Abstract
The deep-sea polychaete Alvinella pompejana colonizes tubes on the sides of black smoker chimneys along the East Pacific Rise. A diverse, yet phylogenetically constrained episymbiotic community is obligately associated with its dorsal surface. The morphologically and phylogenetically distinct dominant episymbionts have not yet been cultured, and there are no clearly defined roles for these bacteria in this symbiosis. A large insert fosmid library was screened for the presence of the two dominant phylotypes. Two fosmids, 35.2 and 38 kb, containing phylotype-specific 16S ribosmal DNA sequences were fully sequenced. Each fosmid had a gene encoding ATP citrate lyase, a key enzyme in the reverse tricarboxylic acid (rTCA) cycle, a CO2 fixation pathway. A selection of episymbiont communities from various geographic locations and vent sites were screened for the presence, diversity, and expression (via reverse transcription-PCR) of the ATP citrate lyase gene. Our results indicate that the ATP citrate lyase gene is not only a consistent presence in these episymbiont communities but is also expressed. Phylogenetically distinct forms of ATP citrate lyase were also found associated with and expressed by bacteria extracted from the tubes of A. pompejana. Utilizing PCR with degenerate primers based on a second key enzyme in the rTCA cycle, 2-oxoglutarate:acceptor oxidoreductase, we also demonstrated the persistent presence and expression of this gene in the episymbiont community. Our results suggest that members of both the episymbiont and the surrounding free-living communities display a chemolithoautotrophic form of growth and therefore contribute fixed carbon to other organisms in the vent community.
The tube-dwelling polychete Alvinella pompejana inhabits high-temperature hydrothermal vent chimneys from 21°N to 32°S along the East Pacific Rise (EPR) (17, 18, 21). A morphologically and phylogenetically diverse episymbiotic bacterial community forms a dense layer on the dorsal surface of A. pompejana (12, 18, 28). This association is specific and obligate, and the dominant epibionts are filamentous and associated with the dorsal epidermal expansions (hair-like projections) of A. pompejana (12, 18). The role of these episymbiotic bacteria in the association has been difficult to decipher. The symbionts have previously been hypothesized to be involved in both the nutrition of the host and in the detoxification of sulfide and heavy metals (1, 19, 43). The δ13C ratios of the A. pompejana worms and fatty acid profiles of the symbiont community indicate potential chemolithoautotrophy within the community (17, 19, 53). However, the dominant bacterial filaments associated with the polysaccharide structures on A. pompejana appear not to assimilate CO2 in any appreciable amount via the Calvin-Benson cycle according to ribulose 1,5-bisphosphate carboxylase-oxygenase, 14C labeling, and bicarbonate uptake assays (17, 19).
Molecular characterizations of the A. pompejana symbiont population indicate that the majority of the symbionts group within a single monophyletic clade within the epsilon subdivision of the Proteobacteria (10, 12, 28). In fact, it is now recognized that the majority of bacteria associated with deep-sea hydrothermal chimneys are members of the epsilon Proteobacteria (15, 28, 35, 39, 45). Little is known about the potential phenotypes within the A. pompejana symbiont community. Although a diverse population of bisulfite reductase genes involved in sulfur metabolism has been demonstrated in the A. pompejana episymbiotic community (16), these have not been linked to any specific phylotype.
Several attempts have been made to isolate the dominant members of the A. pompejana episymbiont community. Various types of bacteria have been cultured, including heavy metal-resistant strains and both heterotrophic and chemolithautotrophic members of the epsilon Proteobacteria (11, 31, 37, 42, 43). None of these isolates are the dominant episymbionts, and they represent <1% of the community (11; B. Campbell and S. Cary, unpublished data).
Current advances in genomic approaches applied to ecological questions have now provided the means to link prokaryotic phylotypes to specific metabolic potentials (8, 9, 51). The expression of metabolic genes has also been tested by using extracted RNA from microbiological communities (41, 55, 58, 59). To utilize a combined approach (16S ribosomal DNA [rDNA] phylogenetic analysis and metabolic gene identification and/or expression) with the episymbiont community, a large-insert fosmid library was constructed from the bacterial biomass found on the dorsal surface of an A. pompejana worm collected from 9°N EPR. Two fosmids were chosen for further analysis based on their DNA hybridization and PCR amplification with probes or primers specific for the two dominant phylotypes (11). One of the genes identified by this approach was ATP citrate lyase, a key enzyme in the reductive tricarboxylic acid (rTCA) pathway of carbon dioxide fixation during autotrophic growth of prokayotes (5, 23). This enzyme catalyzes the cleavage of citrate into acetyl coenzyme A and oxaloacetate, reversing one reaction of the TCA cycle. A reversal of the entire cycle generates one molecule of oxaloacetate from four molecules of CO2. It has been reported, however, that the sulfate reducers Desulfobacter postgatei and Desulfobacter autotrophicum growing on acetate utilize ATP citrate lyase in the TCA cycle (38; S. Sievert, C. Wirsen, and C. Taylor, unpublished data). Moller et al. (38) hypothesize that substrate-level phosphorylation via the TCA cycle with ATP citrate lyase is necessary for growth on acetate with sulfate as an electron acceptor.
Little is known about the utilization and importance of carbon fixation via the rTCA cycle in any microbial ecosystem. The majority of characterized chemlithoautotrophs associated with invertebrates from marine environments, including hydrothermal vents, use the Calvin-Benson pathway for carbon dioxide fixation (13). In deep-sea hydrothermal vent environments, symbiotic bacteria of the tubeworm Riftia pachyptila have been shown by both enzyme-based assays and molecular means to utilize the Calvin-Benson pathway to fix carbon dioxide coupled to the oxidation of sulfide (24, 46). The rTCA cycle is considered an alternate carbon fixation pathway and has been demonstrated in only a few prokaryotes, including thermophiles (i.e., Aquifex pyrophilus, Hydrogenobacter thermophilus, Chlorobium limicola, Desulfobacter hydrogenophilus, and Thermoproteus neutrophilus) (7, 23, 49, 50). Indeed, because of its presence in all three kingdoms, a version of the rTCA cycle has been hypothesized to be the oldest metabolic cycle (47, 54). The microorganisms that utilize the rTCA cycle are characterized by microaerophilic to anaerobic growth and many require various sulfur species for growth (7, 23, 49, 50). Candidatus Arcobacter sulfidicus, a sulfide-oxidizing epsilon Proteobacteria enriched from coastal seawater, forms filamentous sulfur in O2-H2S gradients. It does not use the Calvin cycle and is presumed to use an alternative pathway, most likely the rTCA cycle (57; Sievert et al., unpublished data).
The goals of the present study were to characterize the diversity and expression of two key genes in the rTCA cycle—the ATP citrate lyase and 2-oxoglutarate:acceptor oxidoreductase (OOR) genes—in the episymbiont community associated with A. pompejana. We designed degenerate primers for ATP citrate lyase based on the sequence information generated from the fosmid libraries and several closely related eukaroytic ATP citrate lyase genes. OOR primers were designed based on amino acid and nucleotide alignments of several bacterial OOR genes. We then tested for the presence and expression of these genes in the episymbiotic community associated with A. pompejana.
MATERIALS AND METHODS
Sample collection and nucleic acid extractions.
A. pompejana specimens in their associated tubes were collected during various cruises to the EPR and Guaymas basin sites by the DSV Alvin in 1994 (13°N), 1999 (9°N), 2000 (Guaymas Basin), and 2001 (9°N). These samples were collected by the manipulator arm of the Alvin and placed in an enclosed ice-chilled container that had been previously filled with 0.2-μm (pore-size)-filter-sterilized seawater for transport to the surface. The temperature of the water in the container at the surface was between 4 and 10°C. The worms were then rinsed three times with sterile seawater and frozen at −80°C until extractions could be performed.
For paired DNA-RNA extractions, the standard acid phenol RNA extraction protocol (14) was modified as follows: polyvinylpolypyrrolidone was added to solution D buffer (4 M guanidinium thiocyanate, 25 mM sodium citrate [pH 7], 0.1 M 2-mercaptoethanol, 0.5% Sarkosyl) at a final concentration of 1%). The samples (episymbiont community or tube scrapings) were then homogenized in extraction buffer by five passages through a syringe-20G needle combination. The samples were incubated at 65°C for 5 min and centrifuged at 12,500 rpm for 10 min. Excess insoluble carbohydrates were removed via a 30-min precipitation step with 0.14 M potassium acetate, followed by another centrifugation as described above. Acid phenol-chloroform extractions were performed until the interface was clear, followed by a straight chloroform extraction before isopropanol precipitation of the RNA. The organic layers were saved for DNA extraction by using a modified protocol from Ambion (Austin, Tex.). Briefly, DNA was extracted from the organic and interface layers by using a high-pH buffer (0.1 M NaCl, 10 mM Tris-HCl, 1 mM EDTA, 1% sodium dodecyl sulfate; pH 12), followed by a single extraction with chloroform. The DNA was precipitated with 0.5 M ammonium acetate and 100% ethanol. DNA was eliminated from the RNA samples by a standard DNase I (Invitrogen, Carlsbad, Calif.) treatment, extending the incubation time to 30 min at room temperature. Nucleic acids were quantified by spectrophotometry. For reverse transcription-PCR amplifications from extracted RNA samples, ca. 0.1 to 1 μg of RNA from each sample was used in a first-strand synthesis reaction with Superscript II (Invitrogen) and random hexamers according to the manufacturer's protocol. Negative controls included reactions without added reverse transcriptase. Portions (2 μl) of the first-strand cDNA synthesis reactions were used in the PCRs.
Fosmid library preparation and screening for dominant 16S rDNA phylotypes.
Fosmid libraries were prepared from high-molecular-weight DNA extracted from the episymbiont biomass of A. pompejana collected from 9°N in 1995 by methods similar to those described previously (51). Briefly, bacterial cells were removed from the dorsal surface of Alvinellid worms and washed twice in 2× STE (0.1 M NaCl, 10 mM Tris-Cl, 1 mM EDTA, pH 8.0). Agarose was added to 1%, and the mixture was solidified in 1-ml sterile syringes. Cells were lysed, and proteins were removed as described previously (51). Slices of the plugs were partially digested with HindIII as described previously (51). Vector arms were prepared from pFOS1 as described previously (33). Ligation, packaging, and transfections were also performed as described previously (51).
High-density filters were generated under conditions identical to those outlined by Stein et al. (51). A total of 20 pmol of two oligonucleotides specific to the A. pompejana symbionts (13B and 5A) (12) were end labeled with T4 polynucleotide kinase by using [γ-32P]ATP under standard labeling conditions. Hybridizations were performed at 45°C for 16 h, washed, and exposed to autoradiographic film as described previously (51). Two fosmids (6C6 and 7G3) were chosen for analysis based on their positive hybridizations with the 5A and 13B oligonucleotide probes (12).
Sequencing of fosmids.
6C6 and 7G3 fosmid DNAs were isolated by using a modified standard plasmid preparation (48). After the lysis, neutralization and initial ethanol precipitation, insoluble carbohydrates were removed by incubation with 3.75 M potassium acetate at −80°C for 30 min, followed by centrifugation and ethanol precipitation of the supernatant. RNA was removed by RNase A incubation, and the DNA was further purified by phenol-chloroform extraction, followed by isopropanol precipitation. The sizes of each fosmid were estimated by gel electrophoresis to be 35 kb (7G3) and 40 kb (6C6) after double restriction digestion with HindIII and BamHI. 6C6 and 7G3 fosmids were prepared for sequencing in their entirety by using the transposition reagents F-759 (Entranspsoson [kanamycin resistant]) and F-750 (MuA transposase) from MJ Research (Watertown, Mass.) contained in the MJ Research template generation system (http://www.mjresearch.com/html/enzymes/tgs/index.html). To correct the manufacturer's standard protocol for the larger inserts, ca. 2.1 and 2.4 μg of purified 6C6 and 7G3 fosmid DNAs, respectively, were used in each transposition reaction. Each transposition reaction was carried out under the following conditions: 1 μl of transposase, 1 μl of transposon, 4 μl of 5× buffer, 3 μl of DNA, and 11 μl of water were incubated at 30°C for 1 h, followed by a heat inactivation at 75°C for 10 min. Tranformants were obtained after electroporation of 1 μl of transposed DNA with DH10b electrocompetent cells (Invitrogen) by using the manufacturer's recommended settings. After 1 h of incubation at 37°C with SOC, diluted cells were plated on kanamycin (5 mg/ml)-chloramphenicol (7 mg/ml)-Luria-Bertani agar plates and grown at 37°C overnight. Transformants were picked into 1.5 ml of 2× yeast extract-tryptone-chloramphenicol (7 μg/ml) and then incubated for 18 h at 325 rpm and 37°C. Minipreps of each colony were prepared by using a ProPrep BAC 96 kit (LigoChem, Inc., Fairfield, N.J.). Two primers, SeqA (in the MJ Research Transposase kit) and SeqB (modified from the kit; 5′-GCATTACGCTGACTTGACG) were used for fluorescent dye sequencing (Applied Biosystems). Sequencing reactions contained 4 μl of BigDye, 4 μl of buffer, 3 μl of primer (at 6.4 pmol/μl), 4 μl of water, and 10 μl of DNA (concentration between 70 and 100 ng/μl). Thermocyler conditions were as follows: 95°C for 5 min, followed by 99 cycles of 95°C for 30 s, 50°C for 20 s, and 60°C for 4 min. Reactions were then ethanol precipitated, resuspended in 30 μl of water, and loaded on an ABI 3700 capillary sequencer (Applied Biosystems). Sequences were analyzed and aligned with phred/phrap and viewed in consed (University of Washington). Several gaps were observed in each fosmid sequence after three sets of transposition-transformation-sequencing cycles. Therefore, several sets of primers were generated at the 5′ and 3′ regions of the sequenced regions and were used to generate sequences covering the gaps. All remaining sequences were generated and checked by sequencing the complementary strand, and one contiguous sequence was obtained for each fosmid.
Fosmid and 16S rRNA/rDNA DGGE analysis.
A portion of the 16S rDNA gene was amplified with eubB and 5A1244R or 13B1244R (12) from the indicated fosmids under the following modified touchdown conditions: denaturation at 94°C for 1 min, annealing at 62°C for 1 min, and extension at 72°C for 2 min for 5 cycles, followed by an additional 5 cycles changing the annealing temperature to 60°C, with a final 20 cycles at a 58°C annealing temperature. Denaturing gradient gel electrophoresis (DGGE) analysis of the V3 region of the 16S rDNA (or cDNA) from the extracted episymbiont or A. pompejana tube communities was performed as previously described (10). Several bands were stabbed with sterile pipette tips, reamplified by using the same conditions, and checked for purity (the presence of a single band) on a separate DGGE gel.
Sequences of a portion of the fosmids and selected DGGE fragments were obtained after purification of the PCR by using a column separation protocol (GenElute; Sigma Corp.). Primers used for the fosmid sequencing were eubB, 519F, 519R, and 13B-1244R (4, 12, 28). DGGE fragments were sequenced by using 338F and 519R primers (40). Sequences were generated by using an ABI 310 genetic analyzer (Applied Biosystems, Foster City, Calif.) according to previously published methods (10).
ATP citrate lyase and OOR primer design, PCR amplification, cloning, and sequencing.
Primers for the β ATP citrate lyase gene were designed after CLUSTALW alignments of the 6C6 and 7G3 ATP citrate lyase (translated) genes with a selection of their eukaryotic counterparts (human, mouse, rat, and plant) by using the LaserGene program (DNASTAR, Madison, Wis.) and BLAST (3) (Fig. 2). The ATP citrate lyase gene of Chlorobium limicola was also included in the alignments but was not used for the final design, based on its distance from the rest of the sequences. Degenerate primers for the β gene product were as follows: 892F (5′-TGGACMATGGTDGCYGGKGGT) and 1204R (5′-ATAGTTKGGSCCACCTCTTC).
FIG. 2.
CLUSTALW alignment of a portion of the ATP citrate lyase beta gene from the indicated translated sequence. Sequences are the same as those used in Fig. 1. Numbered amino acids are from the 6C6 and 7G3 fosmid subunits. The regions used to design primers are indicated by boldface type.
Primers for the alpha OOR gene were designed based on alignments of eight phylogenetically distinct bacterial OORs. These included the following (with accession numbers in parentheses): Helicobacter pylori (AF021094), Hydrogenobacter thermophilus (AB046578), Thermotoga maritima (H72323), Thermoanaerobacter tengcongenis (AE013094), Geobacter metallireducens (NZ_AAAS01000001), Desulfovibrio desulfuricans G20 (NZ_AABI01000006), Campylobacter jejuni (NC_002163), and Chlorobium tepidum TLS (NC_002932). The two amino acid regions chosen were FFAGYPITP and QMEDEIA, corresponding to the nucleotide region between 67 and 174 bases of the Campylobacter jejuni alpha OOR gene (Cj0536). The forward primer was OOR67F (5′-TTCTTCGCTGGGTAYCCNATHACNCC), and the reverse primer was OOR177R (5′-CATACCAGCTATYTCRTCYTCCATYTG). The 5′ region of both primers contained invariant nucleotides to help stabilize primer annealing after the initial round(s) of amplification. The 3′ region of both primers contained degeneracies based on the nucleotide alignments of the region. Three random nucleotides were added to the beginning of the reverse primer for further stabilization and to generate a melting temperature closer to that of the forward primer.
Portions of the ATP citrate lyase beta gene and OOR alpha gene were amplified from ca. 5 to 10 ng of the indicated DNA or cDNA samples on a Minicycler (MJ Research) by using Jumpstart Taq polymerase (Sigma, St. Louis, Mo.). The cycling parameters for ATP citrate lyase amplification were as follows: 2 cycles of 94°C for 2 min, 37°C for 2 min, and 72°C for 3 min, followed by 35 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 1 min. The PCR conditions for OOR amplification were as follows: 1 cycle of 94°C for 2 min, followed by 35 cycles of 94°C for 1 min, 49°C for 1 min, and 72°C for 1 min. The final concentrations of the primers and MgCl2 were 0.5 μM and 3 mM, respectively.
PCR products were cloned into the TA TOPO 4.0 vector (Invitrogen) according to the manufacturer's recommendations. To obtain the 40 to 80 ng of amplification product required for sequencing, colonies were picked and PCR amplified with M13F and M13R primers. Amplicons of the correct size were purified with the GenElute kit (Sigma). DNA sequences were obtained by using the M13F and M13R primers and analyzed with a model 310 genetic analyzer (Applied Biosystems) according to the manufacturer's suggested protocol and as previously described (10).
Phylogenetic analysis.
Phylogenetic analysis was performed essentially as described previously (10, 11, 28). DNA distance matrix analysis was performed by using the method of Olsen as previously described (10). Sequences were first aligned with the LaserGene program (DNASTAR) by using CLUSTALW.
Stable isotope analysis and determination of eukaryotic contamination.
A. pompejana specimens were collected from 9°N EPR during the Extreme 2002 cruise (October and November 2002). Worms were frozen at −80°C until they were partially thawed in the laboratory to collect the hair-like projections. A portion of the bacterial symbiont community was placed into glass vials and freeze-dried, while the remainder were saved for DNA extraction. Three freeze-dried samples were placed into tin capsules (in duplicate), and their dry weight was determined. The capsules were then sealed and sent to the University of California at Davis stable isotope facility for stable carbon and nitrogen isotope analysis (http://stableisotopefacility.ucdavis.edu/). Stable isotope ratios of carbon and nitrogen were measured by continuous-flow isotope ratio mass spectrometry (20-20 Mass Spectrometer; PDZEuropa, Northwich, United Kingdom) after sample combustion to CO2 and N2 at 1,000°C in an on-line elemental analyzer (PDZEuropa ANCA-GSL). The gasses were separated on a Carbosieve G column (Supelco, Bellefonte, Pa.) before introduction to the isotope ratio mass spectrometer. Sample isotope ratios were compared to those of standard gases injected directly into the isotope ratio mass spectrometer before and after the sample peaks and δ15N (AIR) and δ13C (PDB) values calculated.
DNA from the three worm episymbiont samples was extracted by using a previously described bacterial genomic extraction protocol (6). The DNA was quantified by spectrophotometry, and a series of dilutions were made (5 to 0.005 ng). These were used in PCR amplifications of eukaryotic DNA with universal β-actin gene primers (Applied Biosystems TaqMan control primers). A parallel dilution series was done with human DNA (Applied Biosystems TaqMan control). The detection limit (i.e., the number of copies able to show a product) was first determined with the control DNA, and the numbers of copies per amount of DNA were then determined with the symbiont DNA. The percentage of eukaryotic DNA in the sample was estimated by calculating the approximate number of genome equilavents of A. pompejana DNA (haploid genome content is 0.8 pg) per total amount of DNA.
Nucleotide sequence accession numbers.
The nucleotide sequence accession numbers of the full-length fosmids (6C6 and 7G3) are AY312990 and AY312991, respectively. The accession numbers of the partial ATP citrate lyase beta gene sequences are AY308587 to AY308641. The accession numbers of the partial OOR alpha gene sequences are AY308642 to AY308651.
RESULTS AND DISCUSSION
Identification of ATP citrate lyase.
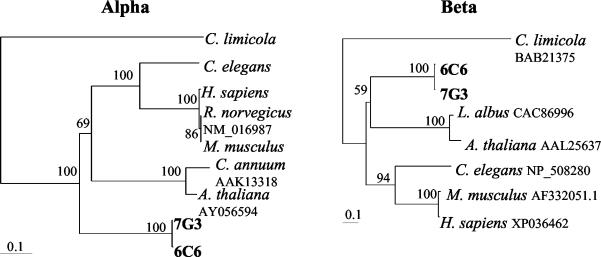
Previous studies in our laboratory demonstrated a unique, phylogenetically constrained bacterial community associated with A. pompejana. More than 80% of the community was dominated by bacteria belonging to a single monophyletic clade within the epsilon subdivision of the Proteobacteria (10, 28). To resolve the metabolic capabilities of the dominant epsilon proteobacterial phylotypes, a fosmid library was generated from an episymbiont community associated with A. pompejana specimens collected from 9°N EPR. The library was probed with primers specific for the two dominant phylotypes identified previously from 13°N EPR (12, 28). Two fosmids were chosen for further analysis based on their DNA hybridizations and subsequent positive PCRs (data not shown). The 6C6 and 7G3 fosmids showed homology to, and amplified with, the 13B and 5A probe-primer sets, respectively. Both fosmids contained two open reading frames with significant homologies to ATP citrate lyases from various species (Fig. 1). The ATP citrate lyase gene from 6C6 and 7G3 consisted of two open reading frames, α and β. The 6C6 fosmid nucleic acid and translated α ATP citrate lyase gene sequences were 98.3 and 99.3% similar to the 7G3 gene, while the β genes were 98.0 and 99.1% similar, respectively. These both were greater than the 16S rRNA similarity, which was 96.2%. Interestingly, they both shared less similarity to the single ATP citrate lyase gene sequenced from a prokaryotic organism (Chlorobium limicola) than to the eukaryotic sequences examined. The calculated distances of the Chlorobium limicola, Homo sapiens, and Caenorhabditis elegans ATP citrate lyase from the 7G3 α gene were 38.8, 55.7, and 52.7%, respectively. Similar results were observed with the β gene, where the distances from 7G3 were 31.6, 42.3, and 39.4%, respectively. Even though the ATP citrate lyase sequences were more similar to their eukaryotic counterparts, they were not from the worm itself, since the 16S rDNA and ATP citrate lyase sequences (as well as several additional bacterial genes [data not shown]) were physically linked on the same fosmid.
FIG. 1.
Phylogenetic amino acid distance trees showing the relationships between the ATP citrate lyase genes (alpha and beta) from 6C6 and 7G3 fosmids with other ATP citrate lyase genes (prokaryotic [Chlorobium limicola] and eukaryotic [Arabidopsis thaliana, Lupinus albus, Capsicum annuum, Caenorhabditis elegans, Mus musculus, Rattus norvegicus, and Homo sapiens]). The trees are based on alignment of ca. 591 and 445 translated amino acids (minus insertions and deletions) from the alpha and beta genes, respectively. Bootstrap values from 100 resamplings are indicated prior to the branch points of the tree. The scale bar represents the calculated number of changes per amino acid position.
The predicted sizes of the 6C6 and 7G3 ATP citrate lyase α and β gene products are 65.7- and 50.5-kDa, respectively. These values are similar to the experimentally determined sizes of the Chlorobium limicola subunits (32). Expression and purification of the recombinant Chlorobium limicola enzyme in Escherichia coli demonstrated that ATP citrate lyase is heteromeric, consisting of 65- and 42-kDa polypeptides, most likely in a α4β4 structure (32). Both fosmid ATP citrate lyases have a region homologous to the eukaryotic and prokaryotic catalytic domain (GHAGA), which also occurs in succinyl coenzyme A synthetases (32).
Diversity and expression of ATP citrate lyase in situ.
One of the major goals of our research is to determine the community structure and functional roles of the symbionts within the community. Clearly, with the discovery of ATP citrate lyase, there is a potential for chemoautotrophy within the community. However, nothing is known about the amount of sequence heterogeneity or expression of ATP citrate lyase in any community in situ. In order to determine ATP citrate lyase sequence heterogeneity and gene expression within the A. pompejana symbiont community, an alignment of various ATP citrate lyase genes was performed. A portion of this alignment is listed in Fig. 2. There were several regions of amino acid identity of the fosmid sequences to eukaryotic ATP citrate lyases. These included two stretches of amino acids, WTMVAGG and RGGPNY, which were used to design the degenerate primers (Fig. 2). The one other prokaryotic sequence (Chlorobium limicola) was not used because of its distance from the other sequences. There were a large number of possible degeneracies based on all of the possible codon combinations. Therefore, wherever possible, invariant nucleic acid sequences were used based on the DNA alignments of the same sequences.
Representative samples from A. pompejana episymbiont communities were chosen from a variety of sources (Table 1). Worms were collected from two geographic sites, 9°N and 13°N EPR. The three 9°N samples were taken from three different vent sites, Bio 9, M vent, and Q vent on separate Alvin dives. Additionally, two A. pompejana tubes were sampled. Scrapings from the interior of the Bio 9 worm (sample 3713) tube were extracted for nucleic acids, whereas the entire tube was extracted from a worm tube collected from M vent (sample 3722). To determine both geographic differences and variation between A. pompejana worms collected from the same site, two worms were sampled from 13°N EPR.
TABLE 1.
Levels of genomic and expressed 7G3-like ATP citrate lyase β gene clones
| Location and vent site | Samplea | No. of clones with 100% amino acid identity to 7G3/total no. of clones
|
|
|---|---|---|---|
| DNA | RNA | ||
| 13°N | 112B | 1/3 | 2/5 |
| 133B | 0/3 | 4/6 | |
| 9°N | |||
| Bio 9 | 3713 | 3/4 | |
| 3713T | 1/3 | 0/5 | |
| M vent | 42 | 0/4 | 3/4 |
| 3722T | 0/4 | 0/4 | |
| Q vent | 64 | 2/4 | 2/6 |
“T” indicates a tube sample.
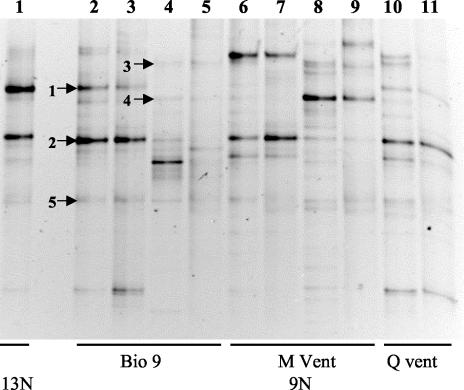
A limited phylogenetic diversity profile was performed on the A. pompejana episymbiont communities by DGGE analysis of the 16S rRNA gene and RNA (Fig. 3). This allowed a comparison of the symbionts to each other, to the active members of the community (via reverse transcription-PCR), and to their surrounding environment (tubes). As indicated by the identically migrating bands in lanes 2 and 3, the symbiont community from Bio 9 was identical to its expressed members. Similar results were obtained from the M vent and Q vent samples (lanes 6 and 7 and lanes 10 and 11, respectively), as well as from the two 13°N communities (data not shown). The sequences of the major 9°N community phylotypes identified by DGGE (as indicated by the arrows 1 and 2) were identical or nearly identical to the two fosmid sequences in the same 16S rDNA region. The 6C6 sequence was 100% identical to GenBank accession numbers AF180315, AF244532, and AF244526. The 7G3 sequence was 99.3% identical to GenBank accession numbers AF244536 and AF244535. These previously identified sequences were obtained from other DGGE analyses of the A. pompejana symbiont communities from 9°N and 13°N (10, 20).
FIG. 3.
DGGE of positive PCR products after amplification with universal primers for 16S rRNA or rDNA. Lanes 1 to 11, separated amplification products obtained from genomic DNA or RNA extracted from various A. pompejana episymbiont (AP) or A. pompejana tube (APT) communities. Lane 1, APG1B (AP, 13°N EPR in 1994); lanes 2 (DNA) and 3 (RNA) from sample 3713 (AP); lanes 4 (DNA) and 5 (RNA) from sample 3713 (APT); lanes 6 (DNA) and 7 (DNA) from sample 42 (AP); lanes 8 (DNA) and 9 (RNA) from sample 3722 (APT); and lanes 10 (DNA) and 11 (RNA) from sample 64 (AP). Samples were obtained from the indicated locations.
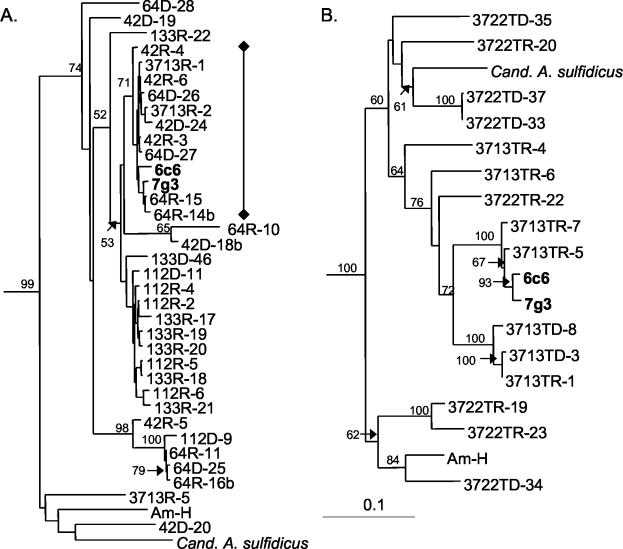
The ATP citrate lyase β gene was present and expressed in all episymbiont communities tested (Table 1, Fig. 4A). We also tested DNA from Persephonella marina (supplied by A. Reseynbach, Portland State University), a chemoautotrophic thermophilic Aquificales sp. isolated from 9°N, and two chemoautotrophic epsilon Proteobacteria—Candidatus Arcobacter sulfidicus (supplied by C. Taylor, Woods Hole Oceanographic Institution) and the deeply branching AM-H (11, 27, 44, 57)—for the presence of ATP citrate lyase with the same primers. The PCR products were sequenced and included in the phylogenetic analysis for reference. There were two major groups of ATP citrate lyase clones within the episymbiont community (as measured by the sequence distances of a portion of the β gene). The clones could by divided into clades according to their geographic differences (Table 1 and Fig. 4A, 42 and 64 versus 112 and 133, from 9°N and 13°N, respectively). Clones within these geographically distinct groups were very similar to each other according to their nucleic acid distances. The most distant clones within the clade containing the fosmid sequences were only ca. 3% apart by distance matrix analysis. In contrast, the DNA distance between the ATP citrate lyase gene sequences of the 7G3 fosmid and Persephonella marina was 34%. There were a few outliers from the episymbiont clones, but they also grouped into the known epsilon proteobacterial subdivision. These results suggest that the ATP citrate lyase from the worm itself or other eukaryotic species were never amplified or sequenced.
FIG. 4.
Phylogenetic DNA distance trees showing the relationships between a section of the ATP citrate lyase β gene from genomic versus expressed amplified products. (A) Phylogenetic DNA distance tree that includes clones amplified from the indicated A. pompejana episymbiont communities. (B) Phylogenetic DNA distance tree that includes clones amplified from samples taken from the inside (sample 3713T) or entire (sample 3722) A. pompejana tubes. The “♦—♦” bracket indicates 100% amino acid identity to the 7G3 ATP citrate lyase β gene within the sequenced region. Genomic samples are indicated by a “D,” expressed samples by are indicated by an “R,” and tube samples are indicated by a “T.” The trees are based on alignment of ca. 311 bases (minus insertions, deletions, and ambiguous bases) from the β ATP citrate lyase gene. Bootstrap values from 100 resamplings are indicated prior to the branch points of the tree. The scale bar represents the calculated number of changes per nucleotide position.
The ATP citrate lyase genes from the two tube communities were different from each other and from those obtained from the episymbiont community (Fig. 4B). Relatively few ATP citrate lyase clones from the 3713 tube sample were similar to either the 3713 sample episymbiont community clones or to either of the fosmids. Interestingly, the sequences of the clones from the 3713 tube sample (3713T) shared more similarities to ones from the episymbiont community; the 3722 tube sample (3722T) clones were much more distantly related to the episymbiont clones and clustered with the Arcobacter and AM-H bacterial strains. These differences reflect where the bacteria were sampled (from the interior of the tube with 3713T and from the entire tube with 3722T) and correspond to presumed physical and chemical differences in their environment (temperature, O2 concentrations, H2S concentrations, etc.). These results also correlate with the different phylotypes observed by 16S rDNA DGGE analysis and indicate that the chemolithoautotrophic members of each community (tube versus symbiont) are different.
As expected, the nucleic acid distances of the ATP citrate lyase β gene were much greater than the predicted amino acid distances in both the episymbiont and tube communities (data not shown). In fact, about half of the episymbiont clones shared 100% amino acid identity to the 7G3 fosmid ATP citrate lyase (Table 1 and Fig. 4A). Unfortunately, the amino acid sequences were too short (ca. 110 amino acids) and too similar to obtain a phylogenetic tree with reliable bootstrap values.
Diversity and expression of 2-oxoglutarate:acceptor oxidoreductase.
OOR (also referred to as α-ketoglutarate synthase) is also a necessary enzyme in the rTCA cycle (7, 23, 49, 50). It catalyzes (via ferredoxin) the reductive carboxylation of succinyl coenzyme A to 2-oxoglutarate in the rTCA cycle. However, it is not unique to the rTCA cycle, where it is found in a variety of anaerobic to microaerophilic chemoautotrophic and heterotrophic bacteria that display a microaerophilic-to-anaerobic mode of growth (the enzyme is highly oxygen sensitive) (26, 29). The same samples screened for the presence and expression of the ATP citrate lyase gene were also screened for the OOR gene. Amplification products of the expected size were obtained from both 9°N and 13°N symbiont populations, as well as cDNAs prepared from the same populations (data not shown). To estimate diversity in the expressed populations, products from 9°N (sample 64) and 13°N (sample 133B) cDNA amplifications were cloned, and five products from each reaction were sequenced. The translated amino acid distances of all of the clones were <7.5%. In fact, six of the clones shared 100% amino acid identity. These results parallel those observed with the more extensive ATP citrate lyase gene diversity analysis.
Isotope analysis of the A. pompejana symbiont community.
Isotopic C and N values of organisms can indicate their trophic level within a community (13). In addition, the stable carbon isotopic ratios of the three major autotrophic pathways are different, so that δ13C values of microorganisms can be a good indication of whether or not they use the rTCA cycle (25). Carbon isotopic analyses of the episymbiotic biomass from three different worms were performed to determine the δ13C values. δ13C ratios for the three samples ranged from −8.9 to −9.3‰. These values fall within the expected range of carbon isotopic ratios of microorganisms that utilize the rTCA cycle (−8 to −12‰) (25), thus supporting the hypothesis that carbon fixation by the episymbiont community is performed via the rTCA cycle. The δ15N values of the symbionts ranged from 2.1 to 2.4‰. These values fall within the expected range for primary producers at hydrothermal vents (0 to 4 ‰) (53). DNA was extracted in parallel from these same samples, and the percentage of eukaryotic DNA was determined to be <1% (data not shown).
Implications.
We have demonstrated that two key genes in the rTCA cycle of autotrophic growth, ATP citrate lyase and OOR, are a diverse and consistent presence within the A. pompejana episymbiont community. They are expressed both in the episymbionts themselves and in bacteria from associated tubes. In addition, we have evidence from a genomic library sequencing effort of the symbiont community that all of the other genes required for the rTCA cycle are present within the community (Campbell and Cary, unpublished). Furthermore, the carbon isotope patterns found within the episymbiont community are consistent with the use of the rTCA cycle. Therefore, these results suggest that at least a portion of the episymbiont community is autotrophic. These results are somewhat different than those found in previous studies that assayed for RuBisCO activity by using bicarbonate uptake and C14 assays, where clear evidence of autotrophy was observed in the tube bacteria but not in the dominant symbionts (1, 2, 52). These discrepancies may be due to a variety of reasons. Bicarbonate uptake assays were performed at a single temperature (40°C), which may not have been optimal for the symbionts (2). In addition, even though expression of the ATP citrate lyase gene was found in both the episymbionts and the tubes, the actual levels of expression were not determined. Studies are now under way to determine the levels of ATP citrate lyase gene expression and protein activity within the symbiont community. This would allow us to expand on the gene expression analyses described here as well as to determine temperature optima on enzymes physically tied to the symbionts.
The present study suggests that the rTCA cycle, as indicated by the presence of the key genes ATP citrate lyase and OOR, is used for chemolithoautotrophic growth by a variety of organisms in extreme environments, as well as in marine sediments. The pathway occurs in both thermophilic and mesophilic autotrophs from different phylogenetic groups (i.e., Persephonella marina and Candidatus Arcobacter sulfidicus). Clearly, it is present in a diverse population of epsilon proteobacteria from the A. pompejana environment. There are a small number of chemolithoautotrophic bacteria that have been cultured from hydrothermal vents (i.e., Persephonella marina, Am-H, Desulfobacterium thermolithoautotrophicum, Thiomicrospira crunogena, and Thiobacillus hydrothermalis) (11, 22, 30, 34, 44, 56), but undoubtedly there are many more chemolithoautotrophs present in these extreme environments that could potentially be discovered by the use of the degenerate primers designed here. Furthermore, the similarities in the ATP citrate lyase sequences between these bacteria and eukaryotes support the hypothesis of the early origin of the rTCA cycle (54).
Several studies indicate that epsilon Proteobacteria predominate at hydrothermal vents (15, 28, 35, 39, 45). The present study indicates that a portion of the epsilon Proteobacteria at hydrothermal vent sites express both the ATP citrate lyase and the OOR genes. Taken together, these results suggest that the rTCA cycle could play a more important role in chemoautotrophic production by free-living microorganisms at hydrothermal vents than the Calvin cycle. Future studies could help clarify the role of the rTCA pathway in CO2 fixation and its contribution to overall carbon cycling on earth.
Acknowledgments
This work was supported by an NSF LexEn grant (9907666) to S.C.C. and J.S. and an NSF Biocomplexity grant (OCE-0120648) to S.C.C. J.S. completed his work at Diversa Corp., San Diego, Calif.
We thank the pilots and crew of the DSV Alvin and R/V Atlantis for help in obtaining samples for these analyses. We also gratefully acknowledge the receipt of DNA samples from C. Taylor and S. Sievert at the Woods Hole Oceanographic Institution (Candidatus Arcobacter sulfidicus) and A. L. Reysenbach at Portland State University (Persephonella marina) for use in the present study. We thank K. Coyne and S. Sievert for critically reviewing the manuscript. We thank M. Dolan and C. Jones at the Dupont Biotechnology Sequencing facility for help in obtaining the fosmid sequences and D. Harris at the Stable Isotope Facility at the University of California at Davis for carbon isotope analysis.
REFERENCES
- 1.Alayse-Danet, A. M., D. Desbruyéres, and F. Gaill. 1987. The possible nutritional or detoxification role of the epibiotic bacteria of Alvinellid polychaetes: review of the current data. Symbiosis 4:51-62. [Google Scholar]
- 2.Alayse-Danet, A. M., F. Gaill, and D. Desbruyéres. 1986. In situ bicarbonate uptake by bacteria-Alvinella associations. Mar. Ecol. Pub. Della Stazione Zool. Napoli I 7:233-240. [Google Scholar]
- 3.Altschul, S., T. Madden, A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1998. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. FASEB J. 12:A1326-A1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antranikian, G., C. Herzberg, and G. Gottschalk. 1982. Characterization of ATP citrate lyase from Chlorobium limicola. J. Bacteriol. 152:1284-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl (ed.). 1988. Current protocols in molecular biology. John Wiley & Sons, Inc., Media, Pa.
- 7.Beh, M., G. Strauss, R. Huber, K. O. Stetter, and G. Fuchs. 1993. Enzymes of the reductive citric acid cycle in the autotrophic eubacterium Aquifex pyrophilus and in the archaebacterium Thermoproteus neutrophilus. Arch. Microbiol. 160:306-311. [Google Scholar]
- 8.Beja, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 9.Beja, O., M. T. Suzuki, J. F. Heidelberg, W. C. Nelson, C. M. Preston, T. Hamada, J. A. Eisen, C. M. Fraser, and E. F. DeLong. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630-633. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, B. J., and S. C. Cary. 2001. Characterization of a novel spirochete associated with the hydrothermal vent polychaete annelid, Alvinella pompejana. Appl. Environ. Microbiol. 67:110-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell, B. J., C. Jeanthon, J. E. Kostka, G. W. Luther, and S. C. Cary. 2001. Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 67:4566-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cary, S. C., M. T. Cottrell, J. L. Stein, F. Camacho, and D. Desbruyères. 1997. Molecular identification and localization of a filamentous symbiotic bacteria associated with the hydrothermal vent annelid, Alvinella pompejana. Appl. Environ. Microbiol. 63:1124-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Childress, J., and C. Fisher. 1992. Biology of hydrothermal vent animals, p. 337-441. In A. D. Ansell, R. N. Gibson, and M. Barnes (ed.), Oceanography and marine biology: an annual review, vol. 30. UCL Press, London, England.
- 14.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 15.Corre, E., A. L. Reysenbach, and D. Prieur. 2001. Epsilon proteobacterial diversity from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge. FEMS Microbiol. Lett. 205:329-335. [DOI] [PubMed] [Google Scholar]
- 16.Cottrell, M. T., and S. C. Cary. 1999. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl. Environ. Microbiol. 65:1127-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desbruyéres, D., P. Chevaldonné, A.-M. Alayse, D. Jollivet, F. H. Lallier, C. Jouin-Toulmond, F. Zal, P.-M. Sarradin, R. Cosson, J.-C. Caprais, C. Arndt, J. O'Brien, J. Guezennec, S. Hourdez, R. Riso, F. Gaill, L. Laubier, and A. Toulmond. 1998. Biology and ecology of the “Pompeii worm” (Alvinella pompejana Desbruyéres and Laubier), a normal dweller of an extreme deep-sea environment: a synthesis of current knowledge and recent developments. Deep-Sea Res. PII 45:383-422. [Google Scholar]
- 18.Desbruyères, D., F. Gaill, L. Laubier, and Y. Fouquet. 1985. Polychaetous annelids from hydrothermal vent ecosystems: an ecological overview. Bull. Biol. Soc. Wash. 6:103-116. [Google Scholar]
- 19.Desbruyères, D., F. Gaill, L. Laubier, D. Prieur, and G. Rau. 1983. Unusual nutrition of the “Pompeii worm” Alvinella pompejana (polychaetous annelid) from a hydrothermal vent environment: SEM, TEM, 13C, and 15N evidence. Mar. Biol. 75:201-205. [Google Scholar]
- 20.DiMeo, C. 2002. Characterization of the physicochemical microhabitat and epibiotic bacterial community associated with Alvinella pompejana, a hydrothermal vent annelid. Ph.D. thesis. University of Delaware, Lewes.
- 21.DiMeo, C., G. Luther, and S. Cary. Physicochemical characterization of the microhabitat of Alvinella pompejana, a hydrothermal vent annelid. Geochim. Cosmo. Acta, in press.
- 22.Durand, P., A. L. Reysenbach, D. Prieur, and N. Pace. 1993. Isolation and characterization of Thiobacillus hydrothermalis sp. nov., a mesophilic obligately chemolithotrophic bacterium isolated from a deep-sea hydrothermal vent in Fiji Basin. Arch. Microbiol. 159:39-44. [Google Scholar]
- 23.Evans, M. C. W., B. B. Buchanan, and D. I. Arnon. 1966. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc. Natl. Acad. Sci. USA 55:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felbeck, H., J. J. Childress, and G. N. Somero. 1981. Calvin-Benson cycle and sulfide oxidation enzymes in animals from sulfide-rich habitats. Nature 293:291-293. [Google Scholar]
- 25.Fuchs, G. 1989. Alternate pathways of autotrophic CO2 fixation, p. 365-382. In H. G. Schlegel and B. Bowien (ed.), Autotrophic bacteria. Springer-Verlag, New York, N.Y.
- 26.Gehring, U., and D. I. Arnon. 1972. Purification and properties of alpha-ketoglutarate synthase from a photosynthetic bacterium. J. Biol. Chem. 247:6963-6969. [PubMed] [Google Scholar]
- 27.Gotz, D., A. Banta, T. J. Beveridge, A. I. Rushdi, B. R. T. Simoneit, and A. Reysenbach. 2002. Persephonella marina gen. nov., sp. nov. and Persephonella guaymasensis sp. nov., two novel, thermophilic, hydrogen-oxidizing microaerophiles from deep-sea hydrothermal vents. Int. J. Syst. Evol. Microbiol. 52:1349-1359. [DOI] [PubMed] [Google Scholar]
- 28.Haddad, M. A., F. Camacho, P. Durand, and S. C. Cary. 1995. Phlogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete Alvinella pompejana. Appl. Environ. Microbiol. 61:1679-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes, N. J., C. L. Clayton, P. A. Chalk, and D. J. Kelly. 1998. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180:1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jannasch, H. W., C. O. Wirsen, D. C. Nelson, and L. A. Robertson. 1985. Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 35:422-424. [Google Scholar]
- 31.Jeanthon, C. 2000. Molecular ecology of hydrothermal vent microbial communities. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 77:117-133. [DOI] [PubMed] [Google Scholar]
- 32.Kanao, T., T. Fukui, H. Atomi, and T. Imanaka. 2001. ATP-citrate lyase from the green sulfur bacterium Chlorobium limicola is a heteromeric enzyme composed of two distinct gene products. Eur. J. Biochem. 268:1670-1678. [PubMed] [Google Scholar]
- 33.Kim, U. J., H. Shizuya, P. J. Dejong, B. Birren, and M. I. Simon. 1992. Stable propagation of cosmid sized human DNA inserts in an F-factor based vector. Nucleic Acids Res. 20:1083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.L'Haridon, S., V. Cilia, P. Messner, G. Raguenes, A. Gambacorta, U. B. Sleytr, D. Prieur, and C. Jeanthon. 1998. Desulfurobacterium thermolithotrophum gen. nov., sp. nov., a novel autotrophic, sulphur-reducing bacterium isolated from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 48(Pt. 3):701-711. [DOI] [PubMed] [Google Scholar]
- 35.Longnecker, K., and A. L. Reysenbach. 2001. Expansion of the geographic distribution of a novel lineage of epsilon Proteobacteria to a hydrothermal vent site on the Southern East Pacific Rise. FEMS Microbiol. Ecol. 35:287-293. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Garcia, P., F. Gaill, and D. Moreira. 2002. Wide bacterial diversity associated with tubes of the vent worm Riftia pachyptila. Environ. Microbiol. 4:204-215. [DOI] [PubMed] [Google Scholar]
- 37.Miroshnichenko, M. L., N. A. Kostrikina, S. l'Haridon, C. Jeanthon, H. Hippe, E. Stackebrandt, and E. A. Bonch-Osmolovskaya. 2002. Nautilia lithotrophica gen. nov., sp. nov., a thermophilic sulfur-reducing epsilon-proteobacterium isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 52:1299-1304. [DOI] [PubMed] [Google Scholar]
- 38.Moller, D., R. Schauder, G. Fuchs, and R. K. Thauer. 1987. Acetate oxidation to CO2 via a citric acid cycle involving an ATP-citrate lyase: a mechanism for the synthesis of ATP via substrate level phosphorylation in Desulfobacter postgatei growing on acetate and sulfate. Arch. Microbiol. 148:202-207. [Google Scholar]
- 39.Moyer, C. L., F. C. Dobbs, and D. M. Karl. 1995. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 61:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul, J. H. 1996. Carbon cycling: molecular regulation of photosynthetic carbon fixation. Microb. Ecol. 32:231-245. [DOI] [PubMed] [Google Scholar]
- 42.Prieur, D., S. Chamroux, P. Durand, G. Erauso, P. Fera, C. Jeanthon, L. Le Borgne, G. Mével, and P. Vincent. 1990. Metabolic diversity in epibiotic microflora associated with the Pompeii worms Alvinella pompejana and A. caudata (Polychaete: Annelida) from deep-sea hydrothermal vents. Mar. Biol. 106:361-367. [Google Scholar]
- 43.Prieur, D., and C. Jeanthon. 1987. Preliminary study of heterotrophic bacteria isolated from two deep-sea hydrothermal vent invertebrates: Alvinella pompejana (polychaete) and Bathymodiolus thermophilus (bivalve). Symbiosis 4:87-98. [Google Scholar]
- 44.Reysenbach, A. L., A. B. Banta, D. R. Boone, S. C. Cary, and G. W. Luther. 2000. Biogeochemistry: microbial essentials at hydrothermal vents. Nature 404:835-836. [DOI] [PubMed] [Google Scholar]
- 45.Reysenbach, A. L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson, J. J., J. L. Stein, and C. M. Cavanaugh. 1998. Cloning and sequencing of a form II ribulose-1,5-biphosphate carboxylase/oxygenase from the bacterial symbiont of the hydrothermal vent tubeworm Riftia pachyptila. J. Bacteriol. 180:1596-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romano, A. H., and T. Conway. 1996. Evolution of carbohydrate metabolic pathways. Res. Microbiol. 147:448-455. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manuel, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Schauder, R., F. Widdel, and G. Fuchs. 1987. Carbon assimilation pathways in sulfate-reducing bacteria. 2. Enzymes of a reductive citric-acid cycle in the autotrophic Desulfobacter hydrogenophilus. Arch. Microbiol. 148:218-225. [Google Scholar]
- 50.Shiba, H., T. Kawasumi, Y. Igarashi, T. Kodama, and Y. Minoda. 1985. The CO2 assimilation via the reductive tricarboxylic-acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch. Microbiol. 141:198-203. [Google Scholar]
- 51.Stein, J. L., T. L. Marsh, K. Y. Wu, H. Shizuya, and E. F. DeLong. 1996. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment front a planktonic marine archaeon. J. Bacteriol. 178:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuttle, J. H. 1985. The role of sulfur-oxidizing bacteria. Bull. Biol. Soc. Wash. 6:335-344. [Google Scholar]
- 53.Van Dover, C. L., and B. Fry. 1989. Stable isotopic compositions of hydrothermal vent organisms. Mar. Biol. 102:257-263. [Google Scholar]
- 54.Wachtershauser, G. 1990. Evolution of the 1st metabolic cycles. Proc. Natl. Acad. Sci. USA 87:200-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wawer, C., M. S. M. Jetten, and G. Muyzer. 1997. Genetic diversity and expression of the [NiFe] hydrogenase large-subunit gene of Desulfovibrio spp. in environmental samples. Appl. Environ. Microbiol. 63:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wirsen, C. O., T. Brinkhoff, J. Kuever, G. Muyzer, S. Molyneaux, and H. W. Jannasch. 1998. Comparison of a new Thiomicrospira strain from the Mid-Atlantic Ridge with known hydrothermal vent isolates. Appl. Environ. Microbiol. 64:4057-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wirsen, C. O., S. M. Sievert, C. M. Cavanaugh, S. J. Molyneaux, A. Ahmad, L. T. Taylor, E. F. DeLong, and C. D. Taylor. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zani, S., M. T. Mellon, J. L. Collier, and J. P. Zehr. 2000. Expression of nifH genes in natural microbial assemblages in Lake George, New York, detected by reverse transcriptase PCR. Appl. Environ. Microbiol. 66:3119-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zehr, J. P., and B. B. Ward. 2002. Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl. Environ. Microbiol. 68:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]