Abstract
Detection of Staphylococcus enterotoxin B (SEB) by biomolecular interaction analysis mass spectrometry (BIA/MS) is presented in this work. The BIA/MS experiments were based on a surface plasmon resonance (SPR) MS immunoassay that detects affinity-captured SEB both via SPR and by means of exact and direct mass measurement by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Experiments were performed with standard samples and food samples to assess the BIA/MS limit of detection for SEB and to set the experimental parameters for proper quantitation. Single and double SPR referencing was performed to accurately estimate the amount of the bound toxin. Reproducible detection of 1 ng of SEB per ml, corresponding to affinity capture and MS analysis of ∼500 amol of SEB, was readily achieved from both the standard and mushroom samples. A certain amount of SEB degradation was indicated by the signals in the mass spectra. The combination of MS with SPR-based methods of detection creates a unique approach capable of quantifying and qualitatively analyzing protein toxins from pathogenic organisms.
Detection of biological warfare agents has become a matter of great concern in the last several years. An environmental exposure to even a subtoxic dose of certain biological agents can lead to serious outcomes, due to the amplification of toxicity via in vivo replication in the human host. The classical approach to pathogen detection involves sampling and growth in suitable media so that higher concentrations of the microorganisms are obtained for subsequent biochemical evaluation. However, in a post-biological incident environment, direct-reading methods and instruments are needed for rapid monitoring and detection of microbial pathogens and their toxins at very low concentration. Furthermore, the detection should be unambiguous and be able to distinguish between pathogenic and similar nonpathogenic microorganisms.
One approach to detection involves recognition of protein phenotypes characteristic of the specific pathogenic microorganisms. This is commonly achieved by immunoassays that utilize antibodies to specific protein antigens or toxins. The immunoassays are most frequently performed in an enzyme-linked immunosorbent assay format, although others have also been developed, including assays based on strip tests and surface plasmon resonance (SPR), piezoelectric, fluorescence, chemiluminometric, and electrochemical detection. The immunoassays either involve indirect detection by optically active reporters that bind to the antibody-retrieved antigen (i.e., amplification detection approach) or use the electrochemical and optical attributes of the material to which the antibodies are immobilized to detect the bound antigen directly (as in surface plasmon resonance). In either case, quantitation of the targeted antigen is readily achieved. However, none of the above-mentioned immunoassay approaches is capable of delivering qualitative (structural) information on the targeted antigens, and they can still suffer from issues such as nonspecific binding, which can lead to false-positive results. Since each protein pathogen or antigen has a distinct molecular mass and structure, there is a clear advantage in designing and using immunoassays that have the ability to delineate the molecular mass of the immunoassayed antigens.
Over the past several years, we have developed several technologies that combine immunoassays (e.g., affinity capture) with matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) (2, 4, 8, 26) for determination of the molecular mass of biomolecules. In one such approach, termed biomolecular interaction analysis MS (BIA/MS) (9, 14, 15, 18, 19), the affinity-interaction analysis between the surface immobilized ligands (e.g., antibodies) and analytes (e.g., proteins) in solutions is monitored and quantified by SPR. SPR is a label-free quantification method that utilizes an interaction of light photons with free electrons (surface plasmons) on a gold surface to quantify the changes in concentration or amount of biomaterial on the surface (1, 6, 10, 12, 20). The SPR detection itself is nondestructive; therefore, the affinity capture-retrieved analyte(s) can be further analyzed (from the same surface where they were captured) via MALDI-TOF MS, yielding the molecular mass of the analyte(s). Signals from other proteins, specifically or nonspecifically retained on the ligand surface, can be also be detected, indicating possible binding of analyte variants, protein complexes, or nonspecific binding.
In this work, we investigated the detection of Staphylococcus enterotoxin B (SEB) via BIA/MS. SEB is one of the several toxins on the National Institute for Allergy and Infectious Diseases Biodefense Priority Pathogens List for which rapid and sensitive methods of detection are needed. SEB is produced by Staphylococcus aureus, and belongs to a family of heat-stable enterotoxins that includes eight other enterotoxins with 50 to 85% sequence homology (27). The amount of enterotoxin needed to cause intoxication is very small (∼1 ng/ml), and hence sensitive and specific detection is essential. In a previous study we showed the general applicability of the BIA/MS approach in detection of staphylococcal toxins in food (17). Presented here are experiments performed with standard samples and food samples to assess the limit of detection of BIA/MS for SEB and to set the experimental parameters for proper quantitation.
MATERIALS AND METHODS
Proteins and antibodies.
Affinity-purified rabbit anti-SEB antibody (1 mg/ml, in 10 mM phosphate buffer [pH 7.5] [Toxin Technology Inc., Sarasota, Fla.]) and SEB (1 mg/ml in water [Sigma, St. Louis, Mo]) were obtained from Avraham Rasooly (Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, College Park, Md.).
Samples.
Solutions with decreasing SEB concentrations were prepared by serial dilution of the SEB stock with HBS-EP buffer (0.01 M HEPES [pH 7.4], 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20). The mushroom sample was prepared by homogenizing the whole content (solid and brine) of a 7-oz can of commercial mushrooms. The homogenate was centrifuged for 5 min at 13,000 rpm (10,000 × g), resulting in ∼65% (vol/vol) clarified supernatant (mushroom stock), which was mixed in a 1:1 ratio with an appropriate SEB stock solution (at 20 and 2 ng/ml) to yield SEB-mushroom samples with 10 and 1 ng of SEB per ml, respectively.
SPR analysis.
A Biacore X instrument (Biacore AB, Uppsala, Sweden) was used for the first dimension of BIA/MS (affinity retrieval and SPR quantification). CM5 research grade sensor chips (carboxymethyldextran-derivatized surface [Biacore AB]) were used in the experiments, with HBS-EP running buffer at a flow rate of 5μl/min. The antibody was immobilized on sections (flow cells, FC) on the chip surface by a standard EDC-NHS [N-ethyl-N′-(dimethylaminopropyl)carbodiimide-N-hydroxysuccinimide] coupling method (7). Regeneration of the antibody surfaces was achieved by short injections of 0.06 N HCl.
MALDI-TOF MS analysts.
Following removal from the biosensor, chips were washed with three 200-μl aliquots of distilled water and prepared for MS by application of a MALDI matrix (aqueous solution of α-cyano-4-hydroxycinnamic acid [ACCA] in 33% [vol/vol] acetonitrile-0.4% [vol/vol] trifluoroacetic acid) with a matrix aerosol application device (16). The MS analysis was performed on a homemade MALDI-TOF mass spectrometer (18).
RESULTS AND DISCUSSION
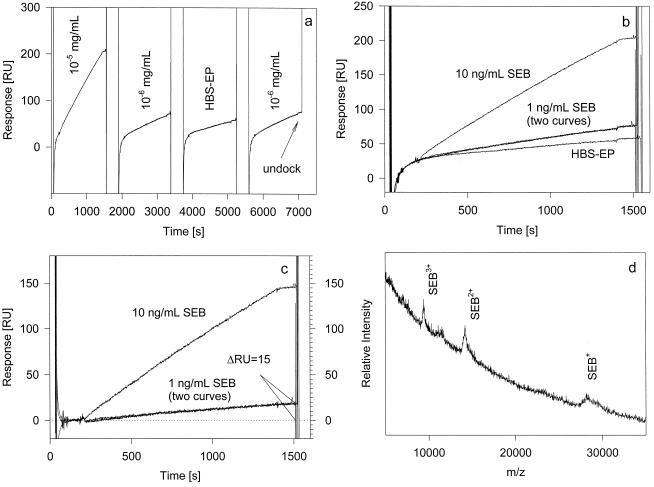
Following the immobilization of ∼90 fmol of anti-SEB in FC1 and ∼180 fmol of anti-TSST-1 (toxic shock syndrome toxin 1) in FC2 (which served as a reference FC), several aliquots of SEB samples were injected over both FCs. Shown in Fig. 1a is a sensorgram with four consecutive, 100-μl injections of 10 ng of SEB per ml in HBS-EP buffer, 1 ng of SEB per ml in HBS-EP buffer, HBS-EP buffer only, and 1 ng of SEB per ml in HBS-EP buffer again. The sensorgrams shown were baseline corrected in real time by subtraction of the signal from the reference (anti-TSST-1) FC2 (i.e., the FC1 − FC2 signal is shown in Fig. 1). Even with this correction, a constant positive bulk refractive index change (positive baseline drift) is noticeable in the sensorgram. Figure 1b shows the overlay of the four response curves, clearly indicating the positive SPR response resulting from the buffer injection. To correctly assess the responses from the SEB injections, the buffer injection curve was subtracted from the SEB curves. From these doubly corrected binding curves (which are shown in Fig. 1c), the true SPR responses resulting from the injections of the 10- and 1-ng/ml SEB aliquots were determined to be 145 and 15 response units (RU), respectively. The two 1-ng/ml SEB binding curves superimpose well, indicating the reproducibility of the experiments. The 15-RU response is indicative of binding of ∼500 amol of SEB (1 RU = 1 pg of protein; MWSEB = 28,366). The mass spectrum taken from the surface of FC1 following the last injection of the 1-ng/ml SEB aliquot is shown in Fig. 1d. Multiply (singly, doubly, and triply) charged SEB signals are observed in the spectrum, confirming the binding of SEB in the flow cell. The signals are somewhat broader, suggesting the existence of multiple SEB forms.
FIG. 1.
(a) SPR sensorgrams (baseline corrected from an anti-TTST-1-derivatized reference FC) resulting from four consecutive 100-μl injections of 10 ng of SEB per ml in HBS-EP buffer, 1 ng of SEB per ml in HBS-EP, HBS-EP buffer, and 1 ng of SEB per ml in HBS-EP. (b) Overlay view of the four binding curves. (c) SEB binding curves corrected by subtraction of the buffer injection sensorgram. (d) MALDI-TOF mass spectrum taken from the surface of FC1 following the last injection of the 1-ng/ml SEB aliquot.
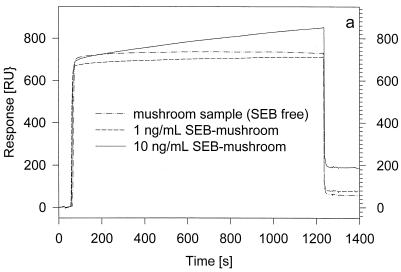
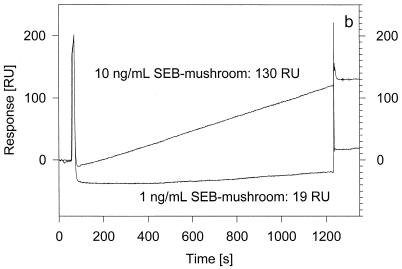
To demonstrate the same level of SEB detection from a food sample, 100-μl aliquots of mushroom extract samples containing 10 or 1 ng of SEB per ml were injected over the surface of an anti-SEB-derivatized FC on a new CM5 sensor chip (Fig. 2a). Also shown and overlaid in the same figure is an injection of a mushroom sample (SEB-free) to determine the amount of non-specific binding to the active/antibody surface. As done in the preceding experiment, the mushroom binding curve was subtracted from the two SEB-mushroom sample curves, yielding the sensorgrams shown in Fig. 2b. The readings taken from these singly corrected binding curves (130 and 19 RU) indicate binding of 130 and 19 pg of material from the injections of the 10- and 1-ng/ml SEB-mushroom sample aliquots, respectively. These responses are similar to those observed in Fig. 1c, suggesting the retrieval of subfemtomole amounts of SEB from the 1-ng/ml sample injection. After regeneration of the antibody surface, another aliquot of the 1-ng/ml SEB-mushroom sample was injected over the anti-SEB-derivatized flow cell and the chip was undocked from the biosensor and prepared for MS. The resulting mass spectrum is shown in Fig. 2c. Signals from the singly, doubly, and triply charged ions of SEB dominate the spectrum, with very few other background signals. The SEB signals are noticeably sharper and stronger than those observed in Fig. 1d. The peak broadening in Fig. 1d can be attributed to SEB degradation during storage; the experiments shown in Fig. 1 were performed after the SEB sample had been stored at 4°C for over 2 years. The existence of degradation products was verified by direct MALDI-TOF MS analysis of the SEB stock sample, which revealed the presence of several truncated SEB forms (containing residues 5 to 239, 5 to 238, 6 to 238, 7 to 238, 8 to 238 [or 7 to 237], and 8 to 237 [or 7 to 236]; native SEB contains 239 residues). This SEB degradation most probably caused the diminished signal intensity in the mass spectrum shown in Fig. 1d (i.e., instead of one strong signal, multiple weaker signals are observed). The detection of these SEB isoforms demonstrates the ability of BIA/MS to delineate the existence of potentially significant posttranslational modification that can go undetected if SPR-only analysis is performed.
FIG. 2.
(a) Overlaid SPR sensorgrams resulting from injections of 10 and 1 ng of SEB-mushroom samples per ml and a mushroom sample (SEB free) over the surface of an anti-SEB-derivatized flow cell. (b) SEB-mushroom binding curves corrected via subtraction of the mushroom sample injection sensorgram. (c) MALDI-TOF mass spectrum taken from the surface of FC1 following an injection of 1 ng of SEB-mushroom sample per ml.
Overall, detection of 1 ng of SEB per ml in buffer and in the mushroom sample was readily achieved in the BIA/MS experiments. SEB signals were observed in the MALDI-TOF mass spectra from all three experiments in which SEB at 1 ng/ml was analyzed, either in buffer or in a food matrix. We have demonstrated the same level of detection for another protein, β2-microglobulin (15). Further optimization (i.e., increased sample volume or decreased flow rates) may lower the SEB limit of detection (LOD) below the 1-ng/ml level. Nevertheless, this limit of detection is on par with (or slightly better than) those reported by authors using other SPR-based methods. Detection of ∼5 ng of SEB per ml was achieved by newly developed miniature fiberoptic (25) and wavelength modulation-based (5) SPR sensors. Another prototype miniature SPR sensor was capable of detecting ∼15 ng of SEB per ml in buffer and urine (13). When SPR was used in the indirect mode of detection (i.e., with signal amplification via binding of secondary antibodies to the affinity capture-retrieved antigens), the LOD was lowered to 0.5 ng/ml with one amplification step (5, 13) and 3 pg/ml (SEB in buffer) with two amplification steps (13). Other (non-SPR) approaches to SEB detection include traditional ELISAs (11, 24) (LOD = 0.05 to 1.0 ng/ml), time-resolved fluorometry (22) (LOD = 4 to 20 pg/ml [SEB in buffer]), and fluorescence-based array sensors (3, 23) (LOD = 4 ng/ml), and all of them use some kind of signal amplification. However, what separates the MS-based immunoassays and BIA/MS from the rest of the immunoassays is the ability to detect proteins exactly by means of direct mass measurement. With the use of polyclonal antibodies, multiple forms of the toxins can be captured and analyzed. In such experiments, cross-reactivities with proteins from similar nonpathogenic microorganisms (21) can readily be delineated form the signals in the mass spectrum (but not from the SPR signal itself) and eliminated from the data analysis. Also implied in the analysis is the detection of mutant forms of the protein that might be (potentially) genetically engineered to escape detection by monoclonal antibody-based (i.e., monospecific) assays. BIA/MS offers a unique capability to phenotypically trace toxins from pathogenic organisms and can serve as a high-performance technology for detecting and quantifying biodefense agents present in potential target media of terrorist attacks.
Acknowledgments
We thank Avraham Rasooly from FDA/CFSAN for providing the proteins and antibodies.
This publication was supported in part by grant 2 R44 CA82079-03A1 from the National Institutes of Health.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Baird, C. L., and D. G. Myszka. 2001. Current and emerging commercial optical biosensors. J. Mol. Recogn. 14:261-268. [DOI] [PubMed] [Google Scholar]
- 2.Cotter, R. J. 1997. Time-of-flight mass spectrometry. Instrumentation and applications in biological research. American Chemical Society, Washington, D.C.
- 3.Delehanty, J. B., and F. S. Ligler. 2002. A microarray immunoassay for simultaneous detection of proteins and bacteria. Anal. Chem. 74:5681-5687. [DOI] [PubMed] [Google Scholar]
- 4.Fenselau, C. 1997. MALDI MS and strategies for protein analysis. Anal. Chem. 69:661A-665A. [DOI] [PubMed]
- 5.Homola, J., J. Dostalck, S. Chen, A. Rasooly, S. Jiang, and S. S. Yee. 2002. Spectral surface plasmon resonance biosensor for detection of staphylococcal enterotoxin B in milk. Int. J. Food Microbiol. 75:61-69. [DOI] [PubMed] [Google Scholar]
- 6.Homola, J., S. S. Yee, and G. Gauglitz. 1999. Surface plasmon resonance sensors: review. Sensors Actuators Ser. B 54:3-15. [Google Scholar]
- 7.Johnsson, B., S. Lofas, and G. Lindquist. 1991. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 198:268-277. [DOI] [PubMed] [Google Scholar]
- 8.Karas, M., and F. Hillenkamp. 1988. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60:2299-2301. [DOI] [PubMed] [Google Scholar]
- 9.Krone, J. R., R. W. Nelson, D. Dogruel, P. Williams, and R. Granzow. 1997. BIA/MS: interfacing biomolecular interaction analysis with mass spectrometry. Anal. Biochem. 244:124-132. [DOI] [PubMed] [Google Scholar]
- 10.McDonnell, J. M. 2001. Surface plasmon resonance: towards an understanding of the mechanisms of biological molecular recognition. Curr. Opin. Chem. Biol. 5:572-577. [DOI] [PubMed] [Google Scholar]
- 11.Morissette, C., J. Goulet, and G. Lamoureux. 1991. Rapid and sensitive sandwich enzyme-linked immunosorbent assay for detection of staphylococcal enterotoxin B in cheese. Appl. Environ. Microbiol. 57:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullett, W. M., E. P. Lai, and J. M. Yeung. 2000. Surface plasmon resonance-based immunoassays. Methods 22:77-91. [DOI] [PubMed] [Google Scholar]
- 13.Naimushin, A. N., S. D. Soelberg, D. K. Nguyen, L. Dunlap, D. Bartholomew, J. Elkind, J. Melendez, and C. E. Furlong. 2002. Detection of Staphylococcus aureus enterotoxin B at femtomolar levels with a miniature integrated two-channel surface plasmon resonance (SPR) sensor. Biosens. Bioelectronics 17:573-584. [DOI] [PubMed] [Google Scholar]
- 14.Nedelkov, D., and R. W. Nelson. 2001. Analysis of human urine protein biomarkers via biomolecular interaction analysis mass spectrometry. Am. J. Kidney Dis. 38:481-487. [DOI] [PubMed] [Google Scholar]
- 15.Nedelkov, D., and R. W. Nelson. 2000. Exploring the limit of detection in biomolecular interaction analysis mass spectrometry (BIA/MS): detection of attomole amounts of native proteins present in complex biological mixtures. Anal. Chim. Acta 423:1-7. [Google Scholar]
- 16.Nedelkov, D., and R. W. Nelson. 2000. Practical considerations in BIA/MS: optimizing the biosensor-mass spectrometry interface. J. Mol. Recogn. 13:140-145. [DOI] [PubMed] [Google Scholar]
- 17.Nedelkov, D., A. Rasooly, and R. W. Nelson. 2000. Multitoxin biosensor-mass spectrometry analysis: a new approach for rapid, real-time, sensitive analysis of staphylococcal toxins in food. Int. J. Food Microbiol. 60:1-13. [DOI] [PubMed] [Google Scholar]
- 18.Nelson, R. W., J. R. Krone, and O. Jansson. 1997. Surface plasmon resonance biomolecular interaction analysis mass spectrometry. 1. Chip-based analysis. Anal. Chem. 69:4363-4368. [DOI] [PubMed] [Google Scholar]
- 19.Nelson, R. W., D. Nedelkov, and K. A. Tubbs. 2000. Biomolecular interaction analysis mass spectrometry. BIA/MS can detect and characterize proteins in complex biological fluids at the low- to subfemtomole level. Anal. Chem. 72:404A-411A. [PubMed]
- 20.Nice, E. C., and B. Catimel. 1999. Instrumental biosensors: new perspectives for the analysis of biomolecular interactions. Bioessays 21:339-352. [DOI] [PubMed] [Google Scholar]
- 21.Park, C. E., M. Akhtar, and M. K. Rayman. 1992. Nonspecific reactions of a commercial enzyme-linked immunosorbent assay kit (TECRA) for detection of staphylococcal enterotoxins in foods. Appl. Environ. Microbiol. 58:2509-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peruski, A. H., L. H. Johnson III, and L. F. Peruski, Jr. 2002. Rapid and sensitive detection of biological warfare agents using time- resolved fluorescence assays. J. Immunol. Methods 263:35-41. [DOI] [PubMed] [Google Scholar]
- 23.Rowe-Taitt, C. A., J. P. Golden, M. J. Feldstein, J. J. Cras, K. E. Hoffman, and F. S. Ligler. 2000. Array biosensor for detection of biohazards. Biosens. Bioelectronics 14:785-794. [DOI] [PubMed] [Google Scholar]
- 24.Schotte, U., N. Langfeldt, A. H. Peruski, and H. Meyer. 2002. Detection of staphylococcal enterotoxin B (SEB) by enzyme-linked immunosorbent assay and by a rapid hand-held assay. Clin. Lab. 48:395-400. [PubMed] [Google Scholar]
- 25.Slavik, R., J. Homola, and E. Brynda. 2002. A miniature fiber optic surface plasmon resonance sensor for fast detection of Staphylococcal enterotoxin B. Biosens. Bioelectronics 17:591-595. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka, K., H. Waki, Y. Ido, S. Akita, Y. Yoshida, and T. Yoshida. 1988. Protein and polymer analyses of up to m/z 100,000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2:151-153. [Google Scholar]
- 27.Van den Bussche, R. A., J. D. Lyon, and G. A. Bohach. 1993. Molecular evolution of the staphylococcal and streptococcal pyrogenic toxin gene family. Mol. Phylogenet. Evol. 2:281-292. [DOI] [PubMed] [Google Scholar]