Abstract
The biogeography of the purple nonsulfur bacterium Rhodopseudomonas palustris on a local scale was investigated. Thirty clones of phototrophic bacteria were isolated from each of five unevenly spaced sampling locations in freshwater marsh sediments along a linear 10-m transect, and a total of 150 clones were characterized by BOX-PCR genomic DNA fingerprinting. Cluster analysis of 150 genomic fingerprints yielded 26 distinct genotypes, and 106 clones constituted four major genotypes that were repeatedly isolated. Representatives of these four major genotypes were tentatively identified as R. palustris based on phylogentic analyses of 16S rRNA gene sequences. The differences in the genomic fingerprint patterns among the four major genotypes were accompanied by differences in phenotypic characteristics. These phenotypic differences included differences in the kinetics of carbon source use, suggesting that there may be functional differences with possible ecological significance among these clonal linages. Morisita-Horn similarity coefficients (CMH), which were used to compare the numbers of common genotypes found at pairs of sampling locations, showed that there was substantial similarity between locations that were 1 cm apart (CMH, ≥0.95) but there was almost no similarity between locations that were ≥9 m apart (CMH, ≤0.25). These calculations showed there was a gradual decrease in similarity among the five locations as a function of distance and that clones of R. palustris were lognormally distributed along the linear 10-m transect. These data indicate that natural populations of R. palustris are assemblages of genetically distinct ecotypes and that the distribution of each ecotype is patchy.
Studies of the spatial distribution of bacteria have demonstrated that populations of the same species found in various ecological habitats have distinctive physiological traits (6, 14, 24, 31). This may be due to the fact that microorganisms are highly mutable (2, 17, 25) and readily acquire traits through adaptive evolution that enable populations to be well suited to persist in a wide array of environments that have different biotic and abiotic characteristics. This is akin to the development of ecotypes within eukaryotic species (8, 13), in which genetic variants possess physiological traits that confer fitness advantages in specific habitats (8). While it is increasingly apparent that ecotypes of prokaryotic species also exist, little is known about their spatial distribution on a local scale.
Studies of the population structure and distribution of the host-associated species Escherichia coli have demonstrated that certain clonal types are better adapted than others for colonization of specific hosts. For example, over an 11-month period Caugant et al. (4) found 53 genotypes of E. coli in a single human host. However, only two of these genotypes were persistent over time, and 46% of all the isolates represented a single genotype; the remaining genotypes were apparently transient members of the gastrointestinal flora. This finding suggests that two specific clones of E. coli were well adapted to the host or to different sites within the host but that the vast majority of genotypes failed to effectively compete and could not become established in the host's gastrointestinal tract. This observation was confirmed and extended by Souza et al. (24), who studied the genetic structure of 202 strains of E. coli that had been isolated from 81 mammalian species on different continents. The strains were characterized genotypically by multilocus enzyme electrophoresis and by determining patterns of sugar utilization, antibiotic resistance, and plasmid profiles. The data showed that the genetic relationships among the strains could best be accounted for the country of origin and the taxonomic order of the host from which they were isolated. These data suggest that the environmental conditions within the gastrointestinal tract of a given host select for specific ecotypes of E. coli and that this is an important factor in determining the distribution of E. coli populations.
Previous studies have also demonstrated the ecotypic structure of non-host-associated (i.e., free-living) bacterial species. For example, Moore et al. (16) have shown that cultured isolates of the cyanobacterium Prochlorococcus vary widely in their pigment compositions and growth responses to light and nutrients, yet exhibit more than 97% identity in their 16S rRNA sequences. Other genetic differences in isolates of these organisms, such as variations in the length and G+C content of the internal transcribed spacer region, have been observed. These results provide further evidence that natural populations of Prochlorococcus consist of multiple coexisting ecotypes that are genetically related but physiologically distinct. Similarly, the genotypic diversity and phenotypic diversity of environmental clones of the purple nonsulfur bacterium Rhodopseudomonas palustris from two ecologically distinct sites have been investigated previously (19). The data from that study showed that the genomic fingerprints of different R. palustris clones isolated from each sampling site were significantly different, and none of the genotypes were found in both sites. These results suggest that natural populations of R. palustris are endemic and that adaptation to a given environment may play an important role in determining the local population structure of this species.
The primary aim of this study was to expand our understanding of the spatial distribution of genetically distinct ecotypes of R. palustris on a local scale. To do this, 30 clones of phototrophic bacteria were isolated from each of five unevenly spaced sampling locations along a linear 10-m transect, and the genotypes of all 150 clones were determined by repetitive element PCR genomic DNA fingerprinting by using the BOX A1R primer. Representatives of the numerically dominant genotypes were used for 16S rRNA gene sequencing and phenotypic characterization based on the kinetics of carbon source use. To further assess the ecotypic structure of R. palustris populations, Morisita-Horn similarity coefficients (CMH) were used to compare the numbers of common genotypes found at pairs of sampling locations.
MATERIALS AND METHODS
Sampling and isolation of phototrophic bacteria.
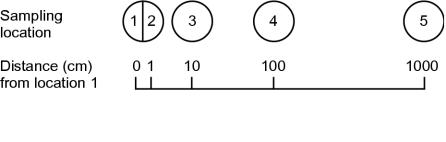
Four cores of sediment samples were collected along a 10-m transect (Fig. 1) on 7 December 1999 from a freshwater marsh in Haren, The Netherlands (19) by using sterile bottomless aluminum cans (500 ml). The first can had a thin partition that divided the core into halves. A total of five samples consisting of 0.3 to 0.8 g (wet weight) of sediment from the surfaces of the cores were taken, and each sample was suspended in 9 volumes of anoxically prepared LCM medium (see below) from which yeast extract had been omitted. The suspensions were sonicated briefly three times for 10 s (B3 sonicator; Branson, Dietzenbach, Germany) and subsequently shaken for 1.5 h at room temperature. After sedimentation, aliquots of the supernatant of each sediment suspension were removed, and a dilution series was plated on LCM agar plates supplemented with 2 mM benzoate and 0.1% sodium bicarbonate. These plates were incubated at 30°C under anoxic conditions (N2 atmosphere) in the light. After 2 weeks of incubation, at least 30 red or red-brown colonies from each sample were streaked on LCMPY-malate plates (see below) and incubated anoxically in the light by using a BBL GasPak anaerobic jar system (Becton Dickinson and Company, Sparks, Md.). Single colonies were then transferred into anoxically prepared liquid LCMPY-malate and incubated in the light.
FIG. 1.
Sampling scheme. Sediment samples were collected at transect locations 1, 2 (1 cm from location 1), 3 (10 cm from location 1), 4 (100 cm from location 1), and 5 (1,000 cm from location 1).
Isolation of DNA.
Genomic DNA was isolated from liquid cultures grown on LCMPY-malate under anoxic conditions in the light. Cells from 2 ml of each culture were harvested by centrifugation and resuspended in 50 mM EDTA (pH 8.0). Genomic DNA was isolated by using a Wizard genomic DNA purification kit (Promega) according to the manufacturer's instructions.
Genomic DNA fingerprinting and computer-assisted cluster analysis of genomic fingerprints.
Aliquots of genomic DNA from various clones were used as templates to generate repetitive element PCR genomic fingerprints with the BOX A1R primer (19, 21). PCR products were separated by electrophoresis on 1.5% agarose gels at 70 V and 4°C for 19 h and stained with ethidium bromide. Images of the stained agarose gels were digitized with an ImageMaster VDS system (Amersham Pharmacia Biotech Benelux, Roosendaal, The Netherlands) and stored as TIFF files. Computer-assisted analysis of the genomic fingerprints was performed by using the GelCompare software program (version 4.1; Applied Maths, Kortrijk, Belgium). Cluster analysis of similarity matrices was performed by the unweighted pair group method using arithmetic averages (23). BOX-PCR fingerprint patterns having r values of more than 0.8 were considered to be the same genotype (19).
The Shannon indices for each sampling location were calculated by using −Σpixlnpi, where pi is the proportion of individuals found in the ith genotype (3, 27). Morisita-Horn similarity coefficients (CMH), which were used to compare the numbers of common genotypes found at pairs of sampling locations, were calculated by using the equation 2 × Σ(Ai × Bi)/[NA × NB × (∑Ai2/NA2 +∑Bi2/NB2)], where Ai and Bi are the numbers of isolates of genotype i in samples A and B, respectively, and NA and NB are the total numbers of isolates in samples A and B, respectively (all ∑ are summed from i = 1 to S, the total number of species).
Amplification of 16S rRNA gene and sequencing.
Nearly complete 16S rRNA genes were amplified by using fD1 and rD1 as the primers (30), and PCR products were purified with a QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany). Sequencing was performed with an ABI PRISM BigDye terminator cycle sequencing Ready Reaction kit (PE Applied Biosystems) and an ABI PRISM 310 genetic analyzer (PE Applied Biosystems). The sequencing primer 536F (E. coli positions 519 to 536) was used (19), and the 16S rRNA gene sequences (500 bp) were compared to sequences in the GenBank database by using the Basic Local Alignment Search Tool (BLAST) (1).
Media and growth conditions.
Batch cultures were routinely grown at 30°C in closed screw-cap tubes (16 ml) containing anoxically prepared LCM medium (29) supplemented with 0.3% peptone and 0.3% yeast extract (LCMPY) and an N2 gas phase. After the LCMPY was autoclaved, 25 ml of a sterile 1 M K(NH4)PO4 solution (pH 7.0) per liter, 2 ml of a filter-sterilized vitamin solution (29) per liter, and 10 ml of an autoclaved 1.5 M malate solution per liter were added to the medium (LCMPY-malate). For experiments done to examine the metabolism of benzoate and 3-chlorobenzoate, carbon sources were added at the concentrations indicated below, and 0.1% sodium bicarbonate was added to the LCM medium. Growth was monitored by measuring the optical densities of cultures at 660 nm. Residual benzoate and 3-chlorobenzoate concentrations were determined by reverse-phase high-performance liquid chromatography by using a MicroSpher C18 column (Chrompack, Middelburg, The Netherlands) with methanol-water-acetic acid (50:49.5:0.5, vol/vol/vol) as the eluent, and the effluent was monitored with a UV-975 Intelligent UV/VIS detector (Jasco, Tokyo, Japan) at 254 nm (18).
RESULTS AND DISCUSSION
Genotypic diversity within each sampling location.
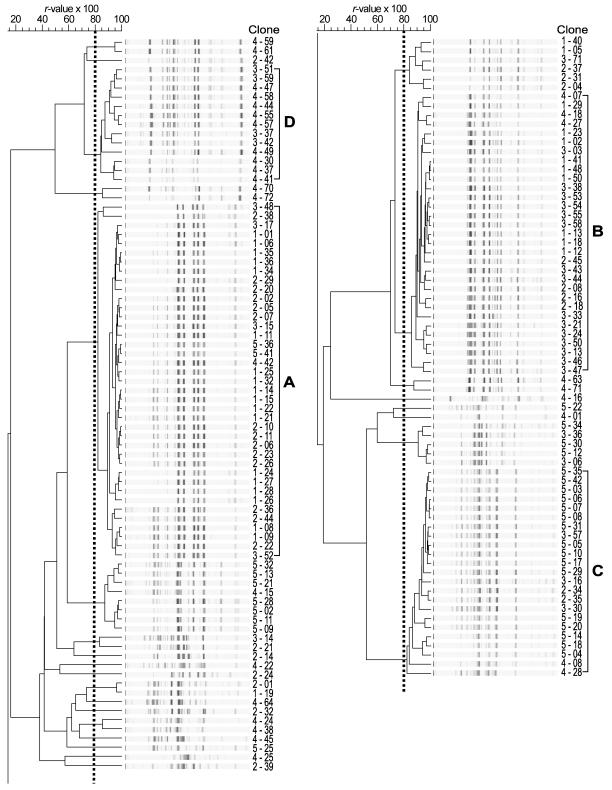
To assess the spatial distribution of environmental clones of the purple nonsulfur bacterium R. palustris on a local scale, 30 clones of phototrophic bacteria from each of five sampling locations along a linear 10-m transect (Fig. 1) were isolated from phototrophically incubated agar plates that had been inoculated with sediment suspensions. Genetic differences among the 150 clones were determined by BOX-PCR genomic DNA fingerprinting. By using computer-assisted cluster analysis, genomic fingerprints having r values of more than 0.8 were considered to be the same genotype (19). A total of 26 genotypes were found at the five locations, and there were 4 to 15 genotypes at each location. Four numerically dominant genotypes, designated genotypes A, B, C, and D, which consisted of more than 10 clones each, were repeatedly isolated from all locations (Fig. 2). Further analyses with representatives of these four major genotypes were performed to more fully characterize the extent of genotypic and phenotypic diversity within the genotypes.
FIG. 2.
Computer-assisted product-moment and unweighted pair group method using arithmetic averages cluster analysis of BOX-PCR genomic DNA fingerprints of 150 clones. Fingerprint patterns having r values of more than 0.8 (dotted line) were considered to be the same genotype. Clone designations (sampling location-clone number) are shown next to the BOX-PCR genomic fingerprints. The four genotypes with more than 10 clones were designated A, B, C, and D.
Biogeography of environmental clones of R. palustris.
Partial sequencing of 16S rRNA genes (500 bp) from two representatives of each genotype was performed to determine the phylogenetic relatedness of the representatives. The overall level of identity of the sequences of these eight clones was more than 98%. The 16S rRNA gene sequences of the clones representing genotypes A and B and of the clones representing genotypes C and D showed 98.4 and 98.2% identity to the 16S rRNA gene sequence of R. palustris strain ATCC 17001 (GenBank accession number D25312), respectively, indicating that each genotype was phylogenetically related to R. palustris (19, 22, 26). A previous study showed that clones with very similar BOX-PCR fingerprints (r values of more than 0.8) had identical 16S rRNA gene sequences (19). We therefore tentatively concluded that 106 of the 150 clones (70.6%) that were classified as genotypes A, B, C, and D were clones of R. palustris.
Only a few studies have focused on the biogeography of free-living bacteria. For example, studies of 3-chlorobenzoate-degrading strains (7) and fluorescent Pseudomonas strains (5) have shown that there is almost no overlap among genotypes between sampling sites at a given location or in continental regions. Similarly, a previous study of the genotypic and phenotypic diversity of environmental clones of R. palustris found at two ecologically distinct sites showed that none of the genotypes was found at both sites (19). These studies suggested that the genotypes of free-living bacteria (based on genomic DNA fingerprinting) are not globally mixed but instead are regionally endemic. In the present study, one or two genotypes were numerically dominant at each location and constituted approximately one-half of the total number of clones obtained (Table 1). The Shannon indices calculated for each sampling location were markedly lower than the index obtained when 106 clones from all locations were included in the calculation. This suggests that endemism that arises as a result of adaptation to local environments is most likely responsible for the observed population structures of environmental clones of R. palustris.
TABLE 1.
Genotypic diversity within the species R. palustris at each sampling location along a linear 10-m transect
| Sample
|
No. of clones in genotype:
|
Shannon indexb | |||||
|---|---|---|---|---|---|---|---|
| Location | Distance (cm)a | Size (g, wet wt) | A | B | C | D | |
| 1 | 0 | 0.32 | 18 | 9 | 0 | 0 | 0.64 |
| 2 | 1 | 0.45 | 14 | 4 | 2 | 0 | 0.80 |
| 3 | 10 | 0.39 | 4 | 15 | 3 | 4 | 1.14 |
| 4 | 100 | 0.54 | 1 | 3 | 2 | 9 | 1.08 |
| 5 | 1,000 | 0.87 | 2 | 0 | 16 | 0 | 0.35 |
| Total | 39 | 31 | 23 | 13 | 1.32 | ||
Distance from sampling location 1.
The Shannon indices were calculated by using −Σpilnpi, where pi is the proportion of individuals found in the ith genotype (derived from Fig. 1).
Phenotypic characteristics of R. palustris genotypes.
To assess differences in the phenotypic properties of R. palustris clones isolated in this study, the rates of growth in LCM media supplemented with four different growth substrates were determined. All 13 genotype D clones grew at about the same rate in medium that contained either a mixture of peptone and yeast extract (LCMPY), malate, or benzoate as the carbon source (data not shown), and they were able to degrade 3-chlorobenzoate in the presence of benzoate. Based on these results, clones previously classified as members of the same genotype were presumed to have similar phenotypic traits. Subsequently, the same eight representatives of the four major genotypes that were used for 16S rRNA gene sequencing were tested to determine their abilities to grow in LCM media supplemented with four different growth substrates (Table 2). While all clones were able to grow in LCMPY, there were significant differences among the genotypes. For example, the growth rates of genotype B clones in LCMPY were significantly lower than those of genotype A, C, and D clones. Genotype B and D clones readily grew on media containing malate, whereas genotype A and C clones did not. Finally, only genotype D clones were able to metabolize benzoate and were able to degrade 3-chlorobenzoate. These data showed that genotype A, B, C, and D clones could be distinguished from one another based on phenotypic characteristics, indicating that the species R. palustris is an assemblage of phenotypically diverse clones.
TABLE 2.
Anaerobic growth of R. palustris clones
| Genotype | Clonea | Growth onb
|
Utilization of 3-chlorobenzoatee
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| LCMPY
|
Malate
|
BA
|
|||||||
| Culture densityc | Growth rated | Culture densityc | Growth rated | Culture densityc | Growth rated | With benzoate | Without benzoate | ||
| A | 3-15 | + | 6.5 (0.5) | + | NC | − | NG | − | − |
| 3-52 | + | 6.8 (0.8) | + | NC | − | NG | − | − | |
| B | 3-50 | +++ | 13.2 (0.6) | +++ | 12.8 (0.8) | − | NG | − | − |
| 3-55 | +++ | 13.0 (1.0) | +++ | 13.3 (0.8) | − | NG | − | − | |
| C | 3-16 | +++ | 7.8 (0.3) | + | NC | − | NG | − | − |
| 3-57 | +++ | 7.8 (0.3) | + | NC | − | NG | − | − | |
| D | 3-37 | +++ | 7.0 (0.5) | +++ | 13.2 (0.8) | ++ | 18.7 (1.3) | + | − |
| 3-59 | +++ | 7.3 (0.8) | +++ | 13.8 (0.6) | ++ | 19.2 (1.3) | + | − | |
Representatives of each genotype that originated from sampling location 3 were used.
The media used were as follows: LCMPY, LCM medium containing a mixture of 0.3% peptone and 0.3% yeast extract; malate, LCM medium containing 15 mM malate; BA, LCM medium containing 2 mM benzoate.
The optical densities at 660 nm after 2 weeks were as follows: +++, >1.0; ++, 0.5 to 1.0; +, 0.1 to 0.5; and −,<0.1.
Doubling time (in hours). The data are averages of three measurements. Standard deviations are indicated in parentheses. NC, not calculated due to the low growth rate; NG, no growth.
Utilization of 1 mM 3-chlorobenzoate in the presence or in the absence of 1 mM benzoate.
Ecotypic structure of an R. palustris population.
Rademaker et al. (20) reported that the similarity coefficients derived from BOX-PCR genomic fingerprints of Xanthomonas strains correlated well with those based on levels of DNA-DNA hybridization. The data of these authors showed that the level of similarity among fingerprints reflects the genetic relatedness of closely related bacteria. In the present study, four distinct genotypes of R. palustris were genetically quite dissimilar (r values, <0.2) based on cluster analysis of BOX-PCR genomic fingerprints (Fig. 2). Furthermore, the differences in genomic fingerprint patterns among the genotypes were accompanied by differences in phenotypic characteristics (Table 2). This implies that each genotype is a distinct clade and that there may be functional differences that have ecological significance among the clonal lineages. The origin of these lineages is unknown; they may represent ecotypes that diverged through sympatric speciation, or they could have evolved in other locations and immigrated to the site.
Like populations of other species, bacterial populations experience periodic selection in which selective sweeps occur in which clones with higher fitness arise and become numerically dominant (12). When a favorable mutation occurs in a population that occupies a homogeneous environment, in theory the clonal variant will eventually replace all other clones in that habitat through competitive exclusion. However, the newly established clones will not replace all other clones if the habitat is spatially structured (28) and will rarely spread outside a given habitat since they probably lack the adaptive traits necessary to successfully compete in other environments (11, 13). In this case, the vast number of individual clones that comprise a species is broken up into patches, each harboring subpopulations that effectively behave and evolve as independent units (11).
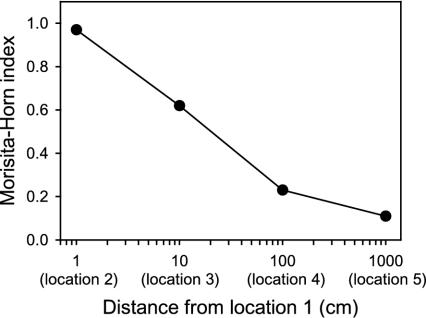
The CMH values for comparisons of populations of the four major genotypes of R. palustris at sampling locations showed that there was a high level of similarity between locations that were 1 cm apart (CMH, ≥0.95) and almost no similarity between locations that were ≥9 m apart (CMH, ≤0.25) (Fig. 3). There was a gradual decrease in similarity as a function of distance along the linear 10-m transect, and there was a roughly linear relationship between the level of similarity between sampling locations and the geographical distance between the locations. These data suggest that at this study site multiple small-scale and ecologically distinct habitats most likely exist in patchy arrangements (9, 10) and that each habitat selects for genetically distinct clones. Soil is generally recognized as a spatially heterogeneous habitat in terms of both its composition and its structure (15, 32). This spatial heterogeneity is characteristic of soils and leads to the development of complex gradients of gases, solutes, nutrients, and other substances that influence microbial growth and survival and ultimately determine the patterns of microbial biodiversity in an environment. Hence, the observed patchy distribution and number of R. palustris ecotypes observed may simply reflect the underlying diversity and number of ecological niches found in the soil environment.
FIG. 3.
CMH values for comparisons of populations of the four major genotypes of R. palustris between sampling location 1 and sampling locations 2, 3, 4, and 5 as a function of distance along a linear 10-m transect.
Acknowledgments
We thank D. M. Pegtel, H. Bolhuis (University of Groningen), and W. G. Meijer (University College Dublin) for valuable discussions and suggestions.
This work was supported by a grant from the Ubbo-Emmius Foundation of the University of Groningen, Groningen, The Netherlands.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arber, W. 2000. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 24:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Begon, M., J. L. Harper, and C. R. Townsend. 1996. Ecology: individuals, populations and communities, 3rd ed. Blackwell Science Ltd., Oxford, United Kingdom.
- 4.Caugant, D. A., B. R. Levin, and R. K. Selander. 1981. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98:467-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, J.-C., and J. M. Tiedje. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fulthorpe, R. R., A. N. Rhodes, and J. M. Tiedje. 1998. High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria. Appl. Environ. Microbiol. 64:1620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Futuyma, D. J. 1998. Evolutionary biology, 3rd ed. Sinauer Associates, Inc., Sunderland, Mass.
- 9.Grundmann, G. L., and D. Debouzie. 2000. Geostatistical analysis of the distribution of NH4+ and NO2−-oxidizing bacteria and serotypes at the millimeter scale along a soil transect. FEMS Microbiol. Ecol. 34:57-62. [DOI] [PubMed] [Google Scholar]
- 10.Hattori, T., and R. Hattori. 1976. The physical environment in soil microbiology: an attempt to extend principles of microbiology to soil microorganisms. Crit. Rev. Microbiol. 4:423-461. [DOI] [PubMed] [Google Scholar]
- 11.Haubold, B., and P. B. Rainey. 1996. Genetic and ecotypic structure of a fluorescent Pseudomonas population. Mol. Ecol. 5:747-761. [Google Scholar]
- 12.Levin, B. R. 1981. Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics 99:1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maynard Smith, J. 1991. The population genetics of bacteria. Proc. R. Soc. Lond. B 245:37-41. [Google Scholar]
- 14.McArthur, J. V., D. A. Kovacic, and M. H. Smith. 1988. Genetic diversity in natural populations of a soil bacterium across a landscape gradient. Proc. Natl. Acad. Sci. USA 85:9621-9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metting, F. B., Jr. 1992. Structure and physiological ecology of soil microbial communities, p. 3-25. In F. B. Metting, Jr. (ed.), Soil microbial ecology. Applications in agricultural and environmental management. Marcel Dekker, Inc., New York, N.Y.
- 16.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 17.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 18.Oda, Y., Y. P. de Vries, L. J. Forney, and J. C. Gottschal. 2001. Acquisition of the ability for Rhodopseudomonas palustris to degrade chlorinated benzoic acids as the sole carbon source. FEMS Microbiol. Ecol. 38:133-139. [Google Scholar]
- 19.Oda, Y., W. Wanders, L. A. Huisman, W. G. Meijer, J. C. Gottschal, and L. J. Forney. 2002. Genotypic and phenotypic diversity within species of purple nonsulfur bacteria isolated from aquatic sediments. Appl. Environ. Microbiol. 68:3467-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rademaker, J. L. W., B. Hoste, F. J. Louws, K. Kersters, J. Swings, L. Vauterin, P. Vauterin, and F. J. de Bruijn. 2000. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int. J. Syst. E vol. Microbiol. 50:665-677. [DOI] [PubMed] [Google Scholar]
- 21.Rademaker, J. L. W., F. J. Louws, and F. J. de Bruijn. 1997. Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting, p. 1-26. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, supplement 3. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 22.Rosselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 23.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. Freeman, San Francisco, Calif.
- 24.Souza, V., M. Rocha, A. Valera, and L. E. Eguiarte. 1999. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65:3373-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spiers, A. J., A. Buckling, and P. B. Rainey. 2000. The causes of Pseudomonas diversity. Microbiology 146:2345-2350. [DOI] [PubMed] [Google Scholar]
- 26.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 27.Stiling, P. D. 1999. Ecology: theories and applications, 3rd ed. Prentice-Hall, Inc., Upper Saddle River, N.J.
- 28.Tilman, D. 1994. Competition and biodiversity in spatially structured habitats. Ecology 75:2-16. [Google Scholar]
- 29.van der Woude, B. J., M. de Boer, N. M. J. van der Put, F. M. van der Geld, R. A. Prins, and J. C. Gottschal. 1994. Anaerobic degradation of halogenated benzoic acids by photoheterotrophic bacteria. FEMS Microbiol. Lett. 119:199-208. [DOI] [PubMed] [Google Scholar]
- 30.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise, M. G., L. J. Shimkets, and J. V. McArthur. 1995. Genetic structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl. Environ. Microbiol. 61:1791-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, J., B. Xia, D. S. Treves, L.-Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]