1% glutaraldehyde, sealed with plastic caps, and stored in the dark until analyzed.
Filter plates were preblocked for 30 min at room temperature by using 250 μl of blocking solution per well (1% BSA in 1× PBS-Tween). Wells were rinsed three times with 250 μl of 1× PBS-Tween per well. Samples were mixed thoroughly, divided into aliquots, and placed into the wells. All analyses (standards and unknowns) were done in triplicate. Therefore, with a standard curve of seven to nine points (21 to 27 wells) and appropriate reagent blanks, 21 to 23 unknowns were analyzed in triplicate on a single plate.
Unknown samples were added to plates at 200 μl. Samples of <200 μl can be used, but variability among replicate wells increased with decreasing sample volume. Liquid was removed by vacuum, and all wells were rinsed with 250 μl of 1× PBS-Tween. A 100-μl portion of the primary antibody working solution (1:25,000 dilution in 1% BSA in 1× PBS-Tween) was added to each well, and the plate was placed on a rotating platform (60 rpm) for 1 h at room temperature. The solution was removed by vacuum, and all wells were rinsed three times with 250 μl of 1× PBS-Tween. Then, 100 μl of the horseradish peroxidase-linked secondary antibody (Peroxidase Affinipure Goat Anti-Mouse IgG+IgM; Jackson Labaratories, 115-035-0440) was added to each well, and the plate returned to the rotating platform (60 rpm) for 1 h at room temperature. The secondary antibody was reconstituted in 2 ml of 1× PBS-Tween and 2 ml of glycerol (1:2 dilution). The final working concentration was 1:10,000 in 1% BSA in 1× PBS-Tween. All wells were rinsed five times with 250 μl of 1× PBS-Tween. The filter plate was placed onto the vacuum manifold with a standard, flat-bottom, 96-well microtiter plate below the unit. Next, 50 μl of the substrate-chromogen (TMB [3,3′,5,5′-tetramethylbenzidine]; Alerchek no. 90101) was added to each well, followed by incubation at room temperature for 30 min for color development. Then, 50 μl of stop solution (0.18 N sulfuric acid) was added, and the solution was vacuumed down into the 96-well plate. The absorbance at 450 nm was read for each well by using a spectrophotometric plate reader.
Determining the abundances of A. anophagefferens in natural samples consisted of correlating absorbance in the wells containing the unknowns to the absorbance in a set of standards with known concentrations of the brown tide alga. The individual standards used to construct the standard curve were obtained by serially diluting a single “stock” culture of A. anophagefferens (preserved in 1% glutaraldehyde). The cell density in the stock culture was determined by direct microscopic counts. The culture was grown to late exponential growth phase, preserved, and stored at 4°C in the dark. Periodic examination indicated that cells in the stock culture remained morphologically stable over a long period of time (>1 year) and that the antigenic character remained stable during that period (i.e., the results with standard curves did not change appreciably during that period). A complete set of standards, consisting of seven to nine concentrations of A. anophagefferens cells, was included on each microtiter plate. The inclusion of standard curves on every plate accounted for slight differences in analytical results resulting from possible minor temperature fluctuations, variable reagent qualities or concentrations, and/or operator performance.
Comparison of the use of MAb with other methods for counting the brown tide alga.
The MAb ELISA method for counting A. anophagefferens was compared directly to the PAb method of Anderson et al. (3) for natural seawater samples containing a wide range of abundances of A. anophagefferens. The PAb method was performed independently by three groups: one at WHOI, one at Suffolk County Department of Health Services (SCDHS), and one at Long Island University (LIU). All counts at the SCDHS and some of the counts at WHOI were performed with the primary PAb at the concentration recommended by Anderson et al. (3). Counts of cultured A. anophagefferens in exponential growth phase were also performed by using the PAb method and compared to counts of the same cultures by using transmitted light microscopy and a hemacytometer. The results from these samples indicated significant underestimation of the abundance of A. anophagefferens by the PAb method relative to the MAb method and direct microscopic counts (see Results). Subsequent counts at WHOI and counts at LIU were performed with concentrations of the PAb at two- to fourfold the concentration recommended by Anderson et al. (3).
Abundances of A. anophagefferens in natural samples containing relatively high abundances of the brown tide alga (>2.5 × 105 ml−1) were determined by the MAb and PAb methods and compared to counts obtained by transmitted light microscopy with a hemacytometer. Hemacytometer counts in samples with high abundances of A. anophagefferens provide a fairly accurate estimate of the alga if the brown tide alga is the dominant species in the 1- to 5-μm size range.
Effects of sample dilution and preservative type.
We empirically determined that standard curves were highly linear up to a concentration of ∼50,000 to 60,000 cells of A. anophagefferens well−1 (see Results). This value corresponded to 250,000 to 300,000 cells ml−1 if 200-μl samples were analyzed. Brown tides with cell abundances of >106 cells ml−1 occur in nature (9). Therefore, it was necessary to extend the dynamic range of our colorimetric method. In practice, we found that filtering aliquots of <200 μl was possible, but smaller volumes progressively increased the amount of variability among replicate subsamples. As an alternative to smaller filtration volumes, we examined the accuracy of diluting samples with filtered seawater, assaying these dilutions by the MAb method and then calculating the abundance of A. anophagefferens in the undiluted samples based on their absorbances and dilution factors. Three natural samples with abundances of A. anophagefferens of >2.5 × 106 cells ml−1 were serially diluted three times, and all four samples (undiluted sample plus three dilutions) were analyzed by the MAb method to examine the degree to which the diluted samples provided accurate estimates of the abundance of brown tide cells in the undiluted samples. Abundances determined by the MAb method were compared to counts obtained by direct microscopy. Counting cells of the appropriate size and shape of A. anophagefferens in these samples provided a reasonable estimate of the abundance of the species because the samples were strongly dominated by the brown tide alga.
The effect of preservative type on the efficacy of the MAb method was examined by comparing counts made on duplicate samples of a culture of A. anophagefferens preserved with different fixatives. Because the MAb was generated with A. anophagefferens cells that were preserved with glutaraldehyde, this is the preferred preservative for this analysis. However, samples for phytoplankton and protozoan enumeration are often preserved with acid Lugol’s solution at concentrations of up to 10% (28). Lugol’s solution provides improved staining of larger phytoplankton species and good preservation of ciliated protozoa. In order to examine the ability of the MAb to recognize and quantify A. anophagefferens cells in samples preserved with Lugol’s solution, we analyzed duplicate sets of a serially diluted culture of A. anophagefferens preserved with either 1% glutaraldehyde as described above or with a 10% acid Lugol’s solution. In addition, we examined the effect of “clearing” the Lugol’s solution by the addition of 6 × 10−4 M sodium thiosulfate (25). This procedure improves the visualization of protistan cells by epifluorescence microscopy, and we reasoned that it might improve the accuracy of the colorimetric analysis. All three types of standard curves were analyzed on a single 96-well microtiter plate to avoid variations among plates.
Field test of the MAb method.
A survey study was conducted to demonstrate the dynamic range and rapidity with which counts of A. anophagefferens can be obtained by using the new MAb method. Samples were collected on 25 May 2000 from 19 sites in the Peconic Bay estuary system, from Shinnecock Bay, from Moriches Bay, and from Great South Bay on Long Island, N.Y. Samples of 5 ml of seawater were collected from the surface at each location and preserved at a final concentration of 1% with 10% glutaraldehyde prepared with filtered natural seawater from a 50% aqueous solution. Samples were stored at 4°C in the dark overnight and then processed the following morning by using the procedure described above.
RESULTS
MAb generation and cross-reactivity.
Numerous hybridoma cell lines were obtained that showed reactivity to A. anophagefferens in the initial screening tests. Many of these cell lines also cross-reacted with other protists or bacteria. MAb LIZ 1F7-1E12, however, had very strong reactivity to the A. anophagefferens strain used to generate the MAb (strain GSB in Table 1) but extremely low cross-reactivity to a wide variety of microalgae, protozoa, bacteria, cyanobacteria, and a prochlorophyte (see Table 1). The 39 strains of protists tested for cross-reactivity encompassed 19 major taxonomic groups of protists. These species represent most of the phylogenetic groups commonly found in coastal phytoplankton assemblages. Several species that co-occur with the brown tide alga were tested. The two unidentified algae (BT3 and BT8) were isolated from Great South Bay, New York, during a brown tide. These algae were similar in size and overall morphology to A. anophagefferens but were not the brown tide alga, i.e., they showed no reactivity to the PAb of Anderson et al. (3). Minutocellus polymorphus is a small diatom that co-occurs with A. anophagefferens during some brown tides (27). In addition, two mixed enrichments of phytoplankton from Great South Bay (LI16a and LI15a) contained high abundances of a variety of minute algae that were present during a brown tide but were not the brown tide alga. None of these cultures reacted positively to the PAb of Anderson et al. (3) after several months of growth in the laboratory in nutrient enriched medium.
Optical densities in all wells with microorganisms that were not A. anophagefferens were at least 2 orders of magnitude less than the optical densities in wells with A. anophagefferens when the cultures were tested with the MAb LIZ 1F7-1E12 during the cross-reactivity test (Table 1). Color development in the wells with A. anophagefferens cells was allowed to proceed to completion before termination of the reactions to ensure that cross-reactivity with nontarget species was low. Color development is not allowed to continue to completion during routine analyses because absorbance in wells with high abundances of the brown tide alga saturates under these conditions. Thus, the differences in optical density between wells with A. anophagefferens and wells containing nontarget species in these tests indicated only minimal differences between the reactivity of target (A. anophagefferens) and nontarget species. These cross-reactivity tests were performed with concentrations of protists and bacteria at biomasses normalized to the biomass of A. anophagefferens in the microtiter plates. The very low reactivities that we observed (in most cases below detection) indicated a very high degree of specificity of the MAb for A. anophagefferens. Two species of pelagophytes (Aureoumbra lagunensis and P. subviridis) that were most closely related phylogenetically to the brown tide alga showed no detectable reactivity to the MAb. The enrichment cultures, the protozoan cultures, and most of the algal cultures used in the cross-reactivity tests contained bacteria. Therefore, the cross-reactivity tests for these cultures also included numerous unidentified bacteria.
Analyses of A. anophagefferens cultures at various growth stages did not show measurable changes in the antigenic nature of A. anophagefferens due to differences in its physiological condition. Moreover, a comparison of strains of A. anophagefferens isolated in three different years and from three different locales in the mid-Atlantic United States (Peconic Bay, New York; Great South Bay, New York; and Barnegat Bay, New Jersey) were quantitatively indistinguishable with respect to their reactivity to the MAb (Table 1). All three strains displayed dramatically higher reactivities relative to nontarget species.
Immunofluorescent staining.
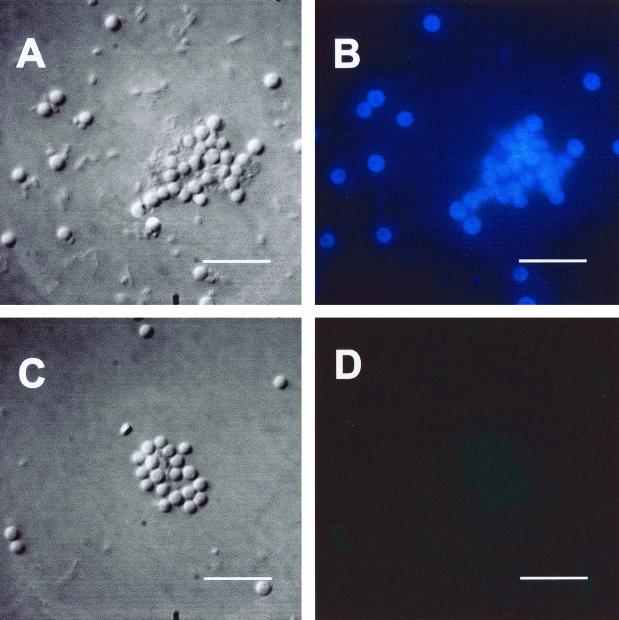
Staining intensity and cellular localization of the MAb was examined by immunofluorescent staining. An Alexa-Fluor-labeled, anti-mouse secondary antibody was used to detect MAb binding to cultured cells of A. anophagefferens (Fig. 1). Cells exposed to the MAb fluoresced intensely, whereas spurious staining with the secondary antibody of cells that were not exposed to the MAb was extremely low (compare Fig. 1B with D). Cells appeared as fluorescent halos when examined by epifluorescence microscopy, indicating localization of the antigen in the cell wall of the alga.
FIG. 1.
Differential interference contrast (A and C) and epifluorescence (B and D) micrographs of A. anophagefferens. Exposure times for both epifluorescence micrographs were the same at 2.4 s. (A and B) Cells stained with LIZ 1F7-1E12 and subsequently with fluorescently labeled goat anti-mouse antibody exhibited strong fluorescence under UV excitation. Localization of fluorescence to the periphery of the cells indicates the location of antigens. (C and D) Cells not stained with the MAb but stained with the fluor-labeled secondary antibody showed virtually no fluorescence. Marker bars, 10 μm.
Standard curves, sample dilution, and preservation.
The range of A. anophagefferens abundances over which the MAb yielded a linear relationship with optical density in the ELISA format was determined empirically. Relative to reagent blanks, the method was capable of detecting cell abundances of A. anophagefferens of ca. 500 cells per well. This corresponds to a lower limit of sensitivity of the method of ca. 1,500 cells ml−1 if 200-μl samples are analyzed (three times the standard deviation of the reagent blank measurement). However, we also found that adding relatively high abundances of nontarget protists and bacteria to cultures of brown tide cells resulted in slight increases in absorbance relative to reagent blanks, presumably due to minute amounts of cross-reactivity of the MAb with nontarget material. Therefore, we conservatively consider 5,000 cells ml−1 to be a practical lower limit for the method. Sensitivity can be increased by increasing the volume of sample filtered, but samples must be added to wells in approximately 200-μl aliquots because of the volume limitation of the wells. This step increases processing time. No adverse ecological effects have been documented in nature or in the laboratory at cell concentrations of <5,000 cells ml−1 (9), so investigators must decide whether the added effort is worth the increased sensitivity.
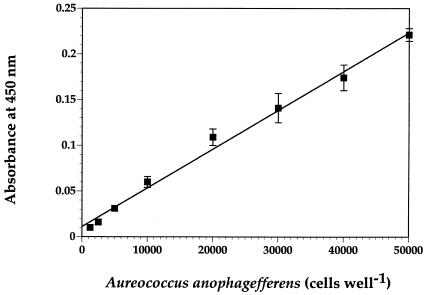
Standard curves were determined for every microtiter plate by using a serially diluted, preserved culture of A. anophagefferens. All concentrations of the brown tide alga were analyzed in triplicate for standard curves, and variability associated with the means typically was small. Regressions of the mean optical density versus known cell concentrations of cultured A. anophagefferens (the latter determined by direct microscopic counts) were highly linear (R2 > 0.97) for cell concentrations of <50,000 to 60,000 cells well−1 by using the ELISA procedure (Fig. 2). Linearity was lost at concentrations above this upper limit due to saturating optical density. Reducing the incubation and/or development times was not desirable because these procedures decreased the sensitivity of the method at the lowest cell abundances. Together with the practical lower limit of sensitivity of the method, the linear portion of the standard curves corresponds to a functional range of detection of A. anophagefferens cells in natural water samples of 5 × 103 to (2.5 to 3) × 105 cells ml−1 if 200-μl samples are analyzed.
FIG. 2.
Typical standard curve generated by the MAb method performed by using an ELISA. The standards of A. anophagefferens were obtained by serially diluting a preserved culture of the alga. Abundances were determined by direct microscopic counts. The optical density (absorbance at 450 nm) is given in arbitrary units. Mean values ± one standard deviation (error bars) for triplicate subsamples are shown.
The range above is acceptable for many natural samples, but A. anophagefferens can attain abundances in excess of 106 cells ml−1 during brown tides (9). Samples with abundances suspected to be >2 × 105 cells ml−1 were analyzed by diluting them by a factor of 5- to 10-fold with natural seawater preserved with glutaraldehyde (final concentration, 1%) and filtered through 0.22-μm-pore-size filters. The diluted samples were analyzed by the MAb method and converted to cell concentration in the undiluted sample based on the optical densities of the diluted samples and their dilution factors. In practice, samples that were suspected to have high abundances of brown tide cells (based on discoloration of the water) were often analyzed as undiluted and diluted subsamples. The subsamples providing readings in the linear portion of the standard curve were used to calculate the abundances of A. anophagefferens in the samples. This type of redundancy is feasible because of the large number of samples that can be processed at one time by the ELISA method.
A test was carried out to ensure that the dilution of samples using the procedure described above resulted in optical-density readings that would accurately and precisely reproduce the abundance of the alga in the undiluted samples. Three natural samples with abundances of A. anophagefferens that were above the upper limit of the linear portion of the standard curve were counted for the abundance of the brown tide alga by using a hemacytometer. Counts of A. anophagefferens made with a hemacytometer provide a reasonable lower-limit estimate of the abundance of A. anophagefferens in samples in which the alga's abundance is high and numerically dominates the phytoplankton assemblage. The accuracy of microscopic counts is dependent on the ability to distinguish brown tide cells from debris and other protistan species of similar size. Direct microscopic counts for the three samples were 2.20 × 106, 1.90 × 106, and 1.60 × 106 cells ml−1 for samples 1, 2, and 3, respectively. Each of these three samples was then serially diluted three times, and the three dilutions were analyzed individually by the MAb method (Fig. 3A). The readings for the three dilutions of each sample were then used to calculate the abundances in the undiluted samples.
FIG. 3.
(A) Effect of sample dilution on the accuracy of counts of A. anophagefferens determined by the MAb method. A standard curve (indicated by the solid line) was generated with a serially diluted culture (see Materials and Methods for details). Open symbols indicate samples from which the standard curve was determined by using linear regression. Three samples (solid symbols) then were serially diluted (each three times), and their absorbances were determined separately for each dilution. Dilution factors and absorbances were then used to calculate the abundance of the brown tide alga in the undiluted samples. Mean values ± one standard deviation are given for each of the three dilutions for all three samples. (B) Comparison of optical densities (absorbance at 450 nm) for duplicate sets of standards preserved with 1% glutaraldehyde versus acid Lugol’s solution (▪) or 1% glutaraldehyde versus acid Lugol’s solution cleared with sodium thiosulfate (•). The dotted line indicates the line of 1-to-1 correspondence between the methods.
Dilution had virtually no effect on the precision of the method to calculate A. anophagefferens abundance in the undiluted samples. A high degree of precision, despite the dilution process, was indicated by very low variability associated with averages obtained for the three dilutions (coefficients of variation about the means for the three undiluted samples were 5.9, 4.3, and 1.3%). Moreover, calculation of the abundances of A. anophagefferens in the three undiluted samples based on the diluted subsamples agreed reasonably well with abundances determined for the undiluted samples by direct microscopic counts obtained with a hemacytometer. The average abundances of the algae in the three samples determined by the MAb method were 2.70 × 106, 2.31 × 106, and 2.32 × 106 cells ml−1 for samples 1, 2, and 3, respectively. These values were obtained by averaging the results of the three diluted subsamples of each sample. The direct microscopic counts constituted 82, 82, and 69%, respectively, of the values determined by the MAb method for the same samples after serial dilution. Given the difficulties associated with distinguishing A. anophagefferens cells by direct microscopy, the MAb estimates agreed well with the microscopic counts. This exercise indicated that accurate counts of the brown tide alga can be obtained for natural samples at abundances up to the highest levels of A. anophagefferens observed in nature.
The method of sample preservation had a significant effect on the ability to count A. anophagefferens cells by using the MAb method (Fig. 3B). Acid Lugol’s solution is a common preservative for phytoplankton and some protozoa that is used for increasing the visibility of cells when viewed by transmitted light microscopy. However, this preservative also greatly diminishes fluorescence when the samples are examined by epifluorescence microscopy (26). We experimentally examined the ability of this method of preservation to interfere with the colorimetric MAb method by using duplicate sets of a serially diluted culture of A. anophagefferens.
Preservation with acid Lugol’s solution significantly decreased optical density readings relative to optical densities obtained with glutaraldehyde-preserved cells (Fig. 3B). The optical densities of samples preserved with Lugol’s solution retained linearity when regressed against glutaraldehyde-preserved cells (R2 = 0.98), but the resulting regression deviated from 1-to-1 correspondence. Based on this analysis, a standard curve generated with A. anophagefferens cells preserved with Lugol’s solution could be used to count the algae in natural samples. However, the sensitivity of the method (ability to detect low abundances of the alga) would presumably be reduced considerably for samples preserved with acid Lugol’s solution.
Removal of the iodine staining by the addition of sodium thiosulfate has been shown to greatly improve the fluorescence of cells preserved with acid Lugol’s solution (26). However, clearing of samples in this manner resulted in poor agreement with optical densities obtained by using glutaraldehyde-preserved cells (Fig. 3B). The slope of the regression (destained Lugol’s solution- versus glutaraldehyde-preserved cells) was quite low (i.e., 0.18), indicating that abundances of the brown tide alga in cultures preserved with acid Lugol’s solution and subsequently destained with sodium thiosulfate were much lower than abundances obtained with glutaraldehyde-preserved cells. Thus, the sensitivity of the method for use with destained Lugol’s solution-preserved cells would be much lower than for glutaraldehyde-preserved cells.
Comparison of the MAb method with other counting methods.
Direct comparisons between the MAb method for counting A. anophagefferens in natural samples and the PAb technique of Anderson et al. (3) were conducted on several occasions and with several working groups. In addition, for samples in which the abundances of A. anophagefferens were high (>3 × 105 cells ml−1), hemacytometer counts were performed by using transmitted light microscopy to estimate the number of brown tide cells in the samples. As noted above, this latter method serves only as an approximate estimate of the density of A. anophagefferens because the accuracy of the counts is dependent on the ability to distinguish brown tide cells from other minute eukaryotic algae.
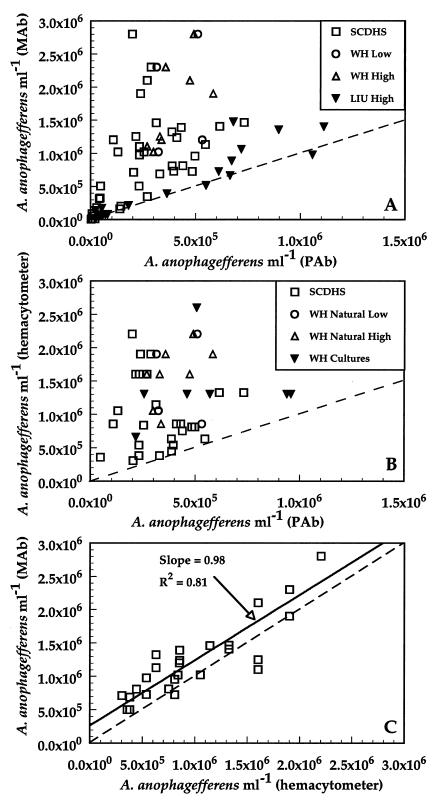
Counts of A. anophagefferens in natural samples by using the MAb method were equal to, or in most cases greater than, counts obtained by using the PAb method (Fig. 4A; note that most datum points occur above the line of one-to-one correspondence). Overall averages of A. anophagefferens abundance for 69 samples as determined by using the PAb method were 39% of the average for the MAb method. This average included sets of samples analyzed by three different working groups using the PAb method at the concentration of the PAb prescribed by Anderson et al. (3) and also at a concentration up to 4-fold that value (“high” values in Fig. 4A and B). Increasing the concentration of the PAb resulted in better agreement between the PAb and MAb methods. For example, all samples analyzed with the recommended concentration of PAb averaged only 28% of the count obtained by using the MAb method, whereas samples analyzed with higher concentrations averaged 78% of the MAb count.
FIG. 4.
Direct comparison of the abundance of A. anophagefferens in natural samples as determined by the MAb method and the PAb method of Anderson et al. (3) (A), by the PAb method and direct microscopic counts with a hemacytometer (B), and by hemacytometer counts and the MAb method (C). SCDHS indicates PAb antibody analyses performed by the Suffolk County Department of Health Services using the PAb at the concentration recommended by Anderson et al. (3). WH indicates analyses performed at the Woods Hole Oceanographic Institution using the PAb at the concentration recommended by Anderson et al. (3) (WH Low) or at 2 or 4 times the recommended PAb concentration (WH High). Counts of natural samples (WH Natural) and cultured A. anophagefferens (WH Cultures) are shown separately in panel B. LIU High indicates analyses performed at Long Island University using the PAb at 2.5 times the recommended PAb concentration. Dotted lines indicate lines of one-to-one correspondence between the methods. The solid line in panel C is the actual linear regression between the MAb method and hemacytometer counts.
The accuracy of the MAb method and the underestimation of brown tide abundances by the PAb method were supported by direct counts of brown tide cells obtained with a hemacytometer for samples with concentrations of A. anophagefferens that were high enough to estimate by direct microscopy. Based on this comparison, the PAb method yielded counts that were much lower than counts obtained by direct microscopy (Fig. 4B). A. anophagefferens counts in natural samples analyzed by the PAb method were only 30% of the counts obtained by light microscopy with a hemacytometer. Analysis of cultures of the brown tide alga indicated that the PAb method was not staining all algal cells. Seven cultures enumerated by the PAb technique yielded counts that averaged 40% of the values obtained by direct microscopical counts (Fig. 4B). Given that only A. anophagefferens was present in the cultures, it is clear that the PAb was not staining all cells. In contrast, there was excellent agreement between hemacytometer counts and the MAb method (Fig. 4C). The slope of the regression between these two counting methods was 0.98 (R2 = 0.81). The overall ratio of brown tide cells between the direct count and the MAb count was 0.81 for 26 samples.
Field study.
A brief survey of the spatial distribution of A. anophagefferens in Long Island coastal estuaries was conducted during late May 2000 to demonstrate the accuracy and rapidity of the new MAb method and its dynamic range. Samples collected at 19 locations in Peconic Bay, Shinnecock Bay, Moriches Bay, and Great South Bay ranged nearly 3 orders of magnitude (<5 × 103 to 1.66 × 106 A. anophagefferens cells ml−1; Fig. 5). High bloom abundances of the algae were present in eastern Great South Bay, with lesser but still substantial abundances (up to 6 × 105 cells ml−1) in Moriches Bay. Densities in Shinnecock Bay were >104 cells ml−1, but abundances at all of the Peconic Bay stations were at or below the practical limit of detection. Samples with visibly discolored water were diluted prior to analysis. All samples were analyzed in triplicate and run on a single microtiter plate together with standards.
FIG. 5.
Abundances of A. anophagefferens at 19 stations within Long Island coastal estuaries on 25 May 2000 as determined by the MAb method. The heights of the black columns indicate the abundances of the brown tide algae at each station location (indicated by the position of the station number). The abundances in the Peconic Bay estuaries (stations 1 to 5) were all <5,000 cells ml−1.
DISCUSSION
Traditional methods for phytoplankton identification (e.g., transmitted light and electron microscopy) have added greatly to our understanding of the community structure and dynamics of these protistan assemblages. However, the abundances and distributions of small (<5-μm) and morphologically nondescript species have been difficult to assess in ecological studies due to the inability of these methods to differentiate among them. Considerable effort has been expended in recent years to develop techniques that can assess the presence and abundance in nature of microorganismal species that cannot be quickly and easily identified by these traditional methods. These efforts have largely been divided between genetic (based on the detection of specific sequences of RNA or DNA) and immunological methods. The former approaches have received tremendous attention in recent years as DNA sequence information has amassed and as technological advances have reduced processing time and increased sensitivity (15). However, immunological approaches possess some inherent advantages that make them useful and sometimes preferable choices as tools for ecological studies.
The application of antibody probes for detecting target species in mixed assemblages does not require the probe to enter the cell or for the cells to be broken open if the antigens are located on the cell surface of the organism. In addition, many genetic approaches rely on amplification of DNA by using the PCR, which may introduce significant bias for quantitative analyses of organismal abundance. Also, the intensity of labeling with rRNA-based probes can be strongly affected by the number of ribosomes in a cell which, in turn, is affected by the physiological state of the target cell. The target number is more stable at least for some antigens. We have not detected notable differences in the reactivity of our MAb to A. anophagefferens cells in exponential growth phase or stationary growth phase. This finding is consistent with a previous study in which a direct comparison of rRNA-based probes and an MAb for the toxic dinoflagellate Alexandrium fundyense indicated that the MAb was less variable with the physiological state of the alga (4). Finally, immunological approaches have great potential for ecological studies if these approaches can be adapted to formats, such as the ELISA, that allow rapid processing of large numbers of samples.
The MAb LIZ 1F7-1E12 and its application in an ELISA format to quantify A. anophagefferens in natural samples and cultures was developed as an improvement to microscopic counts obtained with transmitted light or epifluorescence microscopy. Direct counts of A. anophagefferens by using transmitted light microscopy have been used occasionally to count “A. anophagefferens-like” cells in natural samples (7, 20). However, a number of algal species are similar in size and morphology to the brown tide alga, and thus the estimate obtained by direct microscopy for natural samples provides an accurate count of A. anophagefferens only if the alga numerically dominates the phytoplankton in the 1- to 5-μm size class. Therefore, direct counts are tenuous at best and not possible when A. anophagefferens is not at bloom abundances.
The most widely used method for counting A. anophagefferens has been the immunofluorescent technique with a PAb (3). This technique has been instrumental during the past decade in facilitating survey studies and ecological research on this important harmful bloom-forming species. Unfortunately, the PAb method suffers from a number of shortcomings, most notably lengthy processing and microscopic counting times and the potential for significant variability among investigators (for the preparation of stained samples, as well as microscopic enumeration). In addition, recent applications of the PAb method have significantly underestimated the abundances of A. anophagefferens in natural samples and in pure cultures of the brown tide alga (Fig. 4A).
Comparisons with the MAb method presented here and also with direct microscopic counts (using a hemacytometer) indicated that the PAb underestimated the actual abundance of A. anophagefferens by a factor of 2 to 3. Underestimation of cell number by using the PAb method may stem from problems associated with different batches of the antisera. Increasing the concentration of the PAb resulted in better agreement between the MAb (ELISA format) method and PAb (fluorescence microscopy) methods (although not for all samples; compare WHOI “high” and LIU “high” samples in Fig. 4A). This result may indicate that the reactivity of subsequent batches of serum differed from the original batch.
Given these considerations, the MAb method described here provides a marked improvement over the PAb technique for accurately estimating the abundance of A. anophagefferens in mixed natural assemblages of phytoplankton. In addition, the use of the ELISA format and 96-well microtiter plates allows rapid, simultaneous processing of large numbers of samples as well as a complete set of standards in a relatively short time (3 to 4 h). This rate of sample processing is manyfold greater than the rate that can be achieved by using the PAb method. For example, an experienced investigator might complete six samples per day (without replication and without positive and negative controls) with the PAb epifluorescence microscopic method. In comparison, one person could easily analyze two microtiter plates by using the MAb method during the same period (total of 192 wells). Assuming that 24 to 30 wells per plate are used for standards and controls, 132 to 144 wells can be used for samples. With skill, that number can be increased (i.e., two plates can be processed at the same time).
The MAb exhibited very high specificity for the brown tide alga (Table 1). The degree of specificity of an antibody preparation depends on the immunogenicity and number of antigenic determinants on the molecule(s) used for antibody production. Clearly, molecules present on the surface of whole, preserved A. anophagefferens cells are immunogenic since both polyclonal and MAbs have now been raised against them. Data in Fig. 1 show that the number of determinants on the cell surface must be high because immunofluorescent staining yields a bright, uniform stain over the whole cell. Identical immunofluorescent staining was also performed with MAb LIZ 1F7-1E12 and nontarget cultured protists including I. galbana, H. akashiwo, M. polymorphus, and an unknown minute alga (BT3). None of these nontarget species, either alone or in combination with A. anophagefferens, were stained by the MAb (negative data not shown). The high specificity exhibited by the MAb for A. anophagefferens allowed for its application to the ELISA format. Attempts to adapt the PAb method to this format resulted in very high background absorbances, presumably due to cross-reactivity with nontarget microorganisms or debris. Cross-reactions of MAb LIZ 1F7-1E12 to nontarget species were extremely low at antibody concentrations used to detect the brown tide alga (Table 1). The very high specificity of the MAb is presumably the reason for our success with adapting this antibody to the ELISA format.
All three strains of A. anophagefferens isolated from different water masses and in different years reacted equally well with the MAb. Some differences in reactivity might be expected in comparing strains from different locales. The ability of the MAb to react strongly with all three cultured isolates appears to indicate that A. anophagefferens populations of the mid-Atlantic United States constitute a very closely related group of strains. Very high DNA sequence similarity among these clones supports this conjecture (5).
The dynamic range of the standard curves obtained with our ELISA procedure was sufficient to accommodate water samples with A. anophagefferens ranging up to (2.5 to 3) × 105 cells ml−1 without prior sample manipulation. Samples with abundances greater than this amount were analyzed by dilution of the samples with preserved, filtered seawater. The serial dilution of natural samples provided excellent linearity, as evidenced by the close agreement of abundances calculated for three diluted, natural samples (Fig. 3A). This result allowed the determination of the abundances of A. anophagefferens in the samples based on the absorbances of the diluted samples and their dilution factors. Because of the wide dynamic range of the standard curves (Fig. 2) and the large number of samples that can be processed simultaneously by this method, samples that are suspected of containing high abundances of A. anophagefferens (e.g., samples with discolored water) can be diluted as a routine precaution to avoid having to rerun samples. This additional step added very little to sample processing time. Thus, our procedure can be used effectively to analyze water samples that vary in the abundance of A. anophagefferens over nearly 3 orders of magnitude (∼5 × 103 to >2 × 106 cells ml−1).
The type of preservative was found to be an important factor in the MAb method. Preservation with acid Lugol’s solution resulted in a significant underestimation of the abundance of A. anophagefferens in cultures compared to samples preserved with glutaraldehyde (Fig. 3B). This result may indicate an alteration of the epitope by the Lugol’s solution that makes it less reactive to the MAb. If brown tide cell counts are desired from Lugol’s solution-preserved samples, one strategy would be to construct standard curves with a culture of A. anophagefferens that has been preserved with Lugol’s solution. This approach would decrease the sensitivity of the method but should provide accurate counts of the alga at abundances above the limit of detection. This conclusion is based on the fact that, although counts obtained from the Lugol’s solution-preserved samples were lower than counts from glutaraldehyde-preserved samples, the relationship remained linear (Fig. 3B). This is an encouraging result for obtaining counts of A. anophagefferens from archived plankton samples preserved in Lugol’s solution.
One minor disadvantage of the MAb method relative to the PAb method was the slightly elevated functional lower limit of detection for the method (∼5,000 cells ml−1 for natural samples). Theoretically, the lower limit of detection for the PAb method is a few hundred to several hundred cells ml−1, depending on the volume of water filtered. However, this lower limit is predicated on observing one or a few A. anophagefferens cells during microscopical examination of the preparation. The very low number of cells observed to achieve this lower limit produces very high errors associated with the resulting abundance estimates. Thus, in practice the detection limits of the methods are not substantially different. Moreover, the ecological effects of A. anophagefferens appear to be manifested at abundances well above the limit of detection of either method (8).
Application of the MAb method to a survey of A. anophagefferens in Long Island coastal estuaries demonstrated the utility of this method. A. anophagefferens often occurs over a wide range of abundances within a relatively restricted geographic area, and a range of nearly 3 orders of magnitude was observed in the survey conducted in May 2000 (Fig. 5). During this survey a massive bloom was taking place in Great South Bay, while Peconic Bay estuaries had cell counts that were below the limit of detection. Waters of Moriches Bay and Shinnecock Bay had concentrations that were intermediate to these extremes. The ability of the MAb method to document abundances of the brown tide algae from bloom conditions to near-background demonstrates the versatility of the approach. Moreover, all samples for this survey were analyzed and computations were completed within 4 h, providing the possibility of obtaining information on bloom progression in near-real time. The dynamic range of the method and the speed of analysis make this method an important new tool for ecological studies and monitoring programs of this harmful algal bloom species.
Acknowledgments
We gratefully acknowledge the support of St. Gabriel's Youth House and Katherine Black, who provided housing and access to coastal lagoons for sampling. John Bredemeyer assisted with the PAb staining method at the SCDHS. Donald M. Anderson graciously provided PAb for immunological staining and counting of the brown tide algae by epifluorescence microscopy.
This work was supported by a grant from The Seaver Institute.
REFERENCES
- 1.Andersen, R. A., G. W. Saunders, M. P. Paskind, and J. P. Sexton. 1993. Ultrastructure and 18S rRNA gene sequence for Pelagomonas calceolata gen. et sp. nov. and the description of a new algal class, the Pelagophyceae classis nov. J. Phycol. 29:701-715. [Google Scholar]
- 2.Anderson, D. M., B. A. Keafer, D. M. Kulis, R. M. Waters, and R. Nuzzi. 1993. An immunofluorescent survey of the brown tide chrysophyte Aureococcus anophagefferens along the northeast coast of the United States. J. Plankton Res. 15:563-580. [Google Scholar]
- 3.Anderson, D. M., D. M. Kulis, and E. M. Cosper. 1989. Immunofluorescent detection of the brown tide organism Aureococcus anophagefferens, p. 265-294. In E. M. Cosper, V. M. Bricelj, and E. J. Carpenter (ed.), Novel phytoplankton blooms: causes and impacts of recurrent brown tides and other unusual blooms, vol. 35. Springer-Verlag, Berlin, Germany.
- 4.Anderson, D. M., D. M. Kulis, B. A. Keafer, and E. Berdalet. 1999. Detection of the toxic dinoflagellate Alexandrium fundyense (Dinophyceae) with oligonucleotide and antibody probes: variability in labeling intensity with physiological condition. J. Phycol. 35:870-883. [Google Scholar]
- 5.Bailey, J. C., and R. A. Andersen. 1999. Analysis of clonal cultures of the brown tide algae Aureococcus and Aureoumbra (Pelagophyceae) using 18S rRNA, rbcL, and RUBISCO spacer sequences. J. Phycol. 35:570-574. [Google Scholar]
- 6.Bates, S. S., C. Léger, B. A. Keafer, and D. M. Anderson. 1993. Discrimination between domoic-acid-producing and non-toxic forms of the diatom Pseudonitzschia pungens using immunofluorescence. Mar. Ecol. Prog. Ser. 100:185-195. [Google Scholar]
- 7.Berg, G. M., P. M. Glibert, M. W. Lomas, and M. A. Burford. 1997. Organic nitrogen uptake and growth by the chrysophyte Aureococcus anophagefferens during a brown tide event. Mar. Biol. 129:377-387. [Google Scholar]
- 8.Bricelj, V. M., S. P. MacQuarrie, and R. A. Schaffner. 2001. Differential effects of Aureococcus anophagefferens isolates (“brown tide”) in unialgal and mixed suspensions on bivalve feeding. Mar. Biol. 139:605-615. [Google Scholar]
- 9.Bricelj, W. M., and D. J. Lonsdale. 1997. Aureococcus anophagefferens: causes and ecological consequences of brown tides in U.S. mid-Atlantic coastal waters. Limnol. Oceanogr. 42:1023-1038. [Google Scholar]
- 10.Campbell, L., and R. Iturriaga. 1988. Identification of Synechococcus spp. in the Sargasso Sea by immunofluorescence and fluorescence excitation spectroscopy performed on individual cells. Limnol. Oceanogr. 33:1196-1201. [Google Scholar]
- 11.Campbell, L., L. P. Shapiro, and E. Haugen. 1994. Immunochemical characterization of eukaryotic ultraplankton from the Atlantic and Pacific oceans. J. Plankton Res. 16:35-51. [Google Scholar]
- 12.Caron, D. A., E. L. Lim, H. Kunze, E. M. Cosper, and D. M. Anderson. 1989. Trophic interactions between nano- and microzooplankton and the “brown tide,” p. 265-294. In E. M. Cosper, V. M. Bricelj, and E. J. Carpenter (ed.), Novel phytoplankton blooms: causes and impacts of recurrent brown tides and other unusual blooms, vol. 35. Springer-Verlag, Berlin, Germany.
- 13.Cosper, E. M. 1987. Culturing the “brown tide” alga. Appl. Phycol. Forum 4:3-5. [Google Scholar]
- 14.Cosper, E. M., R. T. Garry, A. J. Milligan, and M. H. Coall. 1993. Iron, selenium and citric acid are critical to the growth of the “brown tide” microalga, Aureococcus anophagefferens, p. 667-673. In G. J. Smayda and Y. Shimizu (ed.), Toxic phytoplankton blooms in the sea. Elsevier Science Publishers, New York, N.Y.
- 15.Coyne, K. J., D. A. Hutchins, C. E. Hare, and S. C. Cary. 2001. Assessing temporal and spatial variability in Pfiesteria piscicida distributions using molecular probing techniques. Aquat. Microb. Ecol. 24:275-285. [Google Scholar]
- 16.Damaj, B. B. 2000. Immunological reagents and solutions: a laboratory handbook. Eaton Publishing, Natick, Mass.
- 17.Gobler, C. J., and S. A. Sañudo-Wilhelmy. 2001. Effects of organic carbon, organic nitrogen, inorganic nutrients and iron additions on the growth of phytoplankton and bacteria during a brown tide bloom. Mar. Ecol. Prog. Ser. 209:19-34. [Google Scholar]
- 18.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 29-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Publishing, Inc., New York, N.Y.
- 19.Lin, S., and E. J. Carpenter. 1996. An empirical protocol for whole-cell immunofluorescence of marine phytoplankton. J. Phycol. 32:1083-1094. [Google Scholar]
- 20.Lomas, M. W., P. M. Glibert, G. M. Berg, and M. Burford. 1996. Characterization of nitrogen uptake by natural populations of Aureococcus anophagefferens (Chrysophyceae) as a function of incubation duration, substrate concentration, light, and temperature. J. Phycol. 32:907-916. [Google Scholar]
- 21.Lopez-Barrerio, T., T. A. Villareal, and S. L. Morton. 1998. Development of an antibody against the Texas brown tide Aureoumbra lagunensis, p. 263-265. In B. Reguera, J. Blanco, M. L. Fernandez, and T. Wyatt (ed.), Harmful algae. Xunta de Galicia and Intergovernmental Oceanographic Commission of UNESCO, Paris, France.
- 22.Mendoza, H., V. López-Rodas, S. González-Gil, A. Aguilera, and E. Costas. 1995. Use of polyclonal antisera and blocking of the antibodies in the identification of marine dinoflagellates: species-specific and clone-specific antisera against Gymnodinium and Alexandrium. J. Exp. Mar. Biol. Ecol. 186:103-115. [Google Scholar]
- 23.Nuzzi, R., and R. Waters. 1989. The spatial and temporal distribution of “brown tide” in eastern Long Island, p. 117-138. In E. M. Cosper, V. M. Bricelj, and E. J. Carpenter (ed.), Novel phytoplankton blooms: causes and impacts of recurrent brown tides and other unusual blooms, vol. 35. Springer-Verlag, Berlin, Germany.
- 24.Shapiro, L. P., L. Campbell, and E. M. Haugen. 1989. Immunochemical recognition of phytoplankton species. Mar. Ecol. Prog. Ser. 57:219-224. [Google Scholar]
- 25.Sherr, E. B., D. A. Caron, and B. F. Sherr. 1993. Staining of heterotrophic protists for visualization via epifluorescence microscopy, p. 213-227. In P. Kemp, B. Sherr, E. Sherr, and J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 26.Sherr, E. B., and B. F. Sherr. 1993. Preservation and storage of samples for enumeration of heterotrophic protists, p. 207-212. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 27.Sieburth, J. M., P. W. Johnson, and P. E. Hargraves. 1988. Ultrastructure and ecology of Aureococcus anophagefferens gen. et sp. nov. (Chrysophyceae): the dominant picoplankter during a bloom in Narragansett Bay, Rhode Island, summer 1985. J. Phycol. 24:416-425. [Google Scholar]
- 28.Stoecker, D. K., D. J. Gifford, and M. Putt. 1994. Preservation of marine planktonic ciliates: losses and cell shrinkage during fixation. Mar. Ecol. Prog. Ser. 110:293-299. [Google Scholar]
- 29.Uchida, A., K. Nagasaki, S. Hiroishi, and Y. Ishida. 1989. The application of monoclonal antibodies to an identification of Chattonella marina and Chattonella antiqua. Nippon Suisan Gakkaishi 55:721-725. [Google Scholar]
- 30.Utermöhl, H. 1958. Zur Vervollkommung der quantitativen Phytoplankton-Methodik. Mitt. Int. Ver. Limnol. 9:1-38.
- 31.Vrieling, E. G., and D. M. Anderson. 1996. Immunofluorescence in phytoplankton research: applications and potential. J. Phycol. 32:1-16. [Google Scholar]
- 32.Vrieling, E. G., G. Vriezekolk, W. W. C. Gieskes, M. Veenhuis, and W. Harder. 1996. Immuno-flow cytometric identification and enumeration of the ichthyotoxic dinoflagellate Gyrodinium aureolum Hulburt in artifically mixed algal populations. J. Plankton Res. 18:1503-1512. [Google Scholar]