Abstract
Variation in gene content among strains of a bacterial species contributes to biomedically relevant differences in phenotypes such as virulence and antimicrobial resistance. Group A Streptococcus (GAS) causes a diverse array of human infections and sequelae, and exhibits a complex pathogenic behavior. To enhance our understanding of genotype-phenotype relationships in this important pathogen, we determined the complete genome sequences of four GAS strains expressing M protein serotypes (M2, M4, and 2 M12) that commonly cause noninvasive and invasive infections. These sequences were compared with eight previously determined GAS genomes and regions of variably present gene content were assessed. Consistent with the previously determined genomes, each of the new genomes is ∼1.9 Mb in size, with ∼10% of the gene content of each encoded on variably present exogenous genetic elements. Like the other GAS genomes, these four genomes are polylysogenic and prophage encode the majority of the variably present gene content of each. In contrast to most of the previously determined genomes, multiple exogenous integrated conjugative elements (ICEs) with characteristics of conjugative transposons and plasmids are present in these new genomes. Cumulatively, 242 new GAS metagenome genes were identified that were not present in the previously sequenced genomes. Importantly, ICEs accounted for 41% of the new GAS metagenome gene content identified in these four genomes. Two large ICEs, designated 2096-RD.2 (63 kb) and 10750-RD.2 (49 kb), have multiple genes encoding resistance to antimicrobial agents, including tetracycline and erythromycin, respectively. Also resident on these ICEs are three genes encoding inferred extracellular proteins of unknown function, including a predicted cell surface protein that is only present in the genome of the serotype M12 strain cultured from a patient with acute poststreptococcal glomerulonephritis. The data provide new information about the GAS metagenome and will assist studies of pathogenesis, antimicrobial resistance, and population genomics.
Introduction
Study of intraspecies variation in chromosomal gene content and sequence diversity has become an area of considerable interest in recent years [1]–[9]. Several factors have prompted research on this topic. First, the genome sequences that are now available for at least one member of many eukaryotic and prokaryotic species provide reference templates for indexing intraspecies diversity (cmr.tigr.org and www.genomesonline.org). Second, analysis of intraspecies genetic diversity is a crucial component of studies designed to understand the molecular basis of phenotypic variation in traits such as organism behavior, disease susceptibility, and response to pharmacologic agents. Third, sequences that are polymorphic within members of the same species are used in molecular epidemiology studies to distinguish among closely related organisms for public health and forensic purposes. Fourth, given that mutation, selection, and inheritance are the basis of evolution, comparison of intraspecies genetic variation provides insights into the molecular processes underlying evolution. A major factor that has contributed to increased interest in species-level genetic diversity among pathogenic microbes is the need to understand the molecular basis of biomedically relevant topics such as strain emergence, virulence differences, disease manifestation, and evolution of pathogenic traits.
The human bacterial pathogen group A Streptococcus (GAS) is an ideal model organism for studying molecular processes generating intraspecies genomic diversity and the contribution of specific genetic differences to host-pathogen interactions. GAS causes a wide range of infections, including pharyngitis, cellulitis, sepsis, necrotizing fasciitis, and post-infection sequelae including acute rheumatic fever (ARF) and acute poststreptococcal glomerulonephritis (APSGN) [10]–[13]. For many decades, GAS strains have been classified based on serologic differences in M protein, a highly polymorphic cell-surface virulence factor [14], [15]. More than 125 M protein types and emm gene types have been identified (for convenience we will use the terms M protein serotype and emm type interchangeably), and the number of subtypes is far higher ([16]; www.cdc.gov/ncidod/biotech/strep/emmtypes.htm). Importantly, epidemiologic studies conducted over many decades have repeatedly found that certain M protein types are non-randomly associated with particular human infections [10], [17]–[22]. For example, serotype M1 and M3 GAS strains commonly cause pharyngitis and invasive infections, and M28 GAS strains are significantly overrepresented among puerperal sepsis and neonatal GAS infections [17], [18], [21]–[25]. Thus, there is a rich phenotypic and clinical framework available for interpreting genome sequence information. Moreover the relatively modest size, low G+C content, single chromosome, rare occurrence of extra-chromosomal elements, and lack of extensive repetitive sequences permits complete GAS genome sequences to be determined at reduced cost, time, and effort relative to many other microbial pathogens [26]–[31]. In addition, the increasing number of reports from many countries describing the emergence of GAS strains resistant to antimicrobial agents such as erythromycin and tetracycline [32]–[44] provides a public health impetus for sequencing the genome of additional strains. In this manuscript we describe new findings based on the genome sequence of serotype M2, M4, and two M12 strains, with a focus on unique gene content encoded by integrated conjugative elements (ICEs).
Results
Strain selection
The strains selected for sequencing were chosen on the basis of a variety of characteristics. The primary criterion was that the strains were of an unsequenced M protein serotype that abundantly cause noninvasive and invasive infections. Consistent with this serotype M2, M4 and M12 strains are all common isolates of noninvasive and invasive infections in the United States and other developed countries (see www.cdc.gov/ncidod/dbmd/abcs/survreports.htm). Strains representing these serotypes have not been previously sequenced.
An additional criterion was an epidemiologic association with a distinct pathogenic character in order to facilitate assessment of gene content influencing strain genotype-patient disease phenotype relationships or epidemic behavior. Serotype M2 strain MGAS10270 was obtained from a patient in Texas with pharyngitis in the late 1990s. Serotype M2 strains are associated with female uritogenital tract infections [18], [45], [46]. Serotype M4 strain MGAS10750 was cultured from a patient with pharyngitis in Florida in 2001. This strain is resistant to erythromycin (MIC 1 µg/ml) and is PCR positive for the erm(A) gene. Erythromycin resistant serotype M4 strains have caused epidemic outbreaks of infection [18], [47], [48]. Serotype M12 strain MGAS2096 was isolated from a patient with poststreptococcal glomerulonephritis in Trinidad in 1960. The isolation of GAS from patients with APSGN is rare as in most cases the infection has cleared prior to glomerulonephritis manifestation. This organism, also known as strain A374, has been studied previously [49]–[51]. Given that serotype M12 strain MGAS2096 was isolated over 45 years ago, and changes in prophage content have been associated with rapid shifts in GAS pathogenesis, we also elected to sequence a contemporary M12 strain for comparison. Serotype M12 strain MGAS9429 was cultured from a pediatric patient with pharyngitis in the Texas in 2001. Strain MGAS9429 has the most common prophage virulence gene profile detected from among 33 contemporary serotype M12 strains studied (J.M.M., unpublished data).
A repeated epidemiological finding dating back to the 1930s is that strains of certain M protein serotypes are nonrandomly associated with the poststreptococcal infection sequela, ARF and APSGN [11], [12], [52], [53]. Observations of distinct differences in disease manifestation from these studies lead to the supposition that rheumatogenicity and nephritogenicity may be independent properties of two separate GAS genetic lineages that broadly correspond with strains most commonly causing throat and skin infections, respectively. The presence or absence of the serum opacity factor (sof) gene encoding for a lipoproteinase that confers the ability to opacify human serum is considered a marker of the two lineages [54], [55]. Excepting serotype M28 strain MGAS6180, all of the previously sequenced GAS strains are sof negative and represent serotypes considered rheumatogenic (2 each of M1 and M3, and one each of M5, M6, and M18) (Table 1). The four strains selected for sequencing and described here are all sof positive representing serotypes considered nephritogenic. Thus in addition to their other disease associations, these four genomes also provide data for assessing genetic differences between the posited GAS skin/nephritogenic and throat/rheumatogenic lineages.
Table 1. Characteristics of Sequenced GAS Strains.
| Strain | M type | MLST | sof | Size, bp | % G+C | CDS | Elements | ||
| No. | % | Phage | ICE | ||||||
| SF370 | 1 | 28 | neg | 1,852,441 | 38.51 | 1,697 | 83.79 | 4 | 1 |
| MGAS5005 | 1 | 28 | neg | 1,838,554 | 38.53 | 1,865 | 86.39 | 3 | 1 |
| MGAS10270 | 2 | 55 | pos | 1,928,252 | 38.43 | 1,987 | 87.18 | 5 | 2 |
| MGAS315 | 3 | 15 | neg | 1,900,521 | 38.59 | 1,865 | 85.76 | 6 | 0 |
| SSI-1 | 3 | 15 | neg | 1,894,275 | 38.55 | 1,861 | 84.61 | 6 | 0 |
| MGAS10750 | 4 | 39 | pos | 1,937,111 | 38.31 | 1,979 | 87.16 | 4 | 2 |
| Manfredo | 5 | 99 | neg | 1,841,271 | 38.62 | 1,803 | 86.72 | 5 | 0 |
| MGAS10394 | 6 | 382 | neg | 1,899,877 | 38.69 | 1,886 | 87.28 | 8* | 0* |
| MGAS2096 | 12 | 36 | pos† | 1,860,355 | 38.73 | 1,898 | 87.39 | 2 | 2 |
| MGAS9429 | 12 | 36 | pos† | 1,836,467 | 38.54 | 1,878 | 87.81 | 3 | 1 |
| MGAS8232 | 18 | 42 | neg | 1,895,017 | 38.55 | 1,845 | 85.23 | 5 | 0 |
| MGAS6180 | 28 | 52 | pos | 1,897,573 | 38.35 | 1,894 | 87.09 | 4 | 3 |
Element 10394.4 is composite element that has a phage-like 5′ end and an ICE-like 3′end.
Strains MGAS2096 and MGAS9429, like most serotype M12 strains, although sof positive by PCR are phenotypically Sof negative due to internal gene mutations.
Overview of general genome features
Consistent with the genomes of eight previously sequenced GAS strains [26]–[31], [56], the serotype M2, M4 and both M12 genomes each is a single circular chromosome of ∼1.9 Mb (Table 1, Fig. 1). The percent G+C content of these genomes is approximately 38.5%, essentially identical to the other eight sequenced GAS genomes (range, 38.31%–38.73%). Each of these genomes has six operons encoding adjacent 5S, 16S, and 23S ribosomal RNAs. Each has the multi-locus sequence type (MLST) that is most common for their M type [57]. Predicted coding sequence composes a similar portion of each of the genomes. Among the 12 sequenced GAS genomes, on average coding sequence constitutes 86.4% of each genome, with a mean gene size of 870 nt.
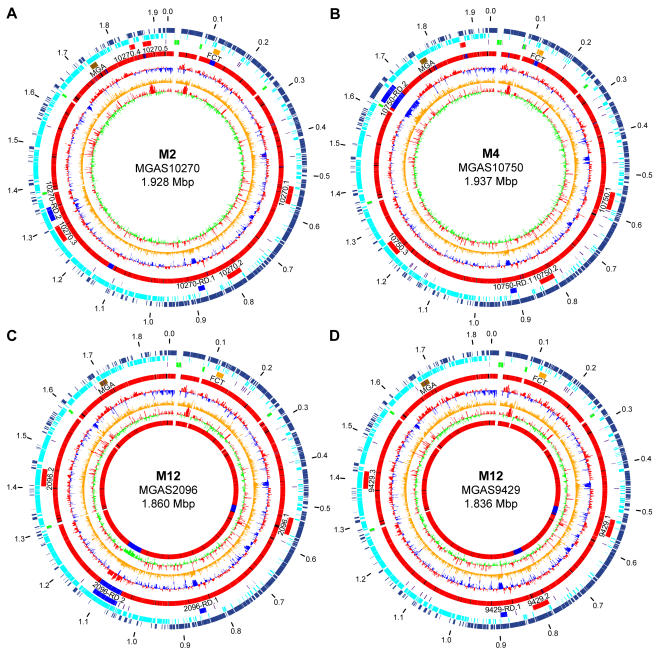
Figure 1. Genome circular atlases.
(A) MGAS10270, (B) MGAS10750, (C) MGAS2096, and (D) MGAS9429. Data from outermost to innermost circles are in the following order. Genome size in mega base pairs (circle 1). Annotated coding sequences on the forward (circle 2) and reverse strands (circle 3) are in dark and light blue, respectively. Reference landmarks (circle 4) illustrated are: ribosomal RNAs in green, FCT region in gold, transposons in purple, prophages in red, ICEs in royal blue, and Mga regulon region in brown. Comparison of gene content to the 11 other sequenced GAS strains (circle 5) is given as a gradient of nucleotide sequence similarity from low in blue to high in red. CDS percent G+C content (circle 6) with greater than and less than average in red and blue, respectively. Net divergence of CDS dinucleotide composition (circle 7) from the average is in orange. Codon adaptation index, that is codon use consistent with that of highly expressed genes (circle 8) with greater than and less than average in red and green, respectively. Additionally for the two serotype M12 strains a comparison of gene content relative to each other (circle 9) is given as a gradient of nucleotide sequence similarity from low in blue to high in red.
The majority of each genome (>85%) is conserved in gene content and context relative to the others (Fig. 1). This core sequence is conserved at greater than 98% nucleotide identity and comprises the endogenous “core” of the GAS metagenome (i.e., the common part of the chromosome that does not include obvious exogenous genetic elements such as prophages and ICEs). The endogenous core encodes many proven or putative secreted virulence factors, including M protein, streptolysin O, streptolysin S, streptokinase, pyrogenic toxin superantigens (SmeZ), collagen-like proteins (SclA and SclB), and proteases (SpeB, Mac, and ScpA) [11], [12] to name a few. The average size of the 12 sequenced genomes is 1,882 kb, and the difference between the smallest and largest genome is 100.6 kb or 5.3% of the average size. The extent of size variation in the GAS genomes is similar to that reported for Staphylococcus aureus genomes, greater than found in Chlamydia trachomatis and Mycobacterium tuberculosis, and considerably less than for certain Escherichia coli strains [1], [2], [4]–[7], [9], [58].
Overview of exogenous genetic elements, prophages, and ICEs
To identify regions of difference among the sequenced GAS genomes they were aligned pair-wise. This revealed regions (5 kb–63 kb) differing in gene content and/or context that disrupted the continuity of the aligned sequences (this is illustrated for the four newly sequenced genomes in Fig. 2). Bioinformatic analysis found that these regions of difference contain gene content similar to prophages and ICEs. Twenty-one exogenous genetic elements (14 prophage-like and 7 ICE-like) ranging from 12 kb to 63 kb in size were identified in the serotype M2 (5 Φ, 2 ICE), M4 (4 Φ, 2 ICE), and M12 strains MGAS2096 (2 Φ, 2 ICE) and MGAS9429 (3 Φ, 1 ICE) genomes (Fig. 1, Table 2). In total, we identified 67 obvious exogenous genetic elements (55 prophages and 12 ICEs) integrated at 21 distinct loci of the core chromosome in the 12 GAS genomes (Fig. 3, Table 2). Based on gene content, some of the smaller elements likely are remnants of ancestral genetic elements that have undergone reductive evolution. However, we cannot exclude the possibility that these elements are mobile and were acquired by lateral gene transfer. As most of these exogenous genetic elements have not been shown experimentally to be transferable we refer to them as putatively-mobile.
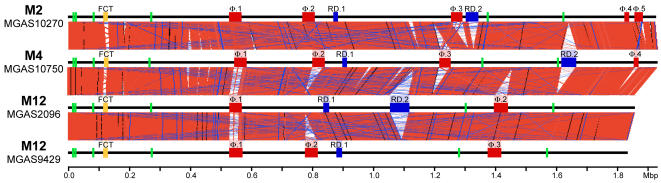
Figure 2. Aligned GAS genomes.
Illustrated are linear diagrams of the four newly determined GAS genome sequences and regions of conserved gene content in pair-wise comparisons. Shown for each genome diagram in green are the six rRNA operons, in red are prophages, and in blue are ICEs. Whole-genome comparisons were made using BLASTN (www.webact.org, e = 1×10−4, word size = 18 nt) and the graphic depictions of the alignments were made using the Artemis Comparison Tool (www.sanger.ac.uk/Software/ACT/). Regions of conserved syntenic gene content are indicated by blocks of salmon linking the stacked genome diagrams. Nearly all regions of discontinuity in the genome alignments are attributable to exogenous genetic elements.
Table 2. Characteristics of GAS Exogenous Elements.
| M-type, Strain | Exogenous Element | Virulence Gene(s) | CDS Start | CDS Stop | Size (kbp) | Percent G+C | |
| M1, SF370 | |||||||
| 370.1 | speC-spd1 | 0655 | 0712 | 40.9 | 39.3 | ||
| 370.2 | speH-speI | 0937 | 1008 | 42.5 | 38.0 | ||
| 370-RD.1 | srtA | 1075 | 1088 | 12.2 | 33.2 | ||
| 370.3 | spd3 | 1436 | 1488 | 33.5 | 38.0 | ||
| 370.4 | none* | 2122 | 2147 | 13.6 | 36.8 | ||
| M1, MGAS5005 | |||||||
| 5005-RD.1 | srtA | 0797 | 0816 | 12.2 | 33.2 | ||
| 5005.1 | speA2 | 0995 | 1052 | 38.1 | 38.0 | ||
| 5005.2 | spd3 | 1168 | 1222 | 33.4 | 37.7 | ||
| 5005.3 | sda | 1414 | 1467 | 40.8 | 39.8 | ||
| M2, MGAS10270 | |||||||
| 10270.1 | speC-spd1 | 0536 | 0598 | 44.1 | 38.0 | ||
| 10270.2 | spd3 | 0796 | 0853 | 37.9 | 38.2 | ||
| 10270-RD.1 | srtA | 0910 | 0932 | 17.5 | 31.9 | ||
| 10270.3 | speK-sla | 1297 | 1361 | 41.8 | 38.5 | ||
| 10270-RD.2 | R28 | 1378 | 1411 | 34.7 | 34.5 | ||
| 10270.4 | none | 1874 | 1896 | 14.3 | 36.9 | ||
| 10270.5 | none | 1917 | 1951 | 11.8 | 36.6 | ||
| M3, MGAS315 | |||||||
| 315.1 | none | 0681 | 0736 | 39.5 | 37.8 | ||
| 315.2 | ssa | 0919 | 0978 | 41.1 | 38.3 | ||
| 315.3 | spd4 | 1094 | 1145 | 34.4 | 38.0 | ||
| 315.4 | speK-sla | 1203 | 1266 | 61.8 | 38.6 | ||
| 315.5 | speA3 | 1300 | 1354 | 38.2 | 38.0 | ||
| 315.6 | sdn | 1408 | 1458 | 40.0 | 39.7 | ||
| M3, SSI-1 | |||||||
| SPsP1 | sdn | 0408 | 0456 | 40.0 | 39.7 | ||
| SPsP2 | speA3 | 0507 | 0561 | 38.2 | 38.1 | ||
| SPsP3 | speK-sla | 0597 | 0659 | 41.8 | 38.6 | ||
| SpsP4 | spd4 | 0717 | 0771 | 34.4 | 38.0 | ||
| SPsP5 | none | 0877 | 0937 | 41.0 | 38.2 | ||
| SPsP6 | ssa | 1118 | 1172 | 39.9 | 37.7 | ||
| M4, MGAS10750 | |||||||
| 10750.1 | speC-spd1 | 0560 | 0622 | 43.8 | 38.0 | ||
| 10750.2 | spd3 | 0831 | 0889 | 37.9 | 38.3 | ||
| 10750-RD.1 | srtA | 0945 | 0967 | 17.6 | 31.9 | ||
| 10750.3 | ssa | 1276 | 1328 | 35.6 | 38.0 | ||
| 10750-RD.2 | erm(A) | 1679 | 1719 | 48.9 | 31.2 | ||
| 10750.4 | none | 1897 | 1921 | 13.9 | 35.2 | ||
| M5, Manfredo | |||||||
| man.1 | spd3 | 0471 | 0535 | 41.4 | 38.6 | ||
| man.2 | spd4 | 0631 | 0692 | 36.0 | 37.9 | ||
| man.3 | speH-speI | 1021 | 1070 | 39.9 | 38.2 | ||
| man.4 | speC-spd1 | 1263 | 1322 | 40.7 | 38.2 | ||
| man.5 | none | 1764 | 1779 | 12.3 | 36.9 | ||
| M6, MGAS10394 | |||||||
| 10394.1 | sdn | 0020 | 0068 | 39.9 | 39.7 | ||
| 10394.2 | speA4 | 0733 | 0741 | 8.8 | 32.2 | ||
| 10394. 3 | speK-sla | 0982 | 1026 | 33.2 | 38.0 | ||
| 10394.4† | mef(A), R6 | 1123 | 1173 | 58.9 | 38.2 | ||
| 10394.5 | speC-spd1 | 1194 | 1242 | 31.9 | 38.4 | ||
| 10394.6 | sda | 1338 | 1366 | 24.5 | 40.4 | ||
| 10394.7 | spd3 | 1540 | 1562 | 21.1 | 39.5 | ||
| 10394.8 | none | 1804 | 1824 | 13.9 | 36.6 | ||
| M12, MGAS2096 | |||||||
| 2096.1 | speC-spd1 | 0553 | 0602 | 41.3 | 39.1 | ||
| 2096-RD.1 | srtA | 0869 | 0890 | 17.5 | 31.9 | ||
| 2096-RD.2 | tet(O) | 1103 | 1159 | 63.0 | 43.1 | ||
| 2096.2 | sda | 1440 | 1492 | 40.8 | 39.8 | ||
| M12, MGAS9429 | |||||||
| 9429.1 | speC-spd1 | 0532 | 0594 | 41.3 | 38.0 | ||
| 9429.2 | speH-speI | 0795 | 0851 | 17.5 | 38.1 | ||
| 9429-RD.1 | srtA | 0911 | 0934 | 63.0 | 31.9 | ||
| 9429.3 | sda | 1415 | 1468 | 40.8 | 39.8 | ||
| M18, MGAS8232 | |||||||
| 8232.1 | speA1 | 0336 | 0394 | 39.0 | 37.7 | ||
| 8232.2 | speC-spd1 | 0716 | 0779 | 41.1 | 38.2 | ||
| 8232.3 | speL-speM | 1238 | 1309 | 46.7 | 37.6 | ||
| 8232.4 | spd3 | 1444 | 1506 | 38.4 | 37.7 | ||
| 8232.5 | sda | 1745 | 1808 | 41.8 | 38.5 | ||
| M28, MGAS6180 | |||||||
| 6180-RD.0 | srtA | 0771 | 0793 | 17.5 | 31.9 | ||
| 6180.1 | speC-spd1 | 0967 | 1033 | 46.3 | 37.2 | ||
| 6180-RD.1 | none | 1079 | 1089 | 11.1 | 30.7 | ||
| 6180.2 | speK-sla | 1220 | 1285 | 42.3 | 38.4 | ||
| 6180-RD.2 | R28 | 1302 | 1337 | 36.3 | 35.0 | ||
| 6180.3 | none | 1789 | 1813 | 14.3 | 36.8 | ||
| 6180.4 | none | 1840 | 1864 | 11.8 | 36.6 | ||
There are no known or obvious virulence genes.
Element 10394.4 is chimeric, composed of a phage-like 5′ end and an ICE-like 3′end.
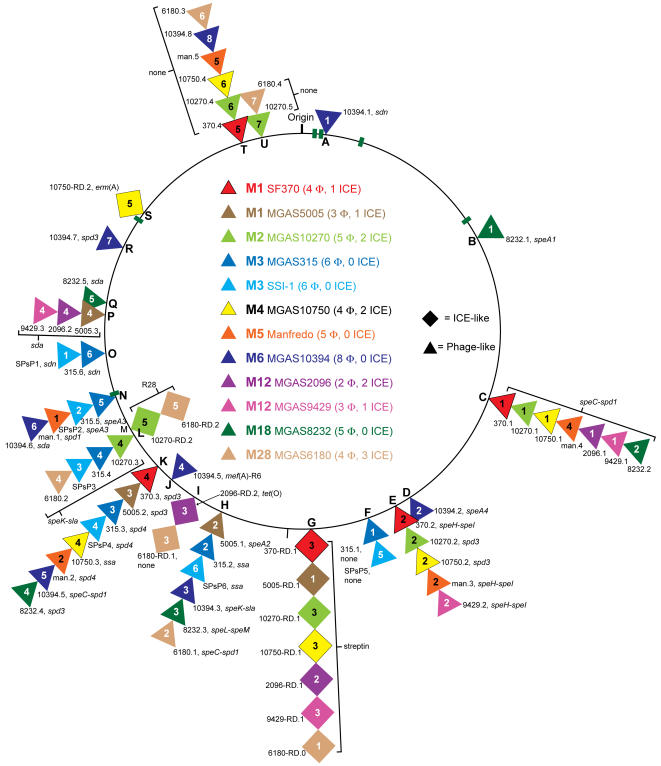
Figure 3. GAS metagenome exogenous elements.
Illustrated are loci of integration of phages and ICEs into the core chromosome. Prophages are indicated with triangles and ICEs with squares. Stacked triangles and squares indicate a common integration site. Elements are color-coded to indicate the source strain. Prophages and ICEs are numbered as they occur clockwise around the core chromosome for each strain. Integration loci are lettered alphabetically as they occur clockwise around the core chromosome. The six rRNA operons are shown as green bars. Gene designations are as follows: 1) secreted pyrogenic-toxin-superantigens: speA, speC, speH, speI, speK, speL, speM, and ssa; 2) secreted DNAses: sda, sdn, spd1, spd3, and spd4; 3) secreted phospholipase: sla; 4) antimicrobial resistance: erm(A), mef(A), and tet(O); 5) cell surface adhesins: R6 and R28; 6) none, these elements lack a known or obvious virulence gene.
All of the sequenced GAS strains have multiple prophages, most of which encode one or two proven or putative secreted virulence factors. Prophages constitute ∼10% of any one genome and are the major contributor to variation in gene content among the sequenced GAS genomes [59]. As such phage have been a major source of virulence factors uniquely present in each of the genomes. In opposition to this prior trend, no new putative secreted virulence factors were encoded by the 14 prophages present in the serotype M2, M4, and M12 genomes sequenced. Importantly however, this does not mean that the prophages in these genomes are identical to those in the other sequenced GAS genomes, or that no new secreted putative virulence genes were identified in these strains. To the contrary, the apparent mobility and highly recombinogenic mosaic structures of the prophages and ICEs in the GAS genomes results in each sequenced GAS genome having a unique complement of exogenous elements and secreted virulence determinants.
In addition to multiple prophages, seven of the sequenced strains also contain large (5 kb–63 kb) regions that have features of ICEs [60], [61]. The presence of ICEs in the newly sequenced strains means that conjugative lateral gene transfer is a second important contributor to GAS metagenome diversification. For example, ICEs, 2096-RD.1 and 2096-RD.2, present in the genome of serotype M12 strain MGAS2096 account for half (49.5%) of the total of 162.6 kb of foreign sequence identified in this strain. Notably, ICEs are more prevalent among the sof positive strains (averaging 2.0 per genome) than the sof negative strains (averaging 0.3 per genome) (Table 1). Each of the five sof positive genomes (M2, M4, 2 M12 and M28) contain one or more ICEs, accounting for 10 of the 12 ICEs among the sequenced strains. In contrast among the 7 sof negative genomes (2 M1, 2 M3, M5, M6, and M18) only the two serotype M1 strains have an ICE. Furthermore, unlike the prophage which on average differ only 0.5 percent in G+C content (ave. = 38.05) from the GAS endogenous core genome (ave. = 38.61), the ICEs differ by an average of 5 percent (ave. = 33.62) (Table 2).
To identify new GAS metagenome gene content, the predicted CDSs of each newly sequenced strain was compared to the genomes of the eight previously sequenced GAS strains using BLASTN. In total 242 genes were identified that shared less than 50% overall nucleotide identity with sequence of any of the previously determined genomes (supplementary Table S1). ICE-like regions accounted for 41% (98/242), the fibronectin-collagen-T antigen encoding (FCT) region 9% (22/242), and prophages 14% (33/242) of this new gene content (supplementary Table S1). Given that ICE-like elements and the FCT region encode half of this newly identified GAS metagenome gene content, the description of the M2, M4, and M12 genome sequences that follows will focus on these components. The ICEs will be presented in order as they occur integrated clockwise around the GAS metagenome (Fig. 3). Specific prophages in these genomes will not be described as no new prophage associated putative virulence genes were found in these genomes, and the contribution of phage to GAS pathogenesis and genome diversification has been the subject of recent reviews [59], [62]–[64].
Streptin production ICE-like region
The genome of the serotype M2, M4, and M12 strains each has an ∼15 kb ICE-like region of difference (designated 10270-RD.1, 10750-RD.1, 2096-RD.1, and 9429-RD.1) composed of 9 or 10 genes (srt genes) encoding proteins mediating production of streptin, a lantibiotic bacteriocin (for a diagram of the srt gene locus see [65]) (Fig. 3 site G). These genes are flanked on the 5′ and 3′ sides by multiple CDSs with similarity to ICE relaxases and site-specific recombinases, respectively. This element is integrated between rpiL and dacA1 (SF370: Spy1073&Spy1093), genes encoding a ribosomal large subunit protein and peptidoglycan synthesis transpeptidase, respectively. The integration of this element appears to result in deletion of a ∼200 nt region located between these two genes. Genes encoding for streptin production are also present at this loci in the two serotype M1 and the M28 genome sequences. However due to multiple internal deletions the streptin ICE-like region present in the genome of the two serotype M1 strains is only ∼10 kb in size. The genome sequences of the serotype M3, M5, M6, and M18 strains (all sof negative) lack an analogous streptin ICE-like region. The G+C content of this element in all 5 strains with the ∼15 kb intact form is 31.9%, a value considerably less that the 38.5% GAS genome average, consistent with interspecies gene horizontal transfer.
Analysis of bacteriocin production by GAS strains of many different serotypes has found that serotype M2, M12, and M28 strains produce a bacteriocin resulting in a P-type 777 growth inhibition profile on indicator strains, whereas serotype M1, M3, M4, M5, M6, and M18 strains do not [65]. Thus, with the exception of serotype M4 strains, the results parallel the distribution of the 15-kb form of this ICE among the sequenced GAS genomes. Serotype M4 strains have a unique bacteriocin growth inhibition profile, P-type 655 attributed to the production of both streptin and salivaricin A [65], [66]. Consistent with this finding, the M4 strain MGAS10750 genome has an intact ∼10 kb locus of seven genes (salR-K-Y-X-T-M-A; MGAS10750_Spy1722-28), encoding for production of salivaricin (for a diagram of the sal gene locus see ref. 116). The salivaricin genes are located 3′ adjacent to 10750-RD.2 (Fig. 3 site S). A homologous gene locus is present in the other sequenced GAS strains but all have deletions in salT and/or salM that preclude SalA production [66]. Although the 30.5% G+C content of this region suggests it is not endogenous to the GAS genome, it lacks gene content characteristic of either phage or ICE.
The streptin element in the 5 strains in which it is intact has an average of 1 SNP every 175 nt. This level of sequence polymorphism is similar to the average of 1 SNP every ∼120 nt frequency present serotype-to-serotype among the sequenced GAS strain core chromosomes. In addition, very few SNPs present in this element were common to strains of different M protein serotypes. These genetic features strongly favor the likelihood that these sof positive serotypes, each possessing the streptin gene locus, share a common ancestor. That is, acquisition of the streptin ICE by lateral transfer occurred before the divergence of the genomes of distinct M protein serotypes that contain this element.
Exogenous genetic element 2096-RD.2 encoding tetracycline resistance
Although most studies of antimicrobial agent resistance in GAS have focused on macrolides such as erythromycin, increasing emphasis is being placed on analysis of strains resistant to tetracycline, either alone or in combination with macrolides [67]. Tetracycline resistance in GAS is mediated either by tet(M) or tet(O), genes which encode proteins that protect the ribosome [67]. The occurrence of the tet(M) gene in GAS has been known for some time, whereas the presence of the tet(O) gene in this species was reported relatively recently [68], [69].
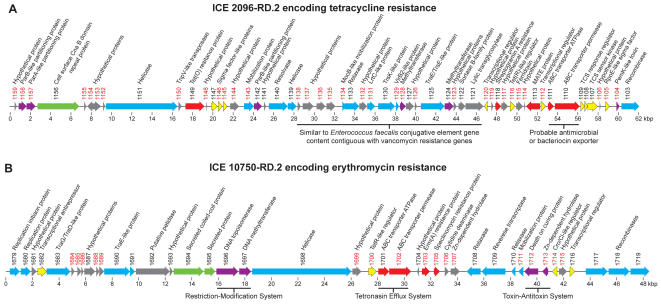
Our primary impetus for sequencing the genome of serotype M12 strain MGAS2096 was its documented association with acute poststreptococcal glomerulonephritis. We found that this genome has a unique 63-kb ICE-like element (designated 2096-RD.2) encoding several antibiotic resistance genes including tet(O) (Fig. 4 panel A). This ICE-like element is both the largest and has the highest G+C content (43%) of the exogenous elements present in the 12 sequenced GAS strains (Table 2). 2096-RD.2 is integrated at the 3′ end of a tRNA uracil methyltransferase gene (SF370: Spy1346). 6180-RD.1 occupies the analogous site in the genome of the sequenced serotype M28 strain, however these two ICEs have almost no genes in common (Table 2, Fig 3). The tet(O) gene (MGAS2096_Spy1149) encoded by 2096-RD.2 is >98% identical to tet(O) gene found in Streptococcus mutans, Streptococcus pneumoniae, and Campylobacter jejuni. 2096-RD.2 also has an acetyltransferase gene (MGAS2096_Spy1118) encoding a product with 47% identity and 66% similarity to Vat(B), a protein conferring resistance to streptogramin A in Staphylococcus aureus [70]. In addition, the 2096-RD.2 element has a gene (MGAS2096_Spy1113) that encodes a hydrophobic protein with ∼65% amino acid similarity to Na+-driven multi-drug efflux pumps (MATE proteins) found in Clostridium tetani, Listeria monocytogenes, and Porphyrmonas gingivalis, and several other species of pathogenic bacteria. Thus 2096-RD.2 has multiple genes which likely confer resistance to antimicrobial agents.
Figure 4. ICEs encoding antimicrobial resistance genes.
(A) 2096-RD.2 encoding Tet(O). (B) 10750-RD.2 encoding Erm(A). Illustrated are predicted coding sequences with gene numbers and predicted functions. Gene numbers given in red denote unique gene content as determined by BLASTP comparison to the GAS metagenome (no hit at e = 1×10−6). CDS are color coded to designate functionally related groups: red, antimicrobial resistance; green, secreted and cell surface; blue, mobilization and transfer; violet, element maintenance; yellow, transcriptional regulation; grey, hypothetical and unclassified.
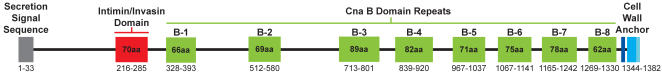
Notably, 2096-RD.2 also has a 4,146-bp gene (MGAS2096_Spy1156) encoding a large predicted exported protein with an aminoterminal secretion signal sequence and a carboxyterminal cell-wall anchoring motif (TPKTG) (Fig. 5). This ∼150 kDa acidic protein has a pI of 4.4, and one-fourth of its 1382 amino acids have charged side-chains. Amino acid residues 328–1330 have similarity to Cna, a collagen-adhesion virulence factor and vaccine candidate made by S. aureus [71]. The similarity is due mainly to eight regions of ∼75 amino acids each resembling the B domain of Cna that form a beta sandwich extended stalk structure [72], [73]. The amino-terminal end of the mature protein (residues 33–300), although lacking significant similarity to proteins of known function, has a 70 amino acid invasin domain as defined by the intimin protein of enterohemorrhagic and enteropathogenic Escherichia coli. Based on these domain similarities MGAS2096_Spy1156 may function as a cell surface adhesin/invasin. Inasmuch as serotype M12 strain MGAS2096 was cultured from a patient with APSGN, and that the 2096-RD.2 ICE is not present in the other sequenced GAS strains including the M12 pharyngitis isolate MGAS9429, the unique proteins encoded by this element warrant further investigation in the context of glomerulonephritis pathogenesis.
Figure 5. Domain architecture of putative cell surface acidic protein MGAS2096_Spy1156.
The protein has a conventional Gram-positive secretion signal sequence and a tripartite (TPKTG, membrane span, positively charged anchor) cell wall attachment domain. The aminoterminal portion of the protein has an intimin/invasin-like domain (Structural Classification of Proteins superfamily: SSF49373) and shares similarity (45% from amino acid ∼50-to-350) with a putative cell surface protein of unknown function (lmo1115) in the genome sequence of the intracellular pathogen Listeria monocytogenes strain EGD-e. The carboxyterminal portion of the protein (∼315-to-1350) has 8 Cna B-type domain repeats (Protein Family: PF05738) and shares similarity with multiple proteins annotated as collagen-binding. These characteristics suggest this protein may function as an adhesin/invasin.
Genetic element 10270-RD.2 and association with puerperal sepsis
The genome of serotype M2 strain MGAS10270 has a 35-kb ICE-like region of difference (designated 10270-RD.2) that is virtually identical to a recently-described exogenous genetic element (6180-RD.2) present in the genome of all serotype M28 strains [29]. An analogous ICE is not present in the genome of the other 10 sequenced GAS strains (Fig 3 site M). ICE 10270-RD.2 is integrated into a tRNA-Thr, is ∼35% G+C, is flanked by 16-bp direct repeats, and has seven genes encoding proteins with predicted secretion signal sequences. Included among these proteins are cognates of Spy1325 and R28 (MGAS10270: Spy1399 and Spy1410, respectively). Spy1325 and R28 are cell surface anchored adhesins that are expressed during the course of human infection, and are immunoprotective in mouse models of infection [74], [75]. Importantly 10270-RD.2 and 6180-RD.2 are closely related to genetic elements present in strains of group B Streptococcus, the leading cause of maternal-neonatal infections in the United States and elsewhere. Including R28, four of the seven inferred extracellular proteins encoded by 6180-RD.2 are made during GAS infection [74], [75]. Excluding differences in the number of repeat domains in the gene encoding R28, 2096-RD.2 and 6180-RD.2 differ by only 8 SNPs. This is one SNP on average every ∼4.4-kb, a frequency 38-fold lower than the core chromosome of strains MGAS2096 and MGAS6180. The very high level of sequence similarity between 2096-RD.2 and 6180-RD.2 means these elements descend from a recent common ancestor and have undergone lateral gene transfer. The occurrence of this ICE in the M2 and M28 clonal lineages is noteworthy because these serotypes have been repeatedly nonrandomly associated with GAS maternal-fetal infections [17], [18], [46], [76]. This epidemiological association and the similarity with sequences of GBS, implicate the 2096-RD.2/6180-RD.2 element in contributing to maternal-fetal infections caused by serotype M2 and M28 strains.
Element 10750-RD.2 encoding erythromycin resistance
Resistance of GAS to macrolide antibiotics has increased dramatically in the last 10 years and is now a worldwide problem [38]. The great majority of macrolide-resistant GAS strains have either the mef(A) gene encoding a drug efflux pump (M resistance phenotype) or the erm(A) gene encoding an erythromycin ribosome dimethyltransferase (MLSB resistance phenotype) that modifies a highly conserved adenine residue located in the target bacterial 23S rRNA [77], [78]. Macrolide resistance has been reported to be transferable by conjugal plasmids, phages, and conjugative transposons [68], [79]–[84]. Consistent with these observations, the mef(A) and erm(A) genes each has been found in association with a very large number of distinct emm types [32]–[34], [43], [44], [85]–[87]. In addition, we recently reported that the mef(A) gene of macrolide resistant M6 strain MGAS10394 is encoded by an unusual 58.8-kb chimeric genetic element (Fig. 3 site J) with conjugative transposon and prophage characteristics [26], [85].
One of our motivations for sequencing the genome of erm(A)-positive serotype M4 strain MGAS10750 was to characterize the genetic element containing the macrolide resistance-conferring gene in this strain. The erm(A) gene was present on a 49-kb exogenous element designated 10750-RD.2 that is integrated into the hsdM gene encoding host DNA restriction-modification methyltransferase (SF370: Spy1906) (Fig. 3 site S, Fig. 4 panel B). The gene content of this ICE is largely unique to the strain MGAS10750 genome. The erm(A) gene in this strain is identical to the erm(TR) sequence initially reported in GAS by Seppala et al. [78], and its product is 81.1% identical to Erm(A) of S. aureus [88]. Just 5′ of erm(A) are two adjacent CDS (SpyM4_1701 and 1702) that encode the ATP-binding and membrane permease components of an ABC transporter. The products of these genes have 66.7% and 44.5% similarity with TnrB2 and TnrB3 respectively, of Streptomyces longisporoflavus a producer of tetronasin, a polyether-ionophore antibiotic. TnrB2 and TnrB3 form an ATP-dependent efflux system that confers resistance to tetronasin [89]. Just 3′ of erm(A) is a CDS (SpyM4_1705) predicted to encode a phosphotransferase. The product of this gene has 25.6% identity and 39.9% similarity with the last 220 amino acids (residues 122–340) of the spectinomycin resistance aminoglycoside phosphotransferase gene, aph, of Legionella pneumophila [90]. This conserved region includes the catalytically important residues defined for Aph(3) and Aph(9) [91].
Two of the 10750-RD.2 element CDSs are predicted to encode proteins with conventional gram-positive secretion signal sequences, suggesting that they are secreted extracellularly (Fig. 4 panel B). SpyM4_1694 is predicted to encode a hydrophilic mature protein (residues 31–783) of 87.7 kDa. This protein is of unknown function but has similarity with many proteins including M protein due to the presence of a central (∼250–525 aa) laminin- and myosin-like coiled-coil domain (PFAM: PF00608 and PF01576 respectively). Notably, the top 10 alignments identified using BLASTP to compare SpyM4_1694 with the NCBI non-redundant sequence database are eukaryotic not prokaryotic proteins. SpyM4_1695 is predicted to encode a mature acidic protein (residues 27–289, pI 4.2) of 29.7 kDa. The function of this protein is unknown. SpyM4_1695 lacks significant similarity to proteins of known function, and to known protein domains as determined using either EMBL InterPro- or NCBI conserved domain-searches. Inasmuch as the majority of secreted and cell surface proteins identified in GAS have proven or putative roles in host-pathogen interaction, these predicted extracellular proteins are candidates for further investigation.
Fibronectin-binding collagen-binding T antigen (FCT) gene region
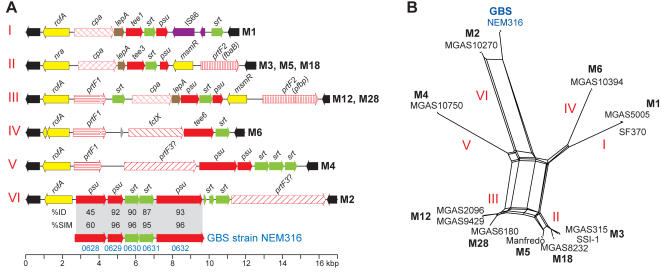
The FCT gene region is an ∼11–16 kb region that encodes global-regulators and extracellular matrix-binding proteins involved in cell adhesion and invasion [92]–[98]. Recently, Mora et al. [99] reported that the FCT region genes encode extended pilus-like structures composed in part of polymerized T-antigen protein subunits. Importantly, immunization of laboratory animals with either GAS or GBS pilin proteins has been shown to provide protection against experimental invasive infections caused by these pathogens [99]–[101]. More recently these pilus components were shown to mediate adhesion to human pharyngeal and skin cells and participate in biofilm formation [102], [103]. We compared the FCT gene regions in the 12 sequenced GAS strains and identified six distinct variants, including four (I, II, III, and IV) that have been previously described (Fig. 6). Two variants, V in serotype M4 strain MGAS10750 and VI in serotype M2 strain MGAS10270, have not been described previously, thereby expanding our understanding of sequence variation in this region. A portion of the FCT region gene content in the serotype M2 strain is more related to genomic islands present in six sequenced GBS strains than to the other GAS variants [104], [105], consistent with the idea that horizontal gene transfer has contributed to diversification in this chromosomal segment. Thus, the serotype M2 clonal lineage has two gene regions (FCT and 10270-RD.2) that are closely related to genetic elements in GBS. The similarity of these elements between these two pathogens provides additional support for the hypothesis that the extracellular products encoded by these two regions contribute to the ability of M2 GAS strains to cause puerperal sepsis infections.
Figure 6. GAS metagenome FCT region variants.
(A) Architecture of the FCT region variants. CDSs are colored to designate the following groups: black, conserved flanking genes (SF370: 5′ Spy_0123 and 3′ Spy_0136); yellow, transcriptional regulators; red, extracellular matrix-binding and/or pilin-subunit proteins; tan, signal peptidases; green, sortases; purple, insertion sequences. Although there are differences both intra- and interserotype indicative of antigenic variation, nearly all of the extracellular matrix-binding proteins and pilin-subunit proteins have predicted secretion signal sequences and cell wall attachment domains in one or more of the genomes. Additionally illustrated is the similarity between the serotype M2 and GBS pilus encoding region proteins in global alignments. (B) Relationships among the FCT region variants. Nucleotide sequences bounded by the flanking conserved genes for the each of the sequenced GAS strains and the five GBS genes in panel A, were aligned with ClustalW and a neighbor-network was generated using SplitsTree.
Discussion
The 12 GAS genomes now available represent serotypes responsible for ∼70% of M protein serotypes that most commonly cause GAS pharyngitis and invasive infections in several countries in the western hemisphere [17], [18], [21], [22], [24], [45], [46], [106]. Although these 12 genome sequences provide extensive information to assist studies of virulence, development of therapeutics and diagnostics, and other aspects of GAS biology, a cautionary note is required. There is considerable variation in prophage content and prophage-associated virulence factor profile among strains of the same M type [26], [27], [29], [45], [46], [59], [106]–[109]. In addition, strains of certain M types are not necessarily clonally related [13], [56], [110]. This intra-M type genetic heterogeneity can mediate significant differences in host-pathogen interactions, as documented recently for distinct clones of serotype M1 and M3 GAS [56], [107]. Similarly, many of the genomes contain large segments of exogenous (foreign), non-prophage DNA acquired by lateral gene transfer events. In the case of serotype M1 GAS, an apparent episode of generalized transduction contributed to the evolution of a new, unusually virulent clone that increased dramatically in frequency since the mid 1980s [56]. Other foreign DNA segments may have been acquired by conjugative transposition as exemplified by the ICE-like elements we have described. Regardless of the exact gene transfer mechanism involved, the key point is that the intraspecies gene content and allelic diversity present in the GAS metagenome is extensive, and can impart important differences in disease character and epidemic behavior. The sequencing of additional GAS strains continues to reveal an unappreciated magnitude of species-level population genomic diversity.
Given the increasing prevalence of drug-resistant strains of GAS, it is important to note that these genome sequences have provided new information about the putatively-mobile genetic elements involved. The ICEs associated with genes conferring resistance to macrolide and tetracycline antibiotics are chimeric structures composed of the multiple drug-resistance genes, genetic machinery to mediate lateral transfer, and genes encoding putative or proven novel extracellular proteins [26], [83], [85]. In the case of the mef(A) element in serotype M6 strain MGAS10394 and other strains, it is known from serologic studies that the extracellular protein designated R6 is expressed during human infections [26], [85]. The putative extracellular proteins encoded by the erm(A) and tet(O) encoding elements, 10750-RD.2 and 2096-RD.2 respectively, have not yet been analyzed in detail. However, since they contain conventional gram-positive secretion sequences and some have carboxyterminal cell wall attachment motifs, we speculate that these proteins are either displayed on the GAS cell surface and function to mediate adherence to host molecules, or are secreted free into the extracellular environment and interact with host molecules. Given that the drug-resistance genes are widespread in Gram negative and positive respiratory tract organisms, and dispersed among many distinct GAS M protein types, we think it likely that further study will identify additional genetic elements associated with drug resistance [33]–[37], [40], [41], [43], [44], [51], [68], [69], [80], [85]–[87], [111]–[117]. Consistent with this hypothesis, an element containing both mef(A) and tet(O) genes was described recently [86]. We note that the various genetic elements associated with tet(O), mef(A), erm(A) likely helps to explain some of the confusing data in the literature regarding the nature and mode of spread of drug-resistant markers in GAS [68], [80]–[82], [84].
One feature of the horizontally transferred regions encoding antibiotic resistance determinants in the GAS metagenome that is of concern is the high level of homology between the genes putatively involved in mobilization and transfer of these elements and genes found in other genera and species of human pathogens such as staphylococci, enterococci, clostridia, and streptococci (S. pneumoniae and S. agalactiae, for example). Additionally the finding that the ICEs present in the GAS genome (more so than the prophages) differ significantly in nucleotide composition from the core chromosome argues that they originate from organisms not closely related to the streptococci. This underscores the potential extensiveness of accessible virulence genes, and the relative lack of barriers to horizontal gene transfer among pathogenic bacteria. The horizontally acquired chimeric elements can provide an immediate selective advantage to recipient bacteria, for example by conferring antibiotic resistance. In addition, the conserved regions of these mobile elements enhance the potential for further future recombination/integration events with horizontally transferred DNA. Thus, these foreign elements likely increase the frequency with which regions of horizontally transferred DNA are retained in the chromosome of recipient bacteria.
The addition of these four complete genome sequences to the eight previously determined makes the GAS metagenome one of the better characterized among important human pathogens. The new gene content found in the ICE-like elements and the FCT regions described in these four genomes encodes proteins providing antimicrobial resistance and proven and putative extracellular adhesin/invasin proteins. Thus the sequencing of these four additional genomes has provided much-needed information about the genetic diversity present in GAS and has revealed factors likely affecting the virulence of these strains. Although medical intervention limits human morbidity and mortality due to GAS in the western countries, globally it has a tremendous toll on human health. This is especially the case in countries with less developed medical systems for which there is relatively little information available about the circulating strains. One future challenge will be to determine to what extent the metagenome of GAS as defined by the twelve currently sequenced strains originating in the western hemisphere is representative of disease causing strains circulating in other areas of the world. Given the considerable role played by mobile exogenous elements in GAS genetic diversity and pathogenesis, this information is crucial to understanding the array of molecular mechanisms used by GAS to cause human disease, and is of paramount importance to vaccine and therapeutics research.
Materials and Methods
Bacterial strains
The four GAS strains sequenced, serotype M2 Strain MGAS10270, M4 strain MGAS10750, and M12 strains MGAS2096 and MGAS9429 were each isolated from human infections of defined disease type. These four strains have been deposited in the American Type Culture Collection under the following accession numbers: MGAS10270 (BAA-1063), MGAS10750 (BAA-1066), MGAS2096 (BAA-1065), and MGAS9429 (BAA-1315).
Genome sequencing
Standard methods were used to determine the complete genome sequence of the M2, M4, and two M12 strains as previously described [26], [29]. Briefly, short sequencing templates were generated from sheared chromosomal DNA fragments cloned into a plasmid vector and sequenced from each end. The resulting random sequence reads were assembled in silico into larger contiguous segments, and contigs were ordered using the GAS metagenome as a scaffold. Sequence gaps were closed by directed sequencing of gap-spanning templates obtained by PCR amplification. Additional directed sequencing was performed as necessary to improve sequence quality genome-wide to a minimum base call error rate of 1 in 10,000 (i.e. Q40). Each genome was tiled by PCR after closure to validate the in silico assembly. Coding sequences were identified with proprietary software (Integrated Genomics, Chicago, IL), annotated, and analyzed with the ERGO bioinformatics suite [118]. The genome sequences have been deposited in the National Center for Biotechnology Information microbial genome database under the following accession numbers: MGAS10270 (CP000260), MGAS10750 (CP000262), MGAS2096 (CP000261), and MGAS9429 (CP000259). An analysis of polymorphisms present in the core chromosomes of these strains and their relationship with the other 8 sequenced GAS genomes has recently been published [119].
Identification of endogenous and exogenous sequences
Sequence common to the GAS genomes constituting the endogenous metagenome core, were identified by a combination of genomic alignment and gene content comparisons. Exogenous putatively-mobile elements such as prophages and integrating conjugative elements (ICEs) were identified by a combination of genomic alignment, gene content, nucleotide composition, and codon usage comparisons. First, by comparative genome sequence alignments (such as MUMmer plots) these regions differ in gene content and/or context relative to one or more of the other GAS genomes. That is, they are an insertion or deletion. Second, they contain modules of genes with similarity to genes previously identified in and considered to be characteristic of mobile genetic elements. For example, prophages have genes encoding coat and tail proteins, and ICEs have genes encoding recombinases, relaxases and excisionases. In addition, prophages and ICEs were differentiated in part by the gene content they lacked, consistent with their known modes of lateral transfer. For example, unlike prophages, ICEs lack genes encoding holins and peptidoglycan lytic enzymes. Third, many ICEs and prophages are flanked by directly repeated attachment sequences, attP-L and attP-R, that are generated as a consequence of the homologous-recombination event mediated by the related prophage and ICE site-specific integrases. Fourth, these elements often contain genes that are most similar in sequence to genes of other bacterial species that sometimes differ from GAS in preferred codon usage, % G+C content, and multimer nucleotide (di-, tri-, tetra-) composition, consistent with intraspecies lateral transfer. Importantly, there is no single distinct gene complement that differentiates among various types of bacterial mobile genetic elements (phage, conjugative plasmids and transposons, insertion sequences, etc.). Thus, ICEs and prophages were identified on the basis of a preponderance of genetic characteristics identified during annotation, rather than use of a single genetic characteristic. For simplicity, integrated foreign elements that have many but not all of the features described above (that is, ICE-like or prophage-like traits) will be referred to in this report as ICEs and prophages. Together, these elements are referred to as exogenous genetic elements.
Sequence alignments and comparisons
Genome alignments and identification of SNPs were performed using MUMmer [120]. Genomic gene content and NCBI non-redundant sequence database comparisons were performed using BLAST [121]. Pair-wise global and local gene and protein alignments were performed using the “needle” (Needleman-Wunsch) and “water” (Smith-Waterman) applications respectively of EMBOSS [122]. Multiple sequence alignments were performed using ClustalW or Muscle [123], [124]. Reconstruction of genetic relationships were performed using SplitsTree [125]. Protein motif searches were performed using the EMBL InterPro scan and NCBI conserved domain servers [126], [127]. Codon usage and nucleotide composition analyses were performed using CodonW (bioweb.pasteur.fr). Various other analyses (M.W., pI, hydrophilicity, etc.) were performed using MacVector [128]. Circular genome atlases were generated using GenomeViz [129]. Schematics of the aligned genomes were generated using the Artemis Comparison Tool [130].
Supporting Information
New GAS Metagenome Genes
(0.37 MB DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported in part by grant UO1-60595 from the National Institute of Allergy and Infectious Diseases (to JMM).
References
- 1.Fitzgerald JR, Musser JM. Evolutionary genomics of pathogenic bacteria. Trends Microbiol. 2001;9:547–553. doi: 10.1016/s0966-842x(01)02228-4. [DOI] [PubMed] [Google Scholar]
- 2.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaser P, Rusniok C, Buchrieser C, Chevalier F, Frangeul L, et al. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol. 2002;45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 4.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindsay JA, Holden MT. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 2004;12:378–385. doi: 10.1016/j.tim.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 7.Subtil A, Dautry-Varsat A. Chlamydia: five years A.G. (after genome). Curr Opin Microbiol. 2004;7:85–92. doi: 10.1016/j.mib.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Tettelin H, Masignani V, Cieslewicz MJ, Eisen JA, Peterson S, et al. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc Natl Acad Sci U S A. 2002;99:12391–12396. doi: 10.1073/pnas.182380799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welch RA, Burland V, Plunkett G, 3rd, Redford P, Roesch P, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisno AL. Group A Streptococcal infections and acute rheumatic fever. N Engl J Med. 1991;325:783–793. doi: 10.1056/NEJM199109123251106. [DOI] [PubMed] [Google Scholar]
- 11.Bisno AL, Brito MO, Collins CM. Molecular basis of group A Streptococcal virulence. Lancet Infect Dis. 2003;3:191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham MW. Pathogenesis of group A Streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musser JM, Krause RM. The revival of group A Streptococcal diseases, with a commentary on staphylococcal toxic shock syndrome. In: Krause RM, editor. Emerging Infections. San Diego: Academic Press; 1998. pp. 185–218. [Google Scholar]
- 14.Fischetti VA. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herwald H, Cramer H, Morgelin M, Russell W, Sollenberg U, et al. M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell. 2004;116:367–379. doi: 10.1016/s0092-8674(04)00057-1. [DOI] [PubMed] [Google Scholar]
- 16.Facklam RF, Martin DR, Lovgren M, Johnson DR, Efstratiou A, et al. Extension of the Lancefield classification for group A Streptococci by addition of 22 new M protein gene sequence types from clinical isolates: emm103 to emm124. Clin Infect Dis. 2002;34:28–38. doi: 10.1086/324621. [DOI] [PubMed] [Google Scholar]
- 17.Chuang I, Van Beneden C, Beall B, Schuchat A. Population-based surveillance for postpartum invasive group A Streptococcus infections, 1995–2000. Clin Infect Dis. 2002;35:665–670. doi: 10.1086/342062. [DOI] [PubMed] [Google Scholar]
- 18.Colman G, Tanna A, Efstratiou A, Gaworzewska ET. The serotypes of Streptococcus pyogenes present in Britain during 1980–1990 and their association with disease. J Med Microbiol. 1993;39:165–178. doi: 10.1099/00222615-39-3-165. [DOI] [PubMed] [Google Scholar]
- 19.Gaworzewska E, Colman G. Changes in the pattern of infection caused by Streptococcus pyogenes. Epidemiol Infect. 1988;100:257–269. doi: 10.1017/s095026880006739x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Sakota V, Jackson D, Franklin AR, Beall B. Array of M protein gene subtypes in 1064 recent invasive group A Streptococcus isolates recovered from the active bacterial core surveillance. J Infect Dis. 2003;188:1587–1592. doi: 10.1086/379050. [DOI] [PubMed] [Google Scholar]
- 21.O'Brien KL, Beall B, Barrett NL, Cieslak PR, Reingold A, et al. Epidemiology of invasive group A Streptococcus disease in the United States, 1995–1999. Clin Infect Dis. 2002;35:268–276. doi: 10.1086/341409. [DOI] [PubMed] [Google Scholar]
- 22.Tyrrell GJ, Lovgren M, Forwick B, Hoe NP, Musser JM, et al. M types of group A Streptococcal isolates submitted to the National Centre for Streptococcus (Canada) from 1993 to 1999. J Clin Microbiol. 2002;40:4466–4471. doi: 10.1128/JCM.40.12.4466-4471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eriksson BK, Norgren M, McGregor K, Spratt BG, Normark BH. Group A Streptococcal infections in Sweden: a comparative study of invasive and noninvasive infections and analysis of dominant T28 emm28 isolates. Clin Infect Dis. 2003;37:1189–1193. doi: 10.1086/379013. [DOI] [PubMed] [Google Scholar]
- 24.Kaul R, McGeer A, Low DE, Green K, Schwartz B. Population-based surveillance for group A Streptococcal necrotizing fasciitis: Clinical features, prognostic indicators, and microbiologic analysis of seventy-seven cases. Ontario Group A Streptococcal Study. Am J Med. 1997;103:18–24. doi: 10.1016/s0002-9343(97)00160-5. [DOI] [PubMed] [Google Scholar]
- 25.Sharkawy A, Low DE, Saginur R, Gregson D, Schwartz B, et al. Severe group A Streptococcal soft-tissue infections in Ontario: 1992–1996. Clin Infect Dis. 2002;34:454–460. doi: 10.1086/338466. [DOI] [PubMed] [Google Scholar]
- 26.Banks DJ, Porcella SF, Barbian KD, Beres SB, Philips LE, et al. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J Infect Dis. 2004;190:727–738. doi: 10.1086/422697. [DOI] [PubMed] [Google Scholar]
- 27.Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci U S A. 2002;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci U S A. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, et al. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192:760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa I, Kurokawa K, Yamashita A, Nakata M, Tomiyasu Y, et al. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 2003;13:1042–1055. doi: 10.1101/gr.1096703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, et al. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc Natl Acad Sci U S A. 2002;99:4668–4673. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bingen E, Leclercq R, Fitoussi F, Brahimi N, Malbruny B, et al. Emergence of group A Streptococcus strains with different mechanisms of macrolide resistance. Antimicrob Agents Chemother. 2002;46:1199–1203. doi: 10.1128/AAC.46.5.1199-1203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandt CM, Honscha M, Truong ND, Holland R, Hovener B, et al. Macrolide resistance in Streptococcus pyogenes isolates from throat infections in the region of Aachen, Germany. Microb Drug Resist. 2001;7:165–170. doi: 10.1089/10766290152045039. [DOI] [PubMed] [Google Scholar]
- 34.De Azavedo JC, Yeung RH, Bast DJ, Duncan CL, Borgia SB, et al. Prevalence and mechanisms of macrolide resistance in clinical isolates of group A Streptococci from Ontario, Canada. Antimicrob Agents Chemother. 1999;43:2144–2147. doi: 10.1128/aac.43.9.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kataja J, Huovinen P, Efstratiou A, Perez-Trallero E, Seppala H. Clonal relationships among isolates of erythromycin-resistant Streptococcus pyogenes of different geographical origin. Eur J Clin Microbiol Infect Dis. 2002;21:589–595. doi: 10.1007/s10096-002-0771-8. [DOI] [PubMed] [Google Scholar]
- 36.Kataja J, Huovinen P, Seppala H. Erythromycin resistance genes in group A Streptococci of different geographical origins. The Macrolide Resistance Study Group. J Antimicrob Chemother. 2000;46:789–792. doi: 10.1093/jac/46.5.789. [DOI] [PubMed] [Google Scholar]
- 37.Kataja J, Huovinen P, Skurnik M, Seppala H. Erythromycin resistance genes in group A Streptococci in Finland. The Finnish Study Group for Antimicrobial Resistance. Antimicrob Agents Chemother. 1999;43:48–52. doi: 10.1128/aac.43.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 39.Martin JM, Green M, Barbadora KA, Wald ER. Erythromycin-resistant group A Streptococci in schoolchildren in Pittsburgh. N Engl J Med. 2002;346:1200–1206. doi: 10.1056/NEJMoa013169. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen HU, Hammerum AM, Ekelund K, Bang D, Pallesen LV, et al. Tetracycline and macrolide co-resistance in Streptococcus pyogenes: co-selection as a reason for increase in macrolide-resistant S. pyogenes? Microb Drug Resist. 2004;10:231–238. doi: 10.1089/mdr.2004.10.231. [DOI] [PubMed] [Google Scholar]
- 41.Palavecino EL, Riedel I, Berrios X, Bajaksouzian S, Johnson D, et al. Prevalence and mechanisms of macrolide resistance in Streptococcus pyogenes in Santiago, Chile. Antimicrob Agents Chemother. 2001;45:339–341. doi: 10.1128/AAC.45.1.339-341.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan JJ, Wu HM, Huang AH, Fu HM, Lee CT, et al. Prevalence of polyclonal mefA-containing isolates among erythromycin-resistant group A Streptococci in Southern Taiwan. J Clin Microbiol. 2000;38:2475–2479. doi: 10.1128/jcm.38.7.2475-2479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zampaloni C, Cappelletti P, Prenna M, Vitali LA, Ripa S. emm Gene distribution among erythromycin-resistant and -susceptible Italian isolates of Streptococcus pyogenes. J Clin Microbiol. 2003;41:1307–1310. doi: 10.1128/JCM.41.3.1307-1310.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vlaminckx B, van Pelt W, Schouls L, van Silfhout A, Elzenaar C, et al. Epidemiological features of invasive and noninvasive group A Streptococcal disease in the Netherlands, 1992–1996. Eur J Clin Microbiol Infect Dis. 2004;23:434–444. doi: 10.1007/s10096-004-1147-z. [DOI] [PubMed] [Google Scholar]
- 46.Vlaminckx BJ, Mascini EM, Schellekens J, Schouls LM, Paauw A, et al. Site-specific manifestations of invasive group A Streptococcal disease: type distribution and corresponding patterns of virulence determinants. J Clin Microbiol. 2003;41:4941–4949. doi: 10.1128/JCM.41.11.4941-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Bouri KW, Lewis AM, Okeahialam CA, Wright D, Tanna A, et al. A community outbreak of invasive and non-invasive group A beta-haemolytic Streptococcal disease in a town in South Wales. Epidemiol Infect. 1998;121:515–521. doi: 10.1017/s095026889800168x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott RJ, Naidoo J, Lightfoot NF, George RC. A community outbreak of group A beta haemolytic Streptococci with transferable resistance to erythromycin. Epidemiol Infect. 1989;102:85–91. doi: 10.1017/s095026880002971x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnston KH, Zabriskie JB. Purification and partial characterization of the nephritis strain-associated protein from Streptococcus pyogenes, group A. J Exp Med. 1986;163:697–712. doi: 10.1084/jem.163.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poon-King R, Bannan J, Viteri A, Cu G, Zabriskie JB. Identification of an extracellular plasmin binding protein from nephritogenic Streptococci. J Exp Med. 1993;178:759–763. doi: 10.1084/jem.178.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villarreal H, Jr, Fischetti VA, van de Rijn I, Zabriskie JB. The occurrence of a protein in the extracellular products of Streptococci isolated from patients with acute glomerulonephritis. J Exp Med. 1979;149:459–472. doi: 10.1084/jem.149.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berrios X, Quesney F, Morales A, Blazquez J, Lagomarsino E, et al. Acute rheumatic fever and poststreptococcal glomerulonephritis in an open population: comparative studies of epidemiology and bacteriology. J Lab Clin Med. 1986;108:535–542. [PubMed] [Google Scholar]
- 53.Stollerman GH. Rheumatogenic and nephritogenic Streptococci. Circulation. 1971;43:915–921. doi: 10.1161/01.cir.43.6.915. [DOI] [PubMed] [Google Scholar]
- 54.Widdowson JP, Maxted WR, Grant DL. The production of opacity in serum by group A Streptococci and its relationship with the presence of M antigen. J Gen Microbiol. 1970;61:343–353. doi: 10.1099/00221287-61-3-343. [DOI] [PubMed] [Google Scholar]
- 55.Widdowson JP, Maxted WR, Grant DL, Pinney AM. The relationship between M-antigen and opacity factor in group A Streptococci. J Gen Microbiol. 1971;65:69–80. doi: 10.1099/00221287-65-1-69. [DOI] [PubMed] [Google Scholar]
- 56.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 57.Enright MC, Spratt BG, Kalia A, Cross JH, Bessen DE. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect Immun. 2001;69:2416–2427. doi: 10.1128/IAI.69.4.2416-2427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 59.Banks DJ, Beres SB, Musser JM. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 2002;10:515–521. doi: 10.1016/s0966-842x(02)02461-7. [DOI] [PubMed] [Google Scholar]
- 60.Burrus V, Pavlovic G, Decaris B, Guedon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 61.Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 62.Boyd EF, Brussow H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002;10:521–529. doi: 10.1016/s0966-842x(02)02459-9. [DOI] [PubMed] [Google Scholar]
- 63.Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. Prophage genomics. Microbiol Mol Biol Rev. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wescombe PA, Tagg JR. Purification and characterization of streptin, a type A1 lantibiotic produced by Streptococcus pyogenes. Appl Environ Microbiol. 2003;69:2737–2747. doi: 10.1128/AEM.69.5.2737-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wescombe PA, Upton M, Dierksen KP, Ragland NL, Sivabalan S, et al. Production of the lantibiotic salivaricin A and its variants by oral Streptococci and use of a specific induction assay to detect their presence in human saliva. Appl Environ Microbiol. 2006;72:1459–1466. doi: 10.1128/AEM.72.2.1459-1466.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–260; second page. doi: 10.1128/MMBR.65.2.232-260.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giovanetti E, Brenciani A, Lupidi R, Roberts MC, Varaldo PE. Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob Agents Chemother. 2003;47:2844–2849. doi: 10.1128/AAC.47.9.2844-2849.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hammerum AM, Nielsen HU, Agerso Y, Ekelund K, Frimodt-Moller N. Detection of tet(M), tet(O) and tet(S) in tetracycline/minocycline-resistant Streptococcus pyogenes bacteraemia isolates. J Antimicrob Chemother. 2004;53:118–119. doi: 10.1093/jac/dkh043. [DOI] [PubMed] [Google Scholar]
- 70.Allignet J, el Solh N. Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new Staphylococcal determinant, vatB. Antimicrob Agents Chemother. 1995;39:2027–2036. doi: 10.1128/aac.39.9.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patti JM, Jonsson H, Guss B, Switalski LM, Wiberg K, et al. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J Biol Chem. 1992;267:4766–4772. [PubMed] [Google Scholar]
- 72.Deivanayagam CC, Rich RL, Carson M, Owens RT, Danthuluri S, et al. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure. 2000;8:67–78. doi: 10.1016/s0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 73.Snodgrass JL, Mohamed N, Ross JM, Sau S, Lee CY, et al. Functional analysis of the Staphylococcus aureus collagen adhesin B domain. Infect Immun. 1999;67:3952–3959. doi: 10.1128/iai.67.8.3952-3959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stalhammar-Carlemalm M, Areschoug T, Larsson C, Lindahl G. The R28 protein of Streptococcus pyogenes is related to several group B Streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol Microbiol. 1999;33:208–219. doi: 10.1046/j.1365-2958.1999.01470.x. [DOI] [PubMed] [Google Scholar]
- 75.Zhang S, Green NM, Sitkiewicz I, Lefebvre RB, Musser JM. Identification and characterization of an antigen I/II family protein produced by group A Streptococcus. Infect Immun. 2006;74:4200–4213. doi: 10.1128/IAI.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Areschoug T, Carlsson F, Stalhammar-Carlemalm M, Lindahl G. Host-pathogen interactions in Streptococcus pyogenes infections, with special reference to puerperal fever and a comment on vaccine development. Vaccine 22 Suppl. 2004;1:S9–S14. doi: 10.1016/j.vaccine.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 77.Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, et al. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 78.Seppala H, Skurnik M, Soini H, Roberts MC, Huovinen P. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother. 1998;42:257–262. doi: 10.1128/aac.42.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clewell DB, Franke AE. Characterization of a plasmid determining resistance to erythromycin, lincomycin, and vernamycin Balpha in a strain Streptococcus pyogenes. Antimicrob Agents Chemother. 1974;5:534–537. doi: 10.1128/aac.5.5.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giovanetti E, Magi G, Brenciani A, Spinaci C, Lupidi R, et al. Conjugative transfer of the erm(A) gene from erythromycin-resistant Streptococcus pyogenes to macrolide-susceptible S. pyogenes, Enterococcus faecalis and Listeria innocua. J Antimicrob Chemother. 2002;50:249–252. doi: 10.1093/jac/dkf122. [DOI] [PubMed] [Google Scholar]
- 81.Hyder SL, Streitfeld MM. Transfer of erythromycin resistance from clinically isolated lysogenic strains of Streptococcus pyogenes via their endogenous phage. J Infect Dis. 1978;138:281–286. doi: 10.1093/infdis/138.3.281. [DOI] [PubMed] [Google Scholar]
- 82.Malke H. Genetics of resistance to macrolide antibiotics and lincomycin in natural isolates of Streptococcus pyogenes. Mol Gen Genet. 1974;135:349–367. doi: 10.1007/BF00271149. [DOI] [PubMed] [Google Scholar]
- 83.Santagati M, Iannelli F, Cascone C, Campanile F, Oggioni MR, et al. The novel conjugative transposon tn1207.3 carries the macrolide efflux gene mef(A) in Streptococcus pyogenes. Microb Drug Resist. 2003;9:243–247. doi: 10.1089/107662903322286445. [DOI] [PubMed] [Google Scholar]
- 84.Ubukata K, Konno M, Fujii R. Transduction of drug resistance to tetracycline, chloramphenicol, macrolides, lincomycin and clindamycin with phages induced from Streptococcus pyogenes. J Antibiot (Tokyo) 1975;28:681–688. doi: 10.7164/antibiotics.28.681. [DOI] [PubMed] [Google Scholar]
- 85.Banks DJ, Porcella SF, Barbian KD, Martin JM, Musser JM. Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A Streptococcus. J Infect Dis. 2003;188:1898–1908. doi: 10.1086/379897. [DOI] [PubMed] [Google Scholar]
- 86.Brenciani A, Ojo KK, Monachetti A, Menzo S, Roberts MC, et al. Distribution and molecular analysis of mef(A)-containing elements in tetracycline-susceptible and -resistant Streptococcus pyogenes clinical isolates with efflux-mediated erythromycin resistance. J Antimicrob Chemother. 2004;54:991–998. doi: 10.1093/jac/dkh481. [DOI] [PubMed] [Google Scholar]
- 87.Tanz RR, Shulman ST, Shortridge VD, Kabat W, Kabat K, et al. Community-based surveillance in the united states of macrolide-resistant pediatric pharyngeal group A Streptococci during 3 respiratory disease seasons. Clin Infect Dis. 2004;39:1794–1801. doi: 10.1086/426025. [DOI] [PubMed] [Google Scholar]
- 88.Murphy E. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J Bacteriol. 1985;162:633–640. doi: 10.1128/jb.162.2.633-640.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Linton KJ, Cooper HN, Hunter IS, Leadlay PF. An ABC-transporter from Streptomyces longisporoflavus confers resistance to the polyether-ionophore antibiotic tetronasin. Mol Microbiol. 1994;11:777–785. doi: 10.1111/j.1365-2958.1994.tb00355.x. [DOI] [PubMed] [Google Scholar]
- 90.Suter TM, Viswanathan VK, Cianciotto NP. Isolation of a gene encoding a novel spectinomycin phosphotransferase from Legionella pneumophila. Antimicrob Agents Chemother. 1997;41:1385–1388. doi: 10.1128/aac.41.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thompson PR, Hughes DW, Cianciotto NP, Wright GD. Spectinomycin kinase from Legionella pneumophila. Characterization of substrate specificity and identification of catalytically important residues. J Biol Chem. 1998;273:14788–14795. doi: 10.1074/jbc.273.24.14788. [DOI] [PubMed] [Google Scholar]
- 92.Bessen DE, Kalia A. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect Immun. 2002;70:1159–1167. doi: 10.1128/IAI.70.3.1159-1167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kreikemeyer B, Klenk M, Podbielski A. The intracellular status of Streptococcus pyogenes: role of extracellular matrix-binding proteins and their regulation. Int J Med Microbiol. 2004;294:177–188. doi: 10.1016/j.ijmm.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 94.Kreikemeyer B, Nakata M, Oehmcke S, Gschwendtner C, Normann J, et al. Streptococcus pyogenes collagen type I-binding Cpa surface protein. Expression profile, binding characteristics, biological functions, and potential clinical impact. J Biol Chem. 2005;280:33228–33239. doi: 10.1074/jbc.M502896200. [DOI] [PubMed] [Google Scholar]
- 95.Kreikemeyer B, Oehmcke S, Nakata M, Hoffrogge R, Podbielski A. Streptococcus pyogenes fibronectin-binding protein F2: expression profile, binding characteristics, and impact on eukaryotic cell interactions. J Biol Chem. 2004;279:15850–15859. doi: 10.1074/jbc.M313613200. [DOI] [PubMed] [Google Scholar]
- 96.Molinari G, Rohde M, Talay SR, Chhatwal GS, Beckert S, et al. The role played by the group A Streptococcal negative regulator Nra on bacterial interactions with epithelial cells. Mol Microbiol. 2001;40:99–114. doi: 10.1046/j.1365-2958.2001.02373.x. [DOI] [PubMed] [Google Scholar]
- 97.Nakata M, Podbielski A, Kreikemeyer B. MsmR, a specific positive regulator of the Streptococcus pyogenes FCT pathogenicity region and cytolysin-mediated translocation system genes. Mol Microbiol. 2005;57:786–803. doi: 10.1111/j.1365-2958.2005.04730.x. [DOI] [PubMed] [Google Scholar]
- 98.Podbielski A, Woischnik M, Leonard BA, Schmidt KH. Characterization of nra, a global negative regulator gene in group A Streptococci. Mol Microbiol. 1999;31:1051–1064. doi: 10.1046/j.1365-2958.1999.01241.x. [DOI] [PubMed] [Google Scholar]
- 99.Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci U S A. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maione D, Margarit I, Rinaudo CD, Masignani V, Mora M, et al. Identification of a universal Group B Streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, et al. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 102.Abbot EL, Smith WD, Siou GP, Chiriboga C, Smith RJ, et al. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell Microbiol. 2007;9:1822–1833. doi: 10.1111/j.1462-5822.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- 103.Manetti AG, Zingaretti C, Falugi F, Capo S, Bombaci M, et al. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol Microbiol. 2007;64:968–983. doi: 10.1111/j.1365-2958.2007.05704.x. [DOI] [PubMed] [Google Scholar]
- 104.Dramsi S, Caliot E, Bonne I, Guadagnini S, Prevost MC, et al. Assembly and role of pili in group B Streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 105.Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 106.Shulman ST, Tanz RR, Kabat W, Kabat K, Cederlund E, et al. Group A Streptococcal pharyngitis serotype surveillance in North America, 2000–2002. Clin Infect Dis. 2004;39:325–332. doi: 10.1086/421949. [DOI] [PubMed] [Google Scholar]
- 107.Beres SB, Sylva GL, Sturdevant DE, Granville CN, Liu M, et al. Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections. Proc Natl Acad Sci U S A. 2004;101:11833–11838. doi: 10.1073/pnas.0404163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Green NM, Beres SB, Graviss EA, Allison JE, McGeer AJ, et al. Genetic diversity among type emm28 group A Streptococcus strains causing invasive infections and pharyngitis. J Clin Microbiol. 2005;43:4083–4091. doi: 10.1128/JCM.43.8.4083-4091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schmitz FJ, Beyer A, Charpentier E, Normark BH, Schade M, et al. Toxin-gene profile heterogeneity among endemic invasive European group A Streptococcal isolates. J Infect Dis. 2003;188:1578–1586. doi: 10.1086/379230. [DOI] [PubMed] [Google Scholar]
- 110.Musser JM, Kapur V, Szeto J, Pan X, Swanson DS, et al. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alberti S, Garcia-Rey C, Dominguez MA, Aguilar L, Cercenado E, et al. Survey of emm gene sequences from pharyngeal Streptococcus pyogenes isolates collected in Spain and their relationship with erythromycin susceptibility. J Clin Microbiol. 2003;41:2385–2390. doi: 10.1128/JCM.41.6.2385-2390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aracil B, Minambres M, Oteo J, Torres C, Gomez-Garces JL, et al. High prevalence of erythromycin-resistant and clindamycin-susceptible (M phenotype) viridans group Streptococci from pharyngeal samples: a reservoir of mef genes in commensal bacteria. J Antimicrob Chemother. 2001;48:592–594. doi: 10.1093/jac/48.4.592. [DOI] [PubMed] [Google Scholar]
- 113.Arpin C, Canron MH, Maugein J, Quentin C. Incidence of mefA and mefE genes in viridans group Streptococci. Antimicrob Agents Chemother. 1999;43:2335–2336. doi: 10.1128/aac.43.9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Luna VA, Coates P, Eady EA, Cove JH, Nguyen TT, et al. A variety of gram-positive bacteria carry mobile mef genes. J Antimicrob Chemother. 1999;44:19–25. doi: 10.1093/jac/44.1.19. [DOI] [PubMed] [Google Scholar]
- 115.Luna VA, Heiken M, Judge K, Ulep C, Van Kirk N, et al. Distribution of mef(A) in gram-positive bacteria from healthy Portuguese children. Antimicrob Agents Chemother. 2002;46:2513–2517. doi: 10.1128/AAC.46.8.2513-2517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Perez-Trallero E, Vicente D, Montes M, Marimon JM, Pineiro L. High proportion of pharyngeal carriers of commensal Streptococci resistant to erythromycin in Spanish adults. J Antimicrob Chemother. 2001;48:225–229. doi: 10.1093/jac/48.2.225. [DOI] [PubMed] [Google Scholar]
- 117.Syrogiannopoulos GA, Grivea IN, Fitoussi F, Doit C, Katopodis GD, et al. High prevalence of erythromycin resistance of Streptococcus pyogenes in Greek children. Pediatr Infect Dis J. 2001;20:863–868. doi: 10.1097/00006454-200109000-00008. [DOI] [PubMed] [Google Scholar]
- 118.Overbeek R, Larsen N, Walunas T, D'Souza M, Pusch G, et al. The ERGO genome analysis and discovery system. Nucleic Acids Res. 2003;31:164–171. doi: 10.1093/nar/gkg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, et al. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci U S A. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 122.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 123.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 126.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 128.Rastogi PA. MacVector. Integrated sequence analysis for the Macintosh. Methods Mol Biol. 2000;132:47–69. doi: 10.1385/1-59259-192-2:47. [DOI] [PubMed] [Google Scholar]
- 129.Ghai R, Hain T, Chakraborty T. GenomeViz: visualizing microbial genomes. BMC Bioinformatics. 2004;5:198. doi: 10.1186/1471-2105-5-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, et al. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
New GAS Metagenome Genes
(0.37 MB DOC)