Abstract
The hyporheic zone of a river is nonphotic, has steep chemical and redox gradients, and has a heterotrophic food web based on the consumption of organic carbon entrained from downwelling surface water or from upwelling groundwater. The microbial communities in the hyporheic zone are an important component of these heterotrophic food webs and perform essential functions in lotic ecosystems. Using a suite of methods (denaturing gradient gel electrophoresis, 16S rRNA phylogeny, phospholipid fatty acid analysis, direct microscopic enumeration, and quantitative PCR), we compared the microbial communities inhabiting the hyporheic zone of six different river sites that encompass a wide range of sediment metal loads resulting from large base-metal mining activity in the region. There was no correlation between sediment metal content and the total hyporheic microbial biomass present within each site. However, microbial community structure showed a significant linear relationship with the sediment metal loads. The abundances of four phylogenetic groups (groups I, II, III, and IV) most closely related to α-, β-, and γ-proteobacteria and the cyanobacteria, respectively, were determined. The sediment metal content gradient was positively correlated with group III abundance and negatively correlated with group II abundance. No correlation was apparent with regard to group I or IV abundance. This is the first documentation of a relationship between fluvially deposited heavy-metal contamination and hyporheic microbial community structure. The information presented here may be useful in predicting long-term effects of heavy-metal contamination in streams and provides a basis for further studies of metal effects on hyporheic microbial communities.
The hyporheic zone is a spatially and temporally dynamic ecotone which provides connectivity between terrestrial, groundwater, and lotic habitats (12, 31, 69, 72, 73). It lies beneath the channel of a stream (46), often extending great distances laterally in the subsurface, and is an essential part of lotic ecosystems (22, 57, 59). The microbial transformations of dissolved and particulate nutrients taking place in the hyporheic zone have been shown to influence both macroinvertebrate and algal assemblages and may play a role in the productivity of riparian vegetation (4, 36, 58). Therefore, alterations in the hyporheic ecosystem that result in changes in the resident microbial community structure may be translated to higher trophic levels. In addition, alterations in the structure of microbial communities may be a useful indicator of the effects and extent of anthropogenic contamination. Previous work in our laboratory has focused on describing the types and seasonal dynamics of microorganisms in the hyporheic zone (21). This investigation explores the effects of heavy-metal contamination on hyporheic-zone microbial community structure.
Heavy metals contaminate numerous aquatic environments worldwide as a result of large-scale mining and other activities (49). Heavy metals reduce water quality and harm many eukaryotic organisms (13, 44, 49, 71). Alteration of streambed geochemistry due to acid mine drainage and the introduction of mine tailings into streams is well documented (for a review, see reference 49); however, the resulting effects on hyporheic microbial communities are poorly understood. Heavy-metal contamination has been shown to alter the activity and composition of microbial communities in terrestrial ecosystems. The majority of these studies have been conducted in systems that were exposed to heavy metals via industrial activities (14, 42, 54, 68, 77), experimental manipulation (18, 28, 29, 41, 70), or acid mine drainage (7, 8, 20, 48). Soils experimentally exposed to increased levels of heavy metals tend to support microbial communities with decreased fungus/bacterium ratios (29), decreased archaeal abundance (64), increased levels of metal tolerance (18), and decreased metabolic potential (40).
The Clark Fork River in western Montana has a legacy of contamination with a mixture of metals from a large copper mine near Butte, Mont. The mining activity in Butte removed approximately 400 million m3 of rock from the subsurface, 90% of which was discarded as tailings (49). It is estimated that 2 million m3 of tailings have been dumped directly into Silver Bow Creek, one of two headwater streams that merge to form the Clark Fork River. The results of this contamination include drastic changes in sediment and pore water geochemistry (76), immediate to long-term effects on local biota (47), and a gradient of elevated sediment metal concentrations which decreases logarithmically with distance and is projected to extend up to 556 km downstream from Butte, Mont. (45, 49). Since the metal contamination introduced into the Clark Fork River came from an ore body that contained a complex mixture of heavy metals, the resulting metal contamination gradient is also comprised of a mixture of metals. Rather than attempt to determine the effects of individual metals on hyporheic microbial communities, we have developed a contamination index (CI) that encompasses the suite of the toxic metals present to relate to measured microbial response variables. This approach increases the environmental relevance of the findings and simplifies interpretation of the results based on microbial community differences related to in situ metal contamination as a whole.
Utilizing a suite of molecular techniques, including 16S rRNA gene phylogenetic analysis, denaturing gradient gel electrophoresis (DGGE), quantitative PCR (qPCR), and phospholipid fatty acid analysis (PLFA), we present the first investigation of the relationship between hyporheic microbial community structure and the concentration of fluvially deposited heavy metals in river sediments. The results demonstrate that total microbial community composition and the abundance of two specific phylogenetic groups were correlated with the sediment metal content, while total bacterial biomass and the abundance of two other phylogenetic groups were not.
MATERIALS AND METHODS
Study sites.
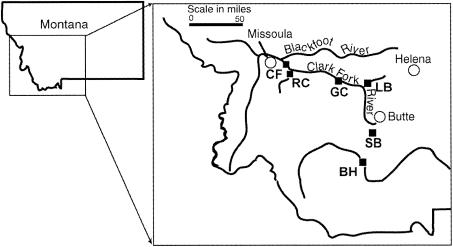
Six different locations were sampled for this study (Fig. 1). These sites were chosen from a comprehensive survey of streams in western Montana (15) to encompass a range of sediment metal concentrations and other physical characteristics such as average discharge, substratum type, and drainage area (Table 1). The U.S. Geological Survey Montana stream-flow website (http://waterdata.usgs.gov/mt/nwis/current?type=flow) was used to obtain general physical characteristics of each stream. The sampled reaches of each stream had riparian vegetation dominated by alder, willow, and cottonwood groves, and channel water flowed freely, unobstructed by any major debris or dams. Three sites were located within the Clark Fork River watershed: Silver Bow Creek (SB), a headwater tributary of the Clark Fork (third-order stream; lat. 46°06′28", long. 112°48′17"; elevation, 4,912 ft); Clark Fork River at Gold Creek (GC) (fourth-order stream; lat. 46°35′26", long. 112°55′40"; elevation, 4,172.8 ft); and Clark Fork River at Rock Creek (CF) (fourth-order stream; lat. 46°49′34", long. 113°48′48"; elevation, 3,320 ft). The other three sites were the Little Blackfoot River (LB), sampled near Garrison, Mont. (third-order stream; lat. 46°31′11", long. 112°47′33"; elevation, 4,344 ft); the Big Hole River (BH), sampled near Glen, Mont. (fourth-order stream; lat. 45°26′26", long. 112°33′20"; elevation, 4,850 ft); and Rock Creek (RC), sampled near Missoula, Mont. (third-order stream; lat. 46°43′21", long. 113°40′56"; elevation, 3,519 ft). The latter three sites were included in the previous study of seasonal dynamics in microbial communities (21).
FIG. 1.
Map of field sampling sites. Each stream sampled is indicated by a square (▪). SB, Silver Bow Creek; GC, Clark Fork at Gold Creek; CF, Clark Fork at Rock Creek; LB, Little Blackfoot River; RC, Rock Creek; BH, Big Hole River.
TABLE 1.
Physical parameters of streams
| Site | Catchment area (square miles) | Steambed gradient | Substratum type | Avg pH | Mean concn (SE) ofa:
|
||||
|---|---|---|---|---|---|---|---|---|---|
| As | Cd | Cu | Pb | Zn | |||||
| LB | 407 | 0.004 | Small cobbles/gravel | 8.3 | 6.01 (0.80) | 0.29 (0.03) | 4.41 (0.40) | 8.26 (0.38) | 21.9 (1.04) |
| RC | 885 | 0.007 | Large cobbles/gravel | 8.1 | 2.01 (0.40) | 0.11 (0.01) | 1.43 (0.13) | 2.59 (0.20) | BDL (N/A) |
| BH | 2,665 | 0.003 | Large cobbles/gravel | 8.0 | 2.85 (0.11) | 0.11 (0.01) | 1.14 (0.10) | 3.71 (0.13) | 6.77 (0.02) |
| CF | 3,641 | 0.006 | Large cobbles/gravel | 7.9 | 3.68 (0.23) | 0.45 (0.01) | 30.1 (1.12) | 7.87 (0.19) | 108 (7.71) |
| GC | 1,760 | 0.004 | Large cobbles/gravel | 8.3 | 5.88 (0.38) | 0.53 (0.04) | 65.9 (5.33) | 13.9 (1.74) | 147 (7.17) |
| SB | 363 | 0.004 | Small cobbles/gravel | 8.2 | 68.9 (19.9) | 1.84 (0.12) | 332 (45.6) | 69.8 (8.91) | 433 (74.7) |
Values are mean concentrations (micrograms of metal/gram [dry wt] of sediment) and standard errors of each metal associated with hyporheic sediments gathered from each stream. Detection limit for Zn, 0.04 μg/g. BDL, below detection limit; N/A, not applicable.
Sampling design.
It has previously been indicated that particle size can affect community structure by altering flow rates, nutrient availability and recharge, and surface area available for colonization (11, 43). Bed materials in freestone rivers in the Rocky Mountain West, including those in this study, are very heterogeneous in nature and distributed patchily, ranging from silt to cobbles. Thus, we chose to compare a single sediment size fraction (2.36 to 1.70 mm in diameter) for all sites and rivers to reduce the potential for high variability between streams resulting from patchy distribution of heterogeneous sediments and to allow direct comparison of microbial communities inhabiting similar environments but in different locations. Further, this same rationale and strategy were employed in prior work at some of these sites (21), making it possible to directly compare and extend on results from different studies.
Three replicate 125-g samples of hand-sieved bulk sediment (diameter, 2.36 to 1.70; depth, 0 to 20 cm) from each of the six sites were packed into sterile, slotted polyvinyl chloride columns and buried in the streambeds at the heads of three separate riffles at each respective site, as described previously (21). Thus, the sediment columns harbored the native community from their respective streams at the time of emplacement and did not require extensive recolonization. Pore water samplers (described previously in reference 52) were placed in groups of two, 30 cm apart, at 5- to 10-cm depths ∼20 cm upstream of the buried columns. After burial, the columns were allowed to equilibrate in place for 6 weeks before they were removed and processed. Handling of sediments at the time of sampling was performed as described previously (21). In addition, fresh sediment samples of the same size fraction were also recovered from each streambed at the time of sampling to determine if the column sediment communities were similar to those associated with previously undisturbed sediments.
Geochemical analyses.
For sediment samples, 5 g of dried sediment was extracted with 12.5 ml of concentrated trace-metal-grade HNO3 and 12.5 ml of concentrated trace-metal-grade HCl. Samples were heated to 95°C (±5°C) and refluxed for 1 h and then cooled for 10 min, diluted to 50 ml with milli-Q water, inverted, and shaken and then allowed to cool and settle overnight. Extracted samples were filtered using a FilterMate filtering device (Environmental Express, Mt. Pleasant, S.C.), and the eluant was analyzed for total dissolved metals on an inductively coupled argon plasma emission spectrometer (ICAPES) (IRIS model, Thermoelemental, Franklin, Mass.) according to U.S. Environmental Protection Agency test method 200.7. Total recoverable metals were measured for each sediment sample, and the concentrations of five metals (As, Cd, Cu, Pb, and Zn) were used to create the CI. This index was used as a measure of contamination relative to the metal content of the sediment at the most pristine site included in the study (RC). The CI was calculated by the formula CI = Σ((log Men/log Men at RC)/number of metals included in index), where n represents As, Cd, Cu, Pb, and Zn.
For water samples, three 10-ml pore water samples were collected from each pore water sampler prior to removal of the columns. One water sample from each sampler was analyzed for total dissolved metals, one was analyzed for dissolved As, and one was analyzed for dissolved NO2−, NO3−, PO42−, F −, Cl−, and SO42−. Water samples were filtered on-site with 0.2-μm-pore-size syringe filters (Supor [Gelman], Ann Arbor, Mich.) directly into autosampler vials. Samples to be analyzed for total dissolved metals were acidified in the field by addition of 150 μl of HNO3 and 60 μl of HCl. Samples analyzed for dissolved As received 30 μl of H2O2 in addition to the acid addition above. Filtered and acidified samples were capped, shaken, and stored on ice or at 4°C until analyzed via ultrasonic nebulization on ICAPES. Dissolved anions were determined from nonacidified samples on a Dionex D500 ion chromatograph (Dionex, Sunnyvale, Calif.) as per U.S. Environmental Protection Agency method 300.0 using an AS14 anion separation column (Dionex).
DNA extraction.
Lyophilized sediment samples (1 g) were extracted based on the direct lysis method of Yu and Mohn (78) with modifications as described previously (21).
DGGE and gel pattern analysis.
PCR amplification for DGGE analysis, details of the DGGE protocol, gel staining, band visualization, and pattern analysis have been described elsewhere (21). Briefly, PCR amplicons were generated using the generally conserved 16S rRNA gene primer pair 536fC (5′-CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC CCA GCM GCC GCG GTA ATW C-3′) and 907r (5′-CCG TCA ATT CMT TTR AGT TT-3′). For DGGE analysis, 400 ng of PCR product generated from each sample was separated on a 6% acrylamide gel with a linear denaturant gradient range of 25 to 60% using the Bio-Rad D-GENE System (Bio-Rad Laboratories, Hercules, Calif.). Gels were stained with SYBERGreen I (BioWhittaker Molecular Applications, Rockland, Maine), and bands were visualized using a Bio-Rad Gel Doc 1000 and Molecular Analyst software (Bio-Rad Laboratories). GelCompar v.4.0 software (Applied Maths, Kortrijk, Belgium) was used to analyze DGGE images for pattern similarities by using the Dice coefficient.
Cloning and DNA sequencing.
Forty-seven individual bands (seven to nine from each site) were recovered from DGGE gels for further sequence analysis and primer development by using modifications of the protocol described by Sanguinetti et al. (65). Briefly, bands were excised from DGGE gels, transferred to 500-μl microcentrifuge tubes, and then macerated and mixed with 100 μl of elution buffer (50 mM KCl, 10 mM Tris, 0.1% Triton X-100 [pH 8.0]). The mixtures were incubated overnight at room temperature to elute the DNA from the acrylamide matrix. The eluted PCR products were again amplified by PCR as described above and then purified using Qiaquick PCR Clean-up columns (Qiagen, Valencia, Calif.). The purified products were cloned into the plasmid vector pT7Blue-3 with the Perfectly Blunt cloning kit (Novagen, Inc., Madison, Wis.). Plasmid clones were identified based on blue-white screening and grown overnight in Luria-Bertani broth amended with ampicillin (300 μg/ml) and tetracycline (15 μg/ml), and plasmid DNA was purified by using Qiagen mini-prep kits as specified. To confirm the identity of each clone, purified plasmid DNA was used as template for PCR using the same 536fC-907r primer pair, and the resulting product was analyzed by DGGE alongside the original total-community PCR products. Bidirectional DNA sequence analysis of confirmed clones was performed by MWGbiotech (High Point, N.C.).
Phylogenetic analysis.
Phylogenetic analysis was performed as described in detail elsewhere (21). Briefly, sequences recovered from excised bands were analyzed for chimeric character by using the Ribosomal Database Project II (RDP II) Chimera Check program (http://rdp.cme.msu.edu/html/). Sequences that appeared chimeric were excluded from further analysis. The closest known relatives of hyporheic microorganisms represented by recovered sequences were identified with the Sequence Match program of RDP II. Sequences were aligned with the Sequence Align program on RDP II, and the resulting alignments were optimized by using SeqPup v.0.8 shareware (http://iubio.bio.indiana.edu/soft/molbio/seqpup/java/seqpup-doc.html). Paup v.4.0b.8.a (Sinauer Associates, Inc., Sunderland, Mass.) was utilized to construct phylogenetic trees from the aligned sequences. The maximum parsimony, maximum likelihood, and neighbor-joining algorithms were each used to generate optimal tree topologies, each of which was confirmed by 100-fold bootstrapping. A consensus tree was generated from these three optimal trees. The four major branches of the consensus tree were used to generate group-specific primers for real-time qPCR analysis as described below.
Real-time qPCR.
Group-specific primers corresponding to the four groups defined by phylogenetic analysis were constructed to monitor group-level abundance across the metal contamination gradient. All primer pairs (Table 2) were generated as described previously (21) except that DNA sequences from all six river sites in the present study were included in the primer development process. Nonetheless, the aligned sequences produced the same consensus sequences and primer sets as in the prior three-site study, although primer pairs Ap, A/R, Ps, and Nfix from reference 21 have been renamed as representing groups I, II, III, and IV, respectively, to reflect their now more general nature.
TABLE 2.
16S rDNA primer pairs generated for qPCR analysis
| Targeted group and product size (nucleotides) | Group most closely affiliated with | Probe name/sequence | Temp (°C) | No. of matches in RDP |
|---|---|---|---|---|
| I (83) | α-Proteobacteria | GIF/forward primer (5′-3′) AACACCAGTGGCGAAGG | 59.61 | 11 |
| GIR/reverse primer 2 (3′-5′) GAGCAAACAGGATTAGATACCC | 60.81 | 17 | ||
| II (171) | β-Proteobacteria | GIIF/forward primer 2 (5′-3′) CGGYAGAGGGGGATGGAA | 62.18 | 0 |
| GIIR/reverse primer 2 (3′-5′) CCCTAAACGATGTCAACTGG | 60.4 | 0 | ||
| III (143) | γ-Proteobacteira | GIIIF/forward primer 2 (5′-3′) GAAATGCGTAGAGATCGGGAG | 62.57 | 0 |
| GIIIR/reverse primer 2 (5′-3′) ACRTCCAGTTCGCATCGTTTAGG | 62.77 | 133 | ||
| IV (86) | Cyanobacteria | GIVF/forward primer F1(5′-3′) CCWGTAGTCCTAGCCGTAA | 60.16 | 0 |
| GIVR/reverse primer 2 (3′-5′) CTAACGCGTTAAGTATCCCG | 60.4 | 208 |
Real-time qPCRs were used to quantify the numbers of copies per gram of sediment representing each of the defined phylogenetic groups by using a Bio-Rad iCycler (Bio-Rad Laboratories) and a SYBERGreen I detection method. PCR mixtures, the construction of qPCR standards, and calculations of target molecule densities have been described in detail previously (21).
Different PCR conditions for each primer pair were as follows: group I, 5 min at 95°C followed by 45 cycles of 15 s at 95°C, 30 s at 58.4°C, and 60 s at 72°C; group II, 5 min at 95°C followed by 45 cycles of 15 s at 95°C, 30 s at 59.4°C, and 60 s at 72°C; group III, 5 min at 95°C followed by 40 cycles of 15 s at 95°C, 30 s at 61.5°C, and 60 s at 72°C; group IV, 5 min at 95°C followed by 40 cycles of 15 s at 95°C, 30 s at 57.3°C, and 60 s at 72°C. SYBER Green fluorescence was measured following the 72°C extension period of each cycle to monitor product accumulation in real time.
PLFA analysis.
PLFAs were extracted and analyzed according to the method of White and Ringelberg (75). Briefly, lipids were removed from samples into chloroform by a modified Bligh and Dyer extraction procedure. Phospholipids were separated from other lipids by silicic acid chromatography and derivatized to fatty acid methyl esters (FAMEs) for analysis by gas chromatography. Two capillary columns of differing polarity (HP-5 [cross-linked 5% phenyl methyl silicon] 50-m by 0.32-mm by 0.52-μm film and HP-225 [50% CNPrPh Me Siloxane] 30-m by 0.32-mm by 0.25-μm film) were used to identify 32 FAMEs by comparison of retention times of suspected FAMEs to retention times of purchased standards. FAME identifications were confirmed by gas chromatography-mass spectrometry.
Microscopic enumeration of bacteria.
Bacterial cell densities associated with 1-g samples of lyophilized hyporheic sediment were determined as described elsewhere (27). Thirty fields of view or 400 cells (whichever was achieved first) were counted from each slide.
Statistical analyses.
All statistical tests for DNA-based measures were run with NCSS 2001 (NCSS, Kaysville, Utah). Statistical analyses used to analyze PLFA data were performed with SPSS software (version 10.0, SPSS Inc.). A P value of 0.05 was set as the significance threshold for all Tukey-Kramer multiple- comparison tests.
RESULTS
CI.
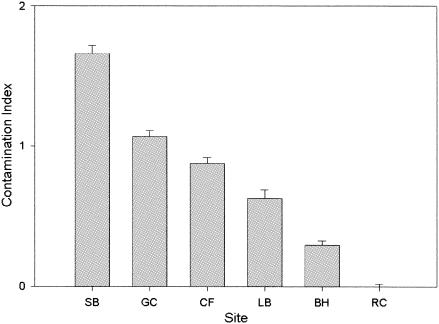
CI values were calculated for the sediment samples gathered from each location (Fig. 2). Concentrations of metals included in the CI differed among sites (FAs = 10.62, P < 0.001; FCd = 147.1, P < 0.001; FCu = 50.15, P < 0.001; FPb = 48.50, P < 0.001; FZn = 28.59, P < 0.001) (Table 1). SB, the location closest to the copper mine in Butte, Mont., had the highest CI value. The GC and CF sites are located downstream in the same drainage as SB and correspondingly had lower CI values. The sites LB and BH are located in drainages that supported mining activities on a smaller scale and thus exhibit lower levels of contamination. The lowest CI values were obtained in RC, a blue-ribbon trout stream that is considered the most pristine of the sites sampled in this study. Together, these six sites represent CI values that decrease linearly across a wide range of sediment metal content values.
FIG. 2.
Bar graph of CI values (mean and standard errors; n = 3) for sediments sieved from each site. Site abbreviations are as described in the legend for Fig. 1.
Direct microscopic enumeration.
Direct microscopy was used to determine bacterial densities associated with the hyporheic sediments. The measured cell densities ranged between 1.73 × 107 and 1.27 × 108 cells g (dry weight) of sediment−1 (data not shown). An analysis of variance indicated that there were significant differences between sites (Fsite = 11.82, P < 0.001). The Tukey-Kramer multiple-comparison test was used to determine significant differences (P < 0.05) between specific sites. The values for log cell number g−1 at GC were significantly higher than those found at LB, BH, and RC. Conversely, the values for log cell number g−1 at BH were significantly lower than those obtained for SB, GC, and CF. Linear regression analysis of log cell number g of sediment−1 versus CI did not reveal a direct relationship between sediment metal content and bacterial biomass (R2 = 0.233, P = 0.33).
Phylogenetic analysis.
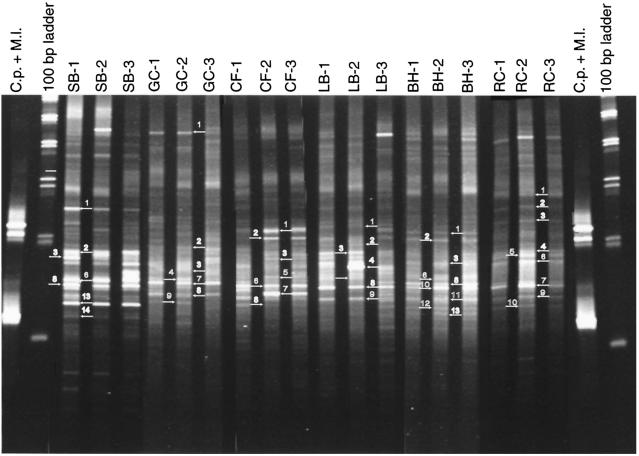
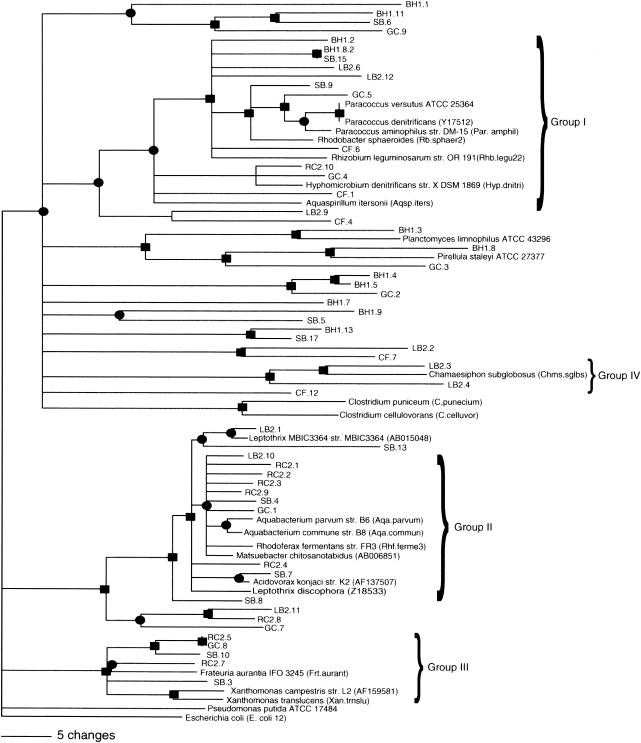
A phylogenetic analysis of the sequences recovered from the DGGE gel (Fig. 3) was performed to determine if there were differences in the distribution of phylogenetic groups along the contamination gradient. The closest matches of the obtained sequences to known species were determined by comparison to the RDP II database (Table 3). The majority of the sequences were most closely related to known aquatic organisms or environmental clones previously recovered from aquatic systems. A phylogenetic tree was generated from the consensus of maximum parsimony, maximum likelihood, and neighbor-joining algorithms as described in Materials and Methods (Fig. 4). There was no clear pattern of species- or group-level distribution along the metal gradient. Rather, sequences recovered from most streams were distributed throughout the tree. Based on this analysis, four phylogenetic groups (I, II, III, and IV) most closely related to α-, β-, and γ-proteobacteria and the cyanobacteria, respectively, were defined for further analysis. The sequences represented in each group from this study formed the basis for the development of the group-specific primers used in the real-time qPCR analyses.
FIG. 3.
DGGE profiles of microbial communities inhabiting each river site. PCR products were synthesized with the universal primer pair 536fC-907r. Labels above each lane indicate which site the pattern represents. Three samples were analyzed from each site. Clostridium perfringens (C.p.) and Micrococcus luteus (M.l.) were used as reference patterns during the normalization procedure in GelCompar. Numbers and arrows indicate which bands were cut and sequenced for the phylogenetic analysis.
TABLE 3.
Closest match of cloned DGGE bands to known species
| Site and clone name | Best match to known species in RDP | Sab score |
|---|---|---|
| SB-1 | Xanthomonas hyacinthi LMG 739 (T) | 0.692 |
| SB-2 | Rhodoferax unidentified proteobacterium arc53 | 0.949 |
| SB-3 | Matsuebacter chitosanotabidus | 0.866 |
| SB-6 | Leptothrix discophora strain SS-1 ATCC 43182 | 0.877 |
| SB-8 | Rhodobacter sphaeroides IFO 12203 | 0.914 |
| SB-13 | Dictyoglomus thermophilum strain H-6-12 DSM 3960 (T) | 0.501 |
| SB-14 | Rhizobium sp. strain CIAM 2927 | 0.763 |
| GC-1 | Aquabacterium commune strain B8 | 0.912 |
| GC-2 | Nitrospira cf. moscoviensis strain SBR2046 | 0.887 |
| GC-3 | Uncultured Pirellula clone 5H12 | 0.704 |
| GC-4 | Hyphomicrobium denitrificans strain X DSM 1869 (T) | 0.765 |
| GC-7 | Thiobacillus aquaesulis | 0.798 |
| GC-8 | Lutelmonas mephitis strain B 1953/27.1 | 0.809 |
| GC-9 | Rhodococcus erythropolis DSM 4318 | 0.993 |
| CF-1 | Sphingomonas subterranea IFO 16086 | 0.919 |
| CF-2 | Calothrix desertica PCC 7102 | 0.713 |
| CF-3 | Desulfobulbus rhabdoformis 16S rRNA gene, complete | 0.541 |
| CF-5 | Bacterial species DNA for 16S rRNA gene (strain IFAM 2074) | 0.699 |
| CF-6 | Methylocystis parvus | 0.711 |
| CF-7 | Acidobacterium subdivision Environmental RB group (clone RB30) | 0.896 |
| CF-8 | Hydrogenophaga palleronii strain S1 DSM 63 (T) | 0.915 |
| LB-1 | Leptothrix strain MBIC3364 | 0.887 |
| LB-2 | Nitrospina subdivision Environmental clone 2027 group (clone 11-25) | 0.725 |
| LB-3 | Chamaesiphon subglobosus PCC 7430 | 0.741 |
| LB-4 | Nostoc strain GSV224 | 0.941 |
| LB-5 | Geobacter sp. strain JW-3 | 0.973 |
| LB-8 | Azospirillum doebereinerae strain 63f | 0.622 |
| LB-9 | Aquabacterium commune strain B8 | 0.922 |
| LB-11 | Unidentified eubacterium from the Amazon 16S rRNA gene | 0.815 |
| BH-1 | Unidentified eubacterium from the Amazon 16S rRNA gene | 0.665 |
| BH-2 | Rhizobium strain CJ5, 24N, USDA 3398 | 0.845 |
| BH-3 | Planctomyces sp. strain Schlesner 642 | 0.741 |
| BH-6 | Rhodoferax unidentified proteobacterium arc53 | 0.904 |
| BH-8 | Pirellula staleyi | 0.637 |
| BH-10 | Pelobacter carbinolicus | 0.624 |
| BH-11 | Janibacter thuringensis | 0.923 |
| BH-12 | Uncultured eubacterium H1.2.f isolated from a deep subsurface paleosol | 0.534 |
| BH-13 | Acidovorax strain G8B1 | 0.842 |
| RC-1 | Acidovorax strain G8B1 | 0.842 |
| RC-2 | Comamonas sp. 16S rRNA gene, isolate 158 | 0.881 |
| RC-3 | Rhodoferax unidentified proteobacterium arc53 | 0.889 |
| RC-4 | Leptothrix strain MBIC3364 | 0.931 |
| RC-5 | Xanthomonas melonis LMG 8670 (T) | 0.894 |
| RC-6 | Frateuria aurantia IFO 3245 (T) | 0.841 |
| RC-7 | Mount Coot-tha region (Brisbane, Australia) 5- to 10-cm-depth soil DNA clone MC 26 | 0.719 |
| RC-9 | Aquabacterium commune strain B8 | 0.942 |
| RC-10 | Hyphomicrobium denitrificans strain X DSM 1869 (T) | 0.846 |
FIG. 4.
Phylogenetic tree of partial 16S rRNA gene sequences amplified with the 536f and 907r universal 16S primers. The symbols and represent branches that are supported by maximum likelihood, maximum parsimony, and neighbor-joining analysis with the following bootstrap values (x): •, 50% < x < 74%; ▪, x > 74%.
This partial phylogenetic survey was undertaken to develop a suite of group-level primers for subsequent analyses of metal effects and should not be taken as a comprehensive assessment of all bacterial populations present. However, the array of phylogenetic groups obtained is in agreement with previous studies in lotic environments (6, 39). Thus, the recovered sequences and identifications appear to be a reasonable representation of the types and diversity of bacteria associated with the hyporheic zone sediments and suitable for the development of group-level primers to monitor effects of metals in this system.
Real-time qPCR.
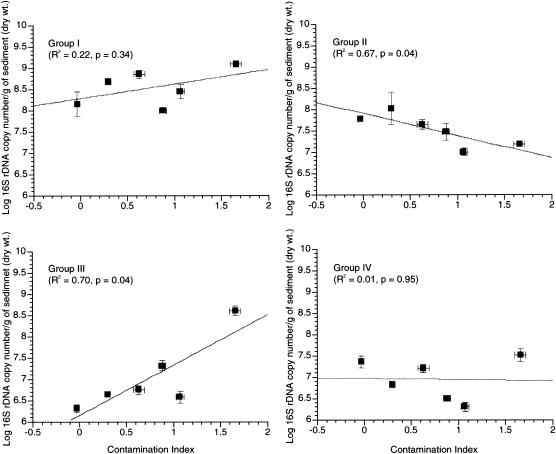
Real-time qPCR was used to quantify the 16S rRNA gene copy number/gram of the four phylogenetic groups defined in this study. The values for copy number/gram of each phylogenetic group were plotted against the CI to determine which groups were affected by the sediment metal content (Fig. 5). The response to the sediment metal content varied among the four groups. Group I abundance was not correlated with CI (R2 = 0.22, P = 0.34), although a general positive trend was observed. The abundance of group II was negatively correlated with the CI (R2 = 0.68, P = 0.043). Group III showed a strong positive correlation with sediment metal content (R2 = 0.70, P = 0.037). Finally, group IV abundance showed no relationship with the sediment metal content (R2 < 0.001, P = 0.954) and no apparent population trends.
FIG. 5.
Abundance of bacterial groups across sites. Linear regressions of mean and standard errors (n = 3) of 16S rRNA gene copy number/gram of sediment versus CI. Each plot represents a separate qPCR response variable (i.e., groups I, II, III, and IV).
DGGE pattern analysis.
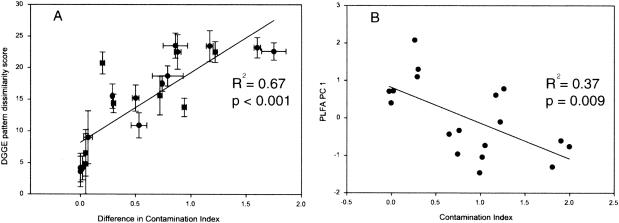
Comparison of fingerprints obtained by DGGE analysis was used as a means of describing the structure of microbial communities at each site. Visual examination of the DGGE patterns (Fig. 3) suggests that each site supports unique molecular species, but a number of bands appear to be common to all sites. In addition, visual inspection suggests that the similarity within a site is greater than the similarity between sites. To better quantify the community structure differences within and between sites, and to test whether there was a relationship between community structure and CI, a similarity matrix was constructed based on the Dice coefficient. Analysis of variance indicated that there was a significant difference between the within-site and between-site similarity scores (Fcomparison type = 109.57, P < 0.001), indicating that there is more variability in community structure between sites than within sites. The mean dissimilarity scores calculated between sites (i.e., SB versus LB, BH versus GC, etc.) for all pairwise combinations were plotted against the difference in CI between the two sites (Fig. 6A). There was a significant positive linear relationship between DGGE pattern dissimilarity and the difference in CI values between sites (R2 = 0.669, P < 0.001).
FIG. 6.
Linear regressions of community structure measures versus CI. (A) DGGE dissimilarity scores versus the difference in CI between all sites. (B) First principal component of the bacterial PLFA analysis versus the CI. Lines represent the best linear fit of the data included in the graph.
PLFA analysis.
Since particular PLFAs can be associated with specific microbial populations or groups, the relative abundance of each identified phospholipid was determined to assess whether there were any identifiable patterns of distribution. Sites with higher CI values (SB, GC, and CF) exhibited a greater proportion of prokaryotic fatty acid markers than the sites with lower CI values (LB, BH, and RC) (data not shown). Conversely, the sites with lower CI values (LB, BH, and RC) had higher relative abundances of markers for certain eukaryotes and actinomycetes (data not shown). Specifically, the low-CI sites tended to have larger amounts of 18:3ω6, 18:2ω6, and 10me16:0, markers for diatoms, fungi, and actinomycetes, respectively. By contrast, SB, GC, and CF tended to have greater amounts of monoenoic, branched, and short-chain fatty acids. However, these are merely qualitative trends and are not consistent along the metal gradient. For example, GC contains a relatively high proportion of the fungal marker 18:2ω6 and other eukaryotic markers (18:3ω5 and 20:5). Thus, the examination of individual fatty acid markers did not reveal a clear relationship between microbial phospholipids and CI.
To further explore possible relationships, we used principal- component analysis and linear modeling. Since some of the eukaryotic markers were not present at all of the sites, we considered only the bacterial phospholipids for the principal- component analysis. The first two principal components of the PLFA analysis were unable to clearly separate the six sites (data not shown). However, a significant negative linear relationship was observed when we plotted the first principal component of the PLFA versus CI (R2 = 0.371, P = 0.009) (Fig. 6B).
Anion analysis.
Linear regression analysis of each dissolved anion versus the community response variables (DGGE community similarity, PLFA PC 1, and real-time qPCR data) revealed no significant relationships (data not shown).
Validation of sediment columns.
Fresh sediment samples gathered at the time of column recovery were analyzed to determine if in situ incubation of sediments in the polyvinyl chloride columns affected the structure of the hyporheic microbial communities. No difference was detected in bacterial cell densities (Fcell density = 1.76, P = 0.19) between freshly sieved sediments and those incubated in the columns. Comparison of DGGE patterns from fresh and column sediments indicated that incubation in the columns did not affect microbial community structure compared to bulk sediment (data not shown). Similarly, there was no significant difference in the patterns of phospholipids recovered from fresh and column sediments.
DISCUSSION
Rivers vary longitudinally (i.e., from headwaters to downstream reaches) in a number of geomorphological factors, including average discharge, substratum type, and drainage area. The river continuum concept (76) states that longitudinal variation in these geomorphological features should control the distribution of biota associated with the river ecosystem. We hypothesized that the influence of fluvial heavy-metal deposition on hyporheic microbial community structure would override the effects of geomorphological variation. If this hypothesis is correct, then there should be a greater similarity among the hyporheic-zone communities in sites with similar CI values. This influence should be apparent regardless of stream size or sampling location, assuming that metal impacts dominate geomorphological differences between streams.
To test this hypothesis, a series of natural stream sites representing a gradient of metal contamination were examined; this can be a useful approach for determining the degree to which metal contaminants influence microbial community structure (2, 18, 29, 54). This field study sampled along a 200-km stretch of the Clark Fork River and several other streams encompassing a range of geomorphological features. Including this geomorphological variation in our sampling regime ensures that any observed metal effects must be dominating to be apparent in light of the other variables in the system. Thus, the correlations discussed herein indicate that the structure of hyporheic microbial communities is controlled to some extent by sediment metal levels and that this heavy- metal influence outweighs the effects of longitudinal geomorphological variation.
This study relates the structure of hyporheic microbial communities to an index of heavy-metal contamination (CI) within each stream. The metals included in the CI are the five most toxic heavy metals present in each stream and are known to influence aquatic and terrestrial biota (1, 17, 32, 34, 35, 70). Metals not known to have toxic effects on aquatic or terrestrial biota were excluded from the CI calculations. All of the heavy metals that were enriched in the contaminated sites covary, and thus determination of the effects of any individual heavy- metal contaminant on hyporheic-zone microbial community structure is confounded (55). Further, owing to the high degree of colinearity in the set of predictor variables (metal concentrations), separate statistical analyses involving individual metal variables would be inappropriate. By relating microbial community structure to CI, we avoided this colinearity issue and developed and utilized an environmentally relevant measure of metal contamination.
It is noted that other metals, such as Fe, can affect the bioavailability of metals in the CI. However, including Fe concentrations in the CI had no effect on the relationships reported here (data not shown). Further, the high degree of correlation between CI and hyporheic microbial community composition and between CI and group-level abundance indicates that the CI is an accurate indicator of bioavailable heavy metals. To our knowledge, there are no prior investigations relating the same types and ranges of metal contaminants used here to changes in river sediment microbial communities. However, previous investigations of terrestrial systems have shown similar community-level responses to heavy-metal contamination (2, 18, 28, 54, 63, 66).
We employed the strategy of analyzing a single sediment size class in emplaced cores to control for the high degree of physical heterogeneity in freestone river sediments, to facilitate future studies on the system (e.g., column transplant experiments), and to maximize our ability to directly compare data from current and future studies.
The absolute size of microbial communities (i.e., standing microbial biomass) has previously been shown to be relatively insensitive to heavy-metal contamination (2, 29, 40, 66). In the current study, the measurements of bacterial cell densities based on direct microscopic enumeration indicated that the total bacterial abundance in the hyporheic zone was unaffected by elevated sediment metal levels. However, it was apparent that the abundance of individual phylogenetic groups inhabiting the hyporheic zone differed along the contamination gradient.
If the group delineations (groups I, II, III, and IV) are extended to encompass the broader phylogenetic groups (i.e., α-, β-, and γ-proteobacteria and cyanobacteria, respectively) suggested by phylogenetic analysis, then these hyporheic-zone data differ from trends seen in prior studies of metal contamination in terrestrial environments. Sandaa et al. (62), studying soils amended with metal-rich sewage sludge, found decreases in α-, β-, and γ-proteobacteria. By contrast, the hyporheic-zone communities exhibited a decrease in α-proteobacteria but no significant change in β-proteobacteria and an increase in γ-proteobacteria with increasing metal contamination. In a different study utilizing ribosomal intergenic spacer analysis, changes in soil microbial communities in response to Hg(II) exposure were attributed to the appearance of previously undetected β-proteobacteria and low-G+C gram-positives (60, 61). The observed differences between these studies suggest that the nature of both the contaminant(s) and the environments in which they are deposited play roles in the response of indigenous microbial communities to heavy-metal contamination. It is also worth noting that these other studies monitored the short-term response of communities to metal amendments, while our study system investigates the long-term effects of fluvially deposited heavy metals decades after their introduction to the environment.
A variety of studies have utilized DNA and phospholipid markers to demonstrate changes in microbial community composition due to heavy-metal contamination (2, 28, 29, 37, 54, 61, 62, 64, 66, 74). Wenderoth and Reber (74), using amplified rDNA restriction analysis (ARDRA), indicated that soils contaminated with Zn and other metals tend to support predominantly gram-negative organisms. Likewise, Sandaa et al. (62) found increased populations of gram-negative α-proteobacteria along with decreases in gram-positive and other gram-negative lineages in soils amended with metal-rich sewage sludge. Phylogenetic analysis of partial 16S rRNA gene sequences recovered from each of the streams sampled in the current study indicated that the hyporheic microbial communities were predominated by gram-negative bacteria (Table 3 and Fig. 4). Several of the sequences obtained were not highly related to known organisms and therefore gave little indication of species-level identifications (Table 3). This was an expected finding, since microbial communities inhabiting the hyporheic zone are relatively unexplored. However, these sequences consistently grouped with known gram-negative microbes (Fig. 4) previously shown to reside in aquatic systems (30).
DGGE and DNA sequence analysis has previously been used to demonstrate changes in soil archaeal communities exposed to increasing levels of heavy metal-contaminated sewage sludge (62). Similarly, the DGGE data in the current study indicated a direct linear relationship between the degree of metal contamination and the similarity of microbial communities between streams.
Prior studies have employed PLFA analysis to demonstrate that Cd, Cu, Ni, Pb, or Zn selects for prokaryotic phospholipid markers over those representative of fungi (29). However, other investigators have found opposite trends in soils contaminated with Cr, Pb, and hydrocarbons (66). The PLFA data presented here indicate an increased proportion of prokaryotic markers at sites with high CI values and increased abundance of fungal and other eukaryotic markers at sites with low CI values. Thus, the preponderance of evidence in this and prior studies indicates that microbial communities in a variety of environments are affected by the presence of heavy metals, but no general conclusions can be made regarding specifically how communities in different environments will respond to various metals.
Based on the data in the current study, it appears that these hyporheic communities do not respond to the presence of metals at the level of total community biomass but rather at the level at which specific bacterial populations or groups comprise the community, and the relative abundances of those populations or groups. It is acknowledged that predation (33, 38) and dissolved-organic-carbon quality and quantity (11, 25, 56, 79) can have effects on the size and structure of microbial communities. Determining the effects of the former was beyond the scope of this investigation, and dissolved-organic-carbon levels were generally below the limit of detection (<5 μg/ml) within the streams sampled (data not shown). However, potential effects of available nutrients on the microbial community structure within each stream were assessed by measuring a suite of dissolved anions in the pore water. The lack of correlation between measurable nutrients and community structure lends support to the conclusion that sediment metal loads play a substantial role in controlling bacterial community structure in the hyporheic zone of the streams studied.
Other studies have suggested that physiological stress caused by the toxic effects of metals leads to selection of less diverse communities comprised of metal-resistant populations (2, 18, 54, 64, 74) and a general suppression of metabolic activity (2, 10, 19, 41, 53). Close examination of the community-level measures described herein and the results of a separate activity-based study (27) do not support those hypotheses. There was no apparent correlation between sediment metal content (CI) and either diversity, as indicated by DGGE pattern complexity in the current study, or total productivity, as indicated by [14C]leucine incorporation (27). These data indicate that metal stress in fluvial environments doesn't reduce biomass, diversity, or productivity, as has previously been suggested for soils (2, 10, 19, 41, 53, 64, 74). Rather, the structure of microbial communities changes (i.e., population and group-level composition and relative abundance). Another prior study suggested that compensatory changes in response to metal toxicity may alter genotypic and/or phenotypic characteristics of a community at metal concentrations below thresholds that impact metabolic activity (2). The results presented here lend support to that hypothesis.
The sediments that underlie a river represent a microbial habitat important to the cycling of nutrients in streams (9, 50). To date, the communities residing in these habitats have generally been characterized at the process level by using activity-based measurements such as total microbial biomass, respiration rates, and bacterial production rates (3, 16, 23-26, 51, 56, 57). Fewer studies have attempted to define or describe the taxa that comprise these communities, especially by using molecular techniques (5, 67). Molecular characterization of these communities may have important implications for river restoration decisions, because without an accurate understanding of the ecology of natural hyporheic populations, it may not be possible to know whether normal community functions have been impacted or subsequently restored in a river ecosystem, since measures with coarse resolution show little or no response. Here, we have utilized moderate- to high-resolution molecular techniques to demonstrate a correlation between bacterial community structure within the hyporheic zone and the sedimentary metal loads therein. The information presented here may guide future experiments and improve our ability to predict long-term effects of metal contamination on river ecosystems and indicate how impacts on microbial communities may be translated to, and thus explain, observed perturbations at higher trophic levels in riverine ecosystems.
Acknowledgments
We thank Justin Harris for his assistance in placing sediment samplers and gathering the dissolved-anion data.
This research was funded by EPA grant no. R-82940001-0.
REFERENCES
- 1.Admiraal, W., H. Blanck, M. Buckert-De Jong, H. Guasch, N. Ivorra, V. Lehmann, B. A. H. Nystrom, M. Paulsson, and S. Sabater. 1999. Short-term toxicity of zinc to microbenthic algae and bacteria in a metal polluted stream. Water Res. 33:1989-1996. [Google Scholar]
- 2.Bååth, E., M. Diaz-Ravina, A. Frostegard, and C. D. Campbell. 1998. Effect of metal-rich sludge amendments on the soil microbial community. Appl. Environ. Microbiol. 64:238-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, M. A., C. N. Dahm, and H. M. Valett. 1999. Acetate retention and metabolism in the hyporheic zone of a mountain stream. Limnol. Oceanogr. 44:1530-1539. [Google Scholar]
- 4.Barlocher, F., and J. H. Murdoch. 1989. Hyporheic biofilms: a potential food source for interstitial animals. Hydrobiologia 184:61-67. [Google Scholar]
- 5.Battin, T. J., A. Wille, B. Sattler, and R. Psenner. 2001. Phylogenetic and functional heterogeneity of sediment biofilms along environmental gradients in a glacial stream. Appl. Environ. Microbiol. 67:799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böckelmann, D. B. U. 2001. Description and characterization of bacteria attached to lotic organic aggregates (river snow) in the Elbe River of Germany and the South Saskatchewan River of Canada. Dissertation. Technischen Universität Berlin, Berlin, Germany.
- 7.Bond, P. L., G. K. Druschel, and J. F. Banfield. 2000. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 66:4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bond, P. L., S. P. Smriga, and J. F. Banfield. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66:3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulton, A., S. Findlay, P. Marmonier, E. Stanley, and H. Valett. 1998. The functional significance of the hyporheic zone in streams and rivers. Annu. Rev. Ecol. Syst. 29:59-81. [Google Scholar]
- 10.Brookes, P. C., and S. P. McGrath. 1986. Effects of heavy metal accumulation in field soils treated with sewage-sludge on soil microbial processes and soil fertility, p. 327-343. In Conference proceedings, FEMS Symposium. Federation of European Microbiological Societies, Delft, The Netherlands.
- 11.Brugger, A., B. Wett, I. Kolar, B. Reitner, and G. Herndl. 2001. Immobilization and bacterial utilization of dissolved organic carbon entering the riparian zone of the alpine Enns River, Austria. Aquat. Microb. Ecol. 24:129-142. [Google Scholar]
- 12.Brunke, M., and T. Gonser. 1997. The ecological significance of exchange processes between rivers and ground water. Freshw. Biol. 37:1-33. [Google Scholar]
- 13.Burton, G. J., A. Drotar, J. Lazorchak, and L. Bahls. 1987. Relationship of microbial activity and Ceriodaphnia responses to mining impacts on the Clark Fork River, Montana. Arch. Environ. Contam. Toxicol. 16:523-530. [DOI] [PubMed] [Google Scholar]
- 14.Chang, Y.-J., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. Mckinley, S. J. Macnaughton, A. K. M. A. Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman, D. C. I. 1992. Assessment of injury to fish populations: Clark Fork River NPL sites. Montana Department of Natural Resources, Helena.
- 16.Craft, J., J. Stanford, and M. Pusch. 2002. Microbial respiration within a floodplain aquifer of a large gravel-bed river. Freshw. Biol. 47:251-261. [Google Scholar]
- 17.Dahlin, S., E. Witter, A. Mart, A. Turner, and E. Baath. 1997. Where's the limit? Changes in the microbiological properties of agricultural soils at low levels of metal contamination. Soil Biol. Biochem. 29:1405-1415. [Google Scholar]
- 18.Diaz-Ravina, M., and E. Bååth. 1996. Development of metal tolerance in soil bacterial communities exposed to experimentally increased metal levels. Appl. Environ. Microbiol. 62:2970-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domsch, K. 1984. Effects of pesticides and heavy metals on biological processes in soil. Conf. Biol. Proc. Soil Fert. 76:367-378.
- 20.Edwards, K. J., T. M. Gihring, and J. F. Banfield. 1999. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl. Environ. Microbiol. 65:3627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feris, K. P., P. W. Ramsey, C. F. Frazar, M. C. Rillig, J. E. Gannon, and W. E. Holben. Structure and seasonal dynamics of hyporheic zone microbial communities in free-stone rivers of the western United States. Microb. Ecol., in press. [Online.] http://link.springer-ny.com/link/service/journals/00248/contents/02/0100/s00248-002-0100-xch002.html. [DOI] [PubMed]
- 22.Findlay, S. 1995. Importance of surface-subsurface exchange in stream ecosystems: the hyporheic zone. Limnol. Oceanogr. 40:159-164. [Google Scholar]
- 23.Findlay, S., and W. Sobczak. 1996. Variability in removal of dissolved organic carbon in hyporheic sediments. J. N. Am. Benthol. Soc. 15:35-41. [Google Scholar]
- 24.Findlay, S., D. Strayer, C. Goumbala, and K. Gould. 1993. Metabolism of streamwater dissolved organic carbon in the shallow hyporheic zone. Limnol. Oceanogr. 38:1493-1499. [Google Scholar]
- 25.Fischer, H., M. Pusch, and J. Schwoerbel. 1996. Spatial distribution and respiration of bacteria in stream-bed sediments. Arch. Hydrobiol. 137:281-300. [Google Scholar]
- 26.Franken, R., R. Storey, and D. Williams. 2001. Biological, chemical and physical characteristics of downwelling and upwelling zones in the hyporheic zone of a north-temperate stream. Hydrobiologia. 444:183-195. [Google Scholar]
- 27.Frazar, C. F. 2002. Microbial community production and tolerance in heavy metals polluted sediment. Masters thesis. The University of Montana—Missoula, Missoula.
- 28.Frostegard, A., A. Tunlid, and E. Bååth. 1996. Changes in microbial community structure during long-term incubation in two soils experimentally contaminated with metals. Soil Biol. Biochem. 28:55-63. [Google Scholar]
- 29.Frostegard, A., A. Tunlid, and E. Bååth. 1993. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl. Environ. Microbiol. 59:3605-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glockner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimm, N. B., and S. G. Fisher. 1984. Exchange between interstitial and surface water: implications for stream metabolism and nutrient cycling. Hydrobiologia 111:219-228. [Google Scholar]
- 32.Guasch, H., M. Paulsson, and S. Sabater. 2002. Effect of copper on algal communities from oligotrophic calcareous streams. J. Phycol. 38:241-248. [Google Scholar]
- 33.Hahn, M. W., and M. G. Hofle. 1999. Flagellate predation on a bacterial model community: interplay of size-selective grazing, specific bacterial cell size, and bacterial community composition. Appl. Environ. Microbiol. 65:4863-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hare, L. 1992. Aquatic insects and trace metals: bioavailability, bioaccumulation, and toxicity. Crit. Rev. Toxicol. 22:327-369. [DOI] [PubMed] [Google Scholar]
- 35.Hughes, M. F. 2002. Arsenic toxicity and potential mechanisms of action. Toxicol. Lett. 133:1-16. [DOI] [PubMed] [Google Scholar]
- 36.Jones, J. B., S. G. Fisher, and N. B. Grimm. 1995. Nitrification in the hyporheic zone of a desert stream ecosystem. J. N. Am. Benthol. Soc. 14:249-258. [Google Scholar]
- 37.Kandeler, E., D. Tscherko, K. Bruce, M. Stemmer, P. Hobbs, R. Bardgett, and W. Amelung. 2000. Structure and function of the soil microbial community in microhabitats of a heavy metal polluted soil. Biol. Fert. Soils 32:390-400. [Google Scholar]
- 38.Kinner, N. E., R. W. Harvey, K. Blakeslee, G. Novarino, and L. D. Meeker. 1998. Size-selective predation on groundwater bacteria by nanoflagellates in an organic-contaminated aquifer. Appl. Environ. Microbiol. 64:618-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kisand, V., R. Cuadros, and J. Wikner. 2002. Phylogeny of culturable estuarine bacteria catabolizing riverine organic matter in the northern Baltic Sea. Appl. Environ. Microbiol. 68:379-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knight, B. P., S. P. Mcgrath, and A. M. Chaudri. 1997. Biomass carbon measurements and substrate utilization patterns of microbial populations from soils amended with cadmium, copper, or zinc. Appl. Environ. Microbiol. 63:39-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komulainen, M., and J. Mikola. 1995. Soil processes as influenced by heavy metals and the composition of soil fauna. J. Appl. Ecol. 32:234-241. [Google Scholar]
- 42.Konopka, A., T. Zakharova, M. Bischoff, L. Oliver, C. Nakatsu, and R. F. Turco. 1999. Microbial biomass and activity in lead-contaminated soil. Appl. Environ. Microbiol. 65:2256-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, W. J., and D. J. Patterson. 2002. Abundance and biomass of heterotrophic flagellates, and factors controlling their abundance and distribution in sediments of Botany Bay. Microb. Ecol. 43:467-481. [DOI] [PubMed] [Google Scholar]
- 44.Lepowski, W. 1998. Arsenic crisis in Bangladesh. Chem. Eng. News 76:27-29. [Google Scholar]
- 45.Lipton, J., H. Bergman, D. Chapman, T. Hillman, M. Kerr, J. N. Moore, and D. Woodward. 1993. Aquatic resource injury assessment report: Upper Clark Fork River Basin. State of Montana Natural Resource Damage Program. Montana Department of Natural Resources, Helena.
- 46.Malard, F., J. V. Ward, and C. T. Robinson. 1999. An expanded perspective of the hyporheic zone. Verh. Internat. Verein. Limnol. 27:1-7. [Google Scholar]
- 47.McCormick, F., B. Hill, L. Parrish, and W. Willingham. 1994. Mining impacts on fish assemblages in the Eagle and Arkansas Rivers, Colorado. J. Freshw. Ecol. 9:175-180. [Google Scholar]
- 48.Mills, A. L., and L. M. Mallory. 1987. The community structure of sessile heterotrophic bacteria stressed by acid mine drainage. Microb. Ecol. 14:219-232. [DOI] [PubMed] [Google Scholar]
- 49.Moore, J. N., and S. N. Luoma. 1990. Hazardous wastes from large-scale metal extraction: a case study. Environ. Sci. Technol. 24:1278-1285. [Google Scholar]
- 50.Mulholland, P. J., E. R. Marzolf, J. R. Webster, D. R. Hart, and S. P. Hendricks. 1997. Evidence that hyporheic zones increase heterotrophic metabolism and phosphorous uptake in forest streams. Limnol. Oceanogr. 42:443-451. [Google Scholar]
- 51.Naegeli, M. W., and U. Uehlinger. 1997. Contribution of the hyporheic zone to ecosystem metabolism in a prealpine gravel-bed river. J. N. Am. Benthol. Soc. 16:794-804. [Google Scholar]
- 52.Nagorski, S. A., and J. N. Moore. 1999. Arsenic mobilization in the hyporheic zone of a contaminated stream. Water Resour. Res. 35:3441-3450. [Google Scholar]
- 53.Obbard, J., D. Sauerbeck, and K. Jones. 1994. Dehydrogenase activity of the microbial biomass in soils from a field experiment amended with heavy metal contaminated sewage sludges. Sci. Total Environ. 142:157-162. [DOI] [PubMed] [Google Scholar]
- 54.Palmborg, C., A. Nordgren, and E. Baath. 1998. Multivariate modelling of soil microbial variables in forest soil contaminated by heavy metals using wet chemical analyses and pyrolysis GC/MS. Soil Biol. Biochem. 30:345-357. [Google Scholar]
- 55.Philippi, T. E. 1993. Multiple regression: herbivory, p. 183-210. In S. M. Scheiner and J. Gurevitch (ed.), Design and analysis of ecological experiments. Chapman and Hall, New York, N.Y.
- 56.Pusch, M. 1997. Community respiration in the hyporheic zone of a riffle-pool sequence, p. 51-56. In J. Gibert, J. Mathieu, and F. Fournier (ed.), Groundwater/surface water ecotones: biological and hydrological interactions and management options. Cambridge University Press, Cambridge, United Kingdom.
- 57.Pusch, M. 1996. The metabolism of organic matter in the hyporheic zone of a mountain stream, and its spatial distribution. Hydrobiologia 323:107-118. [Google Scholar]
- 58.Pusch, M., D. Fiebig, I. Brettar, H. Eisenmann, B. K. Ellis, L. A. Kaplan, M. A. Lock, M. W. Naegeli, and W. Traunspurger. 1998. The role of micro-organisms in the ecological connectivity of running waters. Freshw. Biol. 40:453-495. [Google Scholar]
- 59.Pusch, M., and J. Schwoerbel. 1994. Community respiration in hyporheic sediments of a mountain stream (Steina, Black Forest). Arch. Hydrobiol. 130:35-52. [Google Scholar]
- 60.Ranjard, L., E. Brothier, and S. Nazaret. 2000. Sequencing bands of ribosomal intergenic spacer analysis fingerprints for characterization and microscale distribution of soil bacterium populations responding to mercury spiking. Appl. Environ. Microbiol. 66:5334-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ranjard, L., S. Nazaret, F. Gourbiere, J. Thioulouse, P. Linet, and A. Richaume. 2000. A soil microscale study to reveal the heterogeneity of Hg(II) impact on indigenous bacteria by quantification of adapted phenotypes and analysis of community DNA fingerprints. Microb. Ecol. 31:107-115. [DOI] [PubMed] [Google Scholar]
- 62.Sandaa, R., V. Torsvik, O. Enger, F. Daae, T. Castberg, and D. Hahn. 1999. Analysis of bacterial communities in heavy metal contaminated soils at different levels of resolution. FEMS Microbiol. Ecol. 30:237-251. [DOI] [PubMed] [Google Scholar]
- 63.Sandaa, R.-A., V. Torsvik, and O. Enger. 2001. Influence of long-term heavy-metal contamination on microbial communities in soil. Soil Biol. Biochem. 33:287-295. [Google Scholar]
- 64.Sandaa, R. -A., Ø. Enger, and V. L. Torsvik. 1999. Abundance and diversity of Archaea in heavy-metal-contaminated soils. Appl. Environ. Microbiol. 65:3293-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanguinetti, C. J., E. Dias Neto, and A. J. G. Simpson. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques 17:914-921. [PubMed] [Google Scholar]
- 66.Shi, W., J. Becker, M. Bischoff, R. F. Turco, and A. E. Konopka. 2002. Association of microbial community composition and activity with lead, chromium, and hydrocarbon contamination. Appl. Environ. Microbiol. 68:3859-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smoot, J. C., and R. H. Findlay. 2001. Spatial and seasonal variation in a reservoir sedimentary microbial community as determined by phospholipid analysis. Microb. Ecol. 42:350-358. [DOI] [PubMed] [Google Scholar]
- 68.Southam, G., and T. J. Beveridge. 1992. Enumeration of thiobacilli within pH-neutral and acidic mine tailings and their role in the development of secondary mineral soil. Appl. Environ. Microbiol. 58:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stanford, J. A., and J. V. Ward. 1993. An ecosystem perspective of alluvial rivers: connectivity and the hyporheic corridor. J. N. Am. Benthol. Soc. 12:48-60. [Google Scholar]
- 70.Stephen, J. R., Y.-J. Chang, S. J. Macnaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Traina, S. J., and V. Laperche. 1999. Contaminant bioavailability in soils, sediments, and aquatic environments. Proc. Natl. Acad. Sci. USA 96:3365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valett, H. M., S. G. Fisher, and E. H. Stanley. 1990. Physical and chemical characteristics of the hyporheic zone of a Sonoran Desert stream. J. N. Am. Benthol. Soc. 16:239-247. [Google Scholar]
- 73.Vervier, P., and R. J. Naiman. 1992. Spatial and temporal fluctuations of dissolved organic carbon in subsurface flow of the Stillaguamish (Washington, USA). Arch. Hydrobiol. 123:401-412. [Google Scholar]
- 74.Wenderoth, D., and H. Reber. 1999. Correlation between structural diversity and catabolic versatility of metal affected prototrophic bacteria in soil. Soil Biol. Biochem. 31:345-352. [Google Scholar]
- 75.White, D. C., and D. B. Ringelberg. 1998. Signature lipid biomarker analysis, p. 255-272. In R. S. Burlage (ed.), Techniques in microbial ecology. Oxford University Press, New York, N.Y.
- 76.Wielinga, B., J. K. Lucy, J. N. Moore, O. F. Seastone, and J. E. Gannon. 1999. Microbiological and geochemical characterization of fluvially deposited sulfidic mine tailings. Appl. Environ. Microbiol. 65:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wielinga, B. W., S. G. Benner, C. M. Brick, J. M. Moore, and J. E. Gannon. 1994. Geomicrobiological profile through the hyporheic zone in a historic mining flood plain, p. 267-276. In Proceedings of the Second International Conference on Groundwater Ecology. American Water Resources Association, Herndon, Va.
- 78.Yu, Z., and W. W. Mohn. 1999. Killing two birds with one stone: simultaneous extraction of DNA and RNA from activated sludge biomass. Can. J. Microbiol. 45:269-272. [Google Scholar]
- 79.Zhou, J., B. Xia, D. S. Treves, L.-Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]