Abstract
In clinical staphylococci, the presence of the ica genes and biofilm formation are considered important for virulence. Biofilm formation may also be of importance for survival and virulence in food-related staphylococci. In the present work, staphylococci from the food industry were found to differ greatly in their abilities to form biofilms on polystyrene. A total of 7 and 21 of 144 food-related strains were found to be strong and weak biofilm formers, respectively. Glucose and sodium chloride stimulated biofilm formation. The biofilm-forming strains belonged to nine different coagulase-negative species of Staphylococcus. The icaA gene of the intercellular adhesion locus was detected by Southern blotting and hybridization in 38 of 67 food-related strains tested. The presence of icaA was positively correlated with strong biofilm formation. The icaA gene was partly sequenced for 22 food-related strains from nine different species of Staphylococcus, and their icaA genes were found to have DNA similarities to previously sequenced icaA genes of 69 to 100%. Northern blot analysis indicated that the expression of the ica genes was higher in strong biofilm formers than that seen with strains not forming biofilms. Biofilm formation on polystyrene was positively correlated with biofilm formation on stainless steel and with resistance to quaternary ammonium compounds, a group of disinfectants.
Coagulase-negative staphylococci (CNS) constitute a major component of the normal microfloras of humans, and in contrast to Staphylococcus aureus (31), most of them do not produce exotoxins. Thus, CNS have generally been regarded as saprophytes or organisms with no or very low virulence. However, over the last 3 decades there has been an increase in the documentation of human infections due to CNS, especially with the species S. epidermidis (14, 30, 45). The increase in infections by these organisms has been associated with the wide medical use of prosthetic and indwelling devices. The formation of adherent multilayered biofilms represents an essential factor in the pathogenesis of S. epidermidis biomaterial-related infections (46).
The mechanisms of biofilm formation in clinical strains of S. epidermidis have been investigated in detail and are reported to be a two-step process. The initial bacterial attachment to a surface is related to a cell surface protein (autolysin) encoded by the chromosomal atlE gene (24). The second step of biofilm formation, which includes cell aggregation and biofilm accumulation, is mediated by the products of the chromosomal intercellular adhesion (ica) operon (22, 25). This operon is composed of the icaR regulatory gene and icaADBC biosynthetic genes. Expression of the ica genes leads to production of polysaccharide intercellular adhesin (PIA). PIA consists of β 1,6-linked N-acetylglucosamine (22, 25). Studies have shown that isolates producing PIA are able to form thick biofilms on polystyrene microtiter plates. Not all ica-positive isolates of S. epidermidis produce biofilms. This has been explained by spontaneous insertion of IS256 in the ica operon and by strict regulation of the ica genes (37, 55). Expression of the ica locus and subsequent biofilm formation may be activated via the alternative sigma factor σB in the presence of stress factors (32). Although the presence of atlE seems to be universal in S. epidermidis, the presence of the ica locus is not. For clinical strains, 80 to 90% are ica positive while only 5 to 30% of saprophytic isolates are ica positive (20, 54). This has led to the suggestion that the presence of the ica operon can be used to discriminate virulent strains from normal floras (4, 5, 20, 21). The ica locus has also been found in S. aureus and in eight other species of Staphylococcus (2, 11).
Bacteria in biofilms are generally more resistant to environmental stresses than their free-living counterparts (10). For Listeria monocytogenes, it has been shown that strains that persist in food industry environments form thicker biofilms than isolates only sporadically found (36, 41), indicating that biofilm formation is important for the survival of L. monocytogenes in the food industry. CNS are commonly found on human skin and may be transferred to food or food contact surfaces by food handlers and are thus frequently found in food processing environments (29, 39, 51). To our knowledge, biofilm formation and the presence of the ica locus in staphylococci isolated from the food industry have not been investigated previously. It is of interest to investigate whether food-related staphylococci are strong biofilm formers like the clinical invasive strains or whether they resemble the less harmful saprophytic isolates. In addition to the possible clinical significance of biofilm-forming CNS from the food industry, the ability to form biofilms may also increase the ability of isolates to persist in a food-processing environment. In this work, we investigated biofilm formation on polystyrene (and on stainless steel for selected strains) and the presence of the ica locus among staphylococci (mostly CNS) isolated from food industry.
MATERIALS AND METHODS
Bacterial strains and cultural conditions.
The bacterial strains used in this study are listed in Table 1. The isolates from the food industry were isolated from three separate Norwegian meat- and poultry-processing plants by swabbing food cuttings and other contact surfaces and equipment (51). S. epidermidis RP62A (ATCC 35894), a blood culture isolate, contains the ica locus and is widely used as a positive control in biofilm experiments (7, 13). All strains were routinely cultured in tryptic soy broth (TSB; Oxoid, Basingstoke, United Kingdom), with agitation at 37°C.
TABLE 1.
Staphylococcus spp. used in this study
| Strain(s) | No. of strains | Origin | Reference or source |
|---|---|---|---|
| Food-related strains | |||
| Staphylococcus spp. | 19 | Red-meat-processing planta | 51 |
| Staphylococcus spp. | 111 | Poultry-processing plantsa | 51 |
| Staphylococcus spp. | 2 | Baguettes | 51 |
| Staphylococcus spp. | 12 | Meat | Elisabeth Borch, SIK, Göteborg, Sweden |
| Reference strains | |||
| S. epidermidis Fol89 | Blood culture | 35 | |
| S. epidermidis RP62A | Catheter sepsis | 7 | |
| S. aureus RN4220 | 33 | ||
| S. aureus ATCC 12600 | |||
| S. saprophyticus ATCC 15305 | |||
| S. capitis NCTC 11045 | |||
| S. cohnii NCTC 11041 | |||
| S. intermedius NCTC 11048 | |||
| S. lentus NCTC 12102 | |||
| S. sciuri NCTC 12103 |
The strains were isolates from steel, rubber and different polymers, and chicken and red meat. Examples of sampling sites were cutting boards, conveyor belts, tables, gloves, saws, rubber fingers, different processing machines, trays, pumps, and knifes (51).
Quantitative biofilm assay on polystyrene.
The ability of the strains to adhere to polystyrene microtiter plates was determined as described previously (8), with some modifications. An overnight culture grown in TSB at 37°C was diluted 1:100 in TSB and in TSB with 2% (wt/vol) glucose and 2% (wt/vol) sodium chloride added. A total of 200 μl of these cell suspensions was transferred to each of six parallel wells of a 96-well polystyrene microtiter plate (Nunclon; Nunc, Roskilde, Denmark). After incubation at 37°C for 24 h, the absorbance at 600 nm was recorded on a Micro-ELISA autoreader (Titertek Multiscan Plus; Labsystems, Helsinki, Finland) as a measure of total growth. Furthermore, the culture was removed and plates were washed three times with 200 μl of peptone water (saline with 0.1% Bacto Peptone [Oxoid]) to remove nonadherent cells and dried in an inverted position. Adherent biofilm was stained with 100 μl of 0.1% (wt/vol) safranin (Merck) for 3 min. Then, unbound safranin was removed and the microtiter plate was air dried for 2 h before the absorbance at 492 nm of the biofilm was quantified. All strains were tested in unrelated experiments on two different days. The strains did not form clearly distinct groups (judged on the basis of the distribution of the A492 values), but for convenience they were defined into different categories on the basis of their A492 values. The strains with A492 < 0.20, 0.20 ≤ A492 ≤ 1.0, and A492 > 1.0 were defined as biofilm negative, weak biofilm formers, and strong biofilm formers, respectively.
The effect on biofilm formation was tested in the assay described above with the following additions to TSB: sodium chloride (0.01 to 5% [wt/vol]), glucose (0.01 to 5% [wt/vol]), ethanol (0.005 to 2.5% [vol/vol]), sodium dodecyl sulfate (SDS) (0.00004 to 0.025% [wt/vol]), salicylate (0.025 to 13 mM), tetracycline (0.0025 to 1.25 mg liter−1), and the two disinfectants chlorhexidine (0.005 to 2.5 mg liter−1) and benzalkonium chloride (0.01 to 5 mg liter−1). TSB (Oxoid) without additions contains 0.5% sodium chloride and 0.25% glucose.
Biofilm formation on stainless steel.
The ability to form biofilm on stainless steel was tested in a static model system. Stainless-steel coupons (AISI 304, 2B finish, 75 by 22 by 1 mm) were placed vertically in 150 ml of TSB with 2% sodium chloride and 2% glucose added. The medium was inoculated (0.1% vol/vol) with an overnight culture of the strain (grown in TSB at 25°C) to be tested. After 24 h of incubation at 25°C, the coupons were removed from the culture and gently washed twice with peptone water to remove loosely adherent cells. Then, each coupon was placed in a glass tube containing 45 ml of sterile distilled water and the attached cells were removed by sonication (Bransonic 3510; Branson Ultrasonic B.V., Soest, The Netherlands) at 40°C for 10 min. After sonication, the number of CFU was determined by plating on tryptic soy agar (Oxoid). To ensure that all bacteria were removed from the coupons by sonication, some coupons were stained with Acridine Orange (Sigma) (0.1% wt/vol) and examined by fluorescence microscopy. It was also confirmed that sonication had no negative effect on the number of CFU of cell suspensions.
DNA isolation, PCR amplification, and sequencing.
Cells were lysed by adding lysozyme and lysostaphin (Sigma-Aldrich, St. Louis, Mo.) at 40 and 0.1 mg ml−1, respectively, followed by incubation at 37°C for 30 min. Total DNA from the staphylococci were isolated with an Easy-DNA kit (Invitrogen, Carlsbad, Calif.). For amplification of icaA, a primer pair (ica4f and ica2r) (Table 2) was designed from conserved parts of the sequences of icaA from S. aureus (AF086783), S. epidermidis (U43366), and S. caprae (AF246926) obtained from GenBank. The same ica sequences in the database were also used to design primers for amplification of other parts of the ica operon. Each PCR mixture contained 50 ng of DNA, 2 μM concentrations of each respective primer, 200 μM concentrations of each deoxynucleoside triphosphate, 1× reaction buffer (Dynazym), and Dynazym DNA polymerase (Dynazym) in a 50-μl total volume. The reaction mixtures were subjected to 30 cycles of amplification. The conditions for each cycle were as follows: denaturation for 45 s at 95°C, annealing for 45 s at between 42 and 65°C (depending on the primer set), and primer extension for 2 min at 72°C. Finally, the reaction mixtures were incubated at 72°C for 10 min. Sequencing was performed on an ABI Prism 3100 genetic analyzer using a Big Dye terminator cycle sequencing ready reaction kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.).
TABLE 2.
Primers used in PCR
| Primer | 5′ → 3′ Sequence | Nucleotide position | Accession and/or reference no. |
|---|---|---|---|
| ica4f | TGGGATACTGAYATGATTAC | 1770-1789a | AF246926 |
| ica2r | CCTCTGTCTGGGCTTGACCATG | 2338-2317a | AF246926 |
| ica9f | CTTTCTTACTAGCATGTTACAACG | 1264-1287a | AF246926 |
| altE-F | CAACTGCTCAACCGAGAACA | 3497-3516b | U71377 (20) |
| altE-R | TTTGTAGATGTTGTGCCCCA | 4178-4159b | U71377 (20) |
| ID27f | AGAGTTTGATCCTGGCTCAG | 8-27c | 34 |
| 685r3 | TCTRCGCATTYCACCGCTA | 702-685c | 34 |
| 1492r | TACGGTTACCTTGTTACGACTT | 1513-1492c | 34 |
16S rDNA analysis.
About 1,450 bp of the 16S rRNA genes (16S rDNA) were amplified by PCR using the primers ID27f and 1492r (34). Of the variable region of 16S rDNA, 550 bp were sequenced in both directions using forward (ID27f) and reverse primers (685r3) described previously (34). Database searches were performed with BLASTN 2.1.2 software (3).
Southern blotting and hybridization.
Total DNA was digested with EcoRI and HindIII restriction enzymes (Promega) at 37°C overnight. Digested DNA was separated by agarose gel electrophoresis. HindIII-digested DNA of S. epidermidis RP62A was applied to each gel as a positive control. The restriction enzyme-digested DNA was transferred from agarose gel to a Hybond-N nylon membrane (Amersham Biosciences, Buckinghamshire, United Kingdom) by vacuum blotting according to the manufacturer's instructions (Pharmacia, Uppsala, Sweden). A PCR product specific for the icaA gene (568 bp long; amplified with primers ica4f and ica2r) from S. epidermidis RP62A was used as a probe. In addition, a probe specific for icaA (1,075 bp long; amplified with primers ica9f and ica2r) from the food-related isolate S. capitis St13 was also used. An atlE-specific probe was amplified with PCR using primers atlE-F and atlE-R and DNA from S. epidermidis RP62A. The PCR products were purified with a Qiaquick PCR purification kit (Qiagen GmbH, Hilden, Germany) before being used as probes. The labeling of probes and hybridization was done with an AlkPhos Direct Gene images kit following the manufacturer's instructions (Amersham). Membrane stripping was done as recommended by the manufacturer.
RNA isolation and Northern blotting.
Precultures were grown overnight at 37°C in brain heart infusion (BHI) broth with a final concentration of 2.5% NaCl and 2.5% glucose. These cultures were used to inoculate medium of the same type (1% inoculum) in glass tubes, which were further incubated at 37°C. At an optical density at 600 nm of 0.5 to 0.8, the cultures were added to 2 volumes of RNA Protect bacterial reagent, according to the manufacturer's instructions (Qiagen GmbH), before an RNeasy Mini kit (Qiagen GmbH) was used to isolate RNA from the cell suspensions. For effective lysis, lysozyme and lysostaphin (Sigma) were added at 20 and 0.1 mg ml−1, respectively, and incubated at 37°C for 10 to 20 min. The RNA was separated in 1.2% agarose containing formaldehyde (49) and vacuum blotted onto Hybond N+ filters as described by the manufacturer. Hybridization was performed at 45°C according to standard procedures (49). Probes specific for the icaA gene of S. capitis St13 (food-processing environment), icaA gene of S. epidermidis N15.1 (food-processing environment), and icaA gene of S. aureus RN4220 were amplified by PCR using the primer pair ica4f and ica2r (Table 2) and purified using a Qiaquick PCR purification kit (Qiagen GmbH). Using a RediprimeII kit (Amersham), the DNA probes were labeled with [α-32P]dCTP (Amersham Biosciences).
Phylogenetic constructions.
Separate phylogenetic trees were constructed using the maximum-parsimony (19) and distance methods (48) provided in the PAUP* 4.0 software package developed by D. L. Swoford (Florida State University, Tallahassee, Fla.) and distributed by Sinauer Associates Inc. (Sunderland, Mass.). In addition, maximum-likelihood methods were provided in Puzzle 4.0.2 software (50). The search for the maximum-parsimony tree was done heuristically. Both maximum-likelihood distances with empirically determined base frequencies (18) and log of the determinant of the matrix distances were used for the minimum-evolution distance trees. We used the model developed by Hasegawa (23) for the maximum-likelihood analysis. For the parsimony and the distance methods, consensus trees were constructed from 1,000 bootstrap replicates (17), while for the maximum-likelihood analysis, 1,000 quartet puzzling steps were used (50).
Statistics.
Significance testing was performed with MINITAB for Windows, version 13.3 (Minitab Inc., State College, Pa.), and Fisher's exact test (available at http://www.matforsk.no/ola/fisher.htm).
Nucleotide submissions.
The sequenced parts of icaAD, icaB, and icaR of S. capitis St13 have been deposited in GenBank under accession no. AY146582, AY146583, and AY146584, respectively. Partial icaA sequences of several strains were also deposited as follows: S. aureus H9.3 (AF500262), S. caseolyticus N10.3 (AF500258), S. cohnii O14.9 (AF500260), S. cohnii O5.1H (AF500268), S. condimenti O10.10 (AF500261), S. condimenti O7.1 (AF500266), S. epidermidis H11.6 (AF500265), S. saprophyticus N2.1A (AF500264), S. saprophyticus St28 (AF500270), S. sciuri H14.3 (AF500259), S. simulans O2.5 (AF500263), S. capitis NCTC11045 (AF500269), and S. saprophyticus ATCC15305 (AF500267).
RESULTS
Biofilm formation on polystyrene.
In TSB with 2% sodium chloride and 2% glucose added, biofilm formation measured as A492 after safranin staining ranged between 0.02 and 1.6 (corrected for medium background) for the 144 strains tested. Seven of the strains were strong biofilm formers (A492 > 1.0), and 21 strains were weak biofilm formers (0.20≤ A492 ≤1.0). Of all the tested strains, 71% formed thicker biofilms (>10% increase in A492) in TSB with sodium chloride and glucose added than in TSB without additions. The 16S rDNA was partially sequenced (479 bp) for the 28 biofilm-forming strains together with 39 randomly picked biofilm-negative strains (A492 < 0.20). For all strains, the 479-bp sequences gave matches with >99.5% identity to the 16S rDNA sequences of Staphylococcus spp. in the database. The 67 strains sequenced were assigned to 15 species of Staphylococcus, of which 9 were biofilm formers (Table 3). The strong biofilm formers (A492 > 1.0) belonged to the species S. capitis, S. cohnii, S. epidermidis, and S. saprophyticus. For the biofilm-positive control S. epidermidis RP62A strain grown in TSB cultures with glucose and sodium chloride added, the absorbance at 492 nm after safranin staining was 1.4. The reference strains, S. aureus RN4220, S. aureus ATCC 12600, and S. epidermidis Fol89, were strong biofilm formers and S. intermedius NCTC 11048 and S. sciuri NCTC 12103 were weak biofilm formers, while S. saprophyticus ATCC 15305, S. capitis NCTC 11045, S. cohnii NCTC 11041, and S. lentus NCTC 12102 were biofilm negative.
TABLE 3.
Hybridization to icaA probe among food related staphylococci with different biofilm formation abilities
| Species | Total no. of strains | No. of hybridization-positive strains/ total no. of strains
|
||
|---|---|---|---|---|
| Stronga | Weakb | Negativec | ||
| S. aureus | 3 | 3/3d | ||
| S. capitis | 3 | 3/3d | ||
| S. caseolyticus | 4 | 1/4 | ||
| S. condimenti | 4 | 1/1d | 3/3 | |
| S. cohnii | 10 | 1/1 | 1/7 | 1/2 |
| S. epidermidis | 9 | 1/1 | 4/8 | |
| S. hominis | 1 | 0/1 | ||
| S. intermedius | 4 | 1/1 | 3/3 | |
| S. lentus | 3 | 0/2 | 1/1 | |
| S. piscifermentans | 2 | 1/2 | ||
| S. saprophyticus | 6 | 2/2 | 3/3 | 1/1 |
| S. schleiferi | 1 | 1/1 | ||
| S. sciuri | 2 | 1/2 | ||
| S. simulans | 14 | 3/6 | 1/8 | |
| S. vitulinus | 1 | 0/1 | ||
| Total | 67 | 7/7 | 9/21 | 22/39 |
Strains with A492 > 1.0 in the biofilm assay (number of strains hybridizing to icaA/total number of strains tested).
Strains with 0.20 ≤ A492 ≤ 1.0 in the biofilm assay (number of strains hybridizing to icaA/total number of strains tested).
Strains with A492 < 0.20 in the biofilm assay (number of strains hybridizing to icaA/total number of strains tested).
Number of strains which hybridized at 55°C to the S. epidermidis RP62A icaA-specific probe/total number of strains.
Effect of stress conditions on biofilm formation.
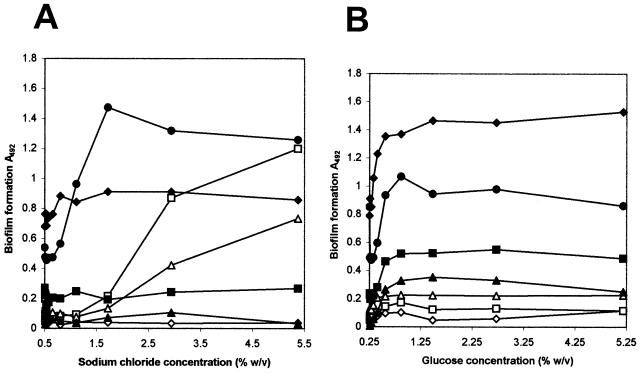
For four of the seven strains tested, the addition of sodium chloride clearly stimulated biofilm formation (Fig. 1A). The two S. capitis strains formed the thickest biofilm at the highest sodium chloride concentration tested (5.4%). The addition of glucose had a positive effect on biofilm formation for all seven strains tested. The glucose concentration needed for maximal biofilm formation ranged between 0.3 and 1.3%, depending on the strain (Fig. 1B). The addition of ethanol, benzalkonium chloride, chlorhexidine, salicylate, SDS, or tetracycline had no effect on biofilm formation at the concentrations tested (data not shown). Growth (measured as absorbance at 600 nm before the removal of the fluid culture) was little affected by the concentration range used for all additions except for inhibition at the highest concentration of tetracycline (data not shown).
FIG. 1.
Effect of addition of sodium chloride (A) and glucose (B) to TSB on biofilm formation (A492) of Staphylococcus spp. (TSB without additions contains 0.5% sodium chloride and 0.25% glucose.) Biofilm formation was measured after 24 h of growth at 37°C. ♦, S. epidermidis RP62A; ⋄, S. epidermidis N5.5; □, S. capitis St6; ▵, S. capitis St13; ▪, S. saprophyticus N16.7; •, S. saprophyticus St28; ▴, S. simulans O2.5.
Biofilm formation on stainless steel.
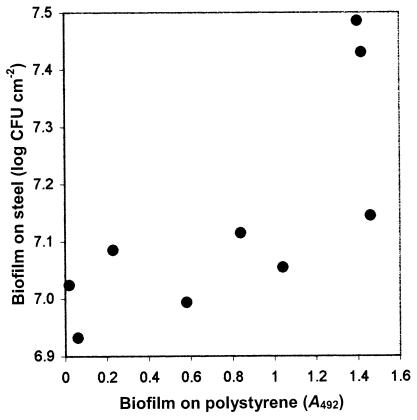
Nine staphylococci selected for having different abilities to form biofilms on polystyrene microtiter plates were tested for their ability to form biofilms on stainless steel in TSB with 2.25% glucose and 2.5% sodium chloride. For the different strains, biofilm formation on steel after 24 h of incubation ranged from 8.6 × 106 to 3.1 × 107 adherent cells cm−2 (Fig. 2). Biofilm formation on polystyrene and stainless steel showed a weak but significant correlation (R2 = 0.56; P = 0.02) (Fig. 2).
FIG. 2.
Relationship between biofilm formation on stainless steel at 25°C (log CFU cm−2) and biofilm formation on polystyrene (A492) at 37°C by staphylococci grown in cultures for 24 h in TSB with 2% glucose and 2% sodium chloride added. Each point represents the mean for an individual strain of two independent experiments each on steel and polystyrene.
Presence and sequence of the icaA gene.
The 67 food-related strains from which 16S rDNAs were sequenced were also investigated by Southern blotting followed by hybridization for the presence of icaA. A total of 38 strains hybridized with the S. epidermidis RP62A icaA-specific probe at 55°C (Table 3). Except for S. hominis and S. vitulinus, at least one strain from each of the 15 species hybridized to the icaA probe. All seven strong biofilm formers hybridized to icaA, and hybridization to the icaA probe was significantly higher (P = 0.02) among strong biofilm formers (A492 > 1.0) compared to the other strains. The prevalence of icaA among the weak biofilm formers (0.20≤ A492 ≤1.0) was not significantly higher than among biofilm-negative strains (A492 < 0.20). When hybridization was performed at 65°C, only strains of S. aureus, S. capitis, S. cohnii, S. condimenti, S. epidermidis, and S. saprophyticus hybridized to the S. epidermidis RP62A icaA-specific probe (data not shown). Hybridization at 55°C with an S. capitis St13 icaA-specific probe gave results similar to those of hybridization at 55°C with the S. epidermidis RP62A icaA-specific probe (data not shown). The reference strains S. epidermidis RP62A (positive control), S. epidermidis Fol89, S. aureus ATCC 12600, S. capitis NCTC 11045, S. saprophyticus ATCC 15305, S. intermedius NCTC 11048, and S. aureus RN4220 all hybridized to the S. epidermidis RP62A-specific icaA probe at 55°C, while S. sciuri NCTC 12103, S. cohnii NCTC 11041, and S. lentus NCTC 12102 showed no hybridization signal.
For the 38 food-related strains that hybridized to the icaA probe at 55°C, an attempt was made to amplify a part of the icaA gene by PCR with the primer pair ica4f and ica2r. For 22 of the 38 strains, icaA-specific PCR products of 568 bp were amplified and sequenced with both primers used for the amplification. With the rest of the food-related strains, no PCR product was obtained or no sequence was obtained for the PCR product. icaA from S. aureus, S. capitis, S. caseolyticus, S. cohnii, S. condimenti, S. epidermidis, S. saprophyticus, S. sciuri, and S. simulans was partly sequenced. The sequences obtained with the two primers were aligned, and for each strain, a database search was performed with a 372-bp-long consensus sequence. The DNA similarities of the partially sequenced icaA genes to icaA sequences available in the database were 69 to 100%. Sequences of icaA from the reference strains S. aureus RN4220, S. epidermidis Fol89, S. saprophyticus ATCC15305, and S. capitis NCTC11045 were also obtained.
For S. capitis St13 and S. saprophyticus St28, the icaA genes were amplified and completely sequenced using primers designed from the S. caprae icaA sequence in the database (AF246926). For these two strains, their icaA genes were found to be identical at the nucleotide level. According to the results of a database search, the complete icaA sequences of S. capitis St13 and S. saprophyticus St28 showed the closest match to the icaA gene of S. caprae (AF246926), with 85% DNA homology and 92% amino acid homology. Using primers designed from the S. caprae ica sequence, PCR products specific for icaB, icaD, and icaR were also amplified and sequenced from S. capitis St13. The sequenced products of icaB, icaD, and icaR corresponded to the regions 2998 to 3337, 2318 to 2614, and 892 to 625 in the S. caprae ica sequence (AF246926), respectively. The sequenced parts of the ica genes of S. capitis St13 had DNA and amino acid homologies to the corresponding sequence in S. caprae (AF246926) of 84 and 88% (icaB), 88 and 94% (icaD), and 90 and 93% (icaR).
Four of eight biofilm-negative food isolates of S. epidermidis strains hybridized to icaA (Table 3). PCR mapping of the entire ica locus of these strains revealed that the lengths of the PCR products were as anticipated (data not shown); thus, no insertion element was present in the ica locus and the orientation of the icaRABCD genes was similar to that of S. epidermidis RP62A.
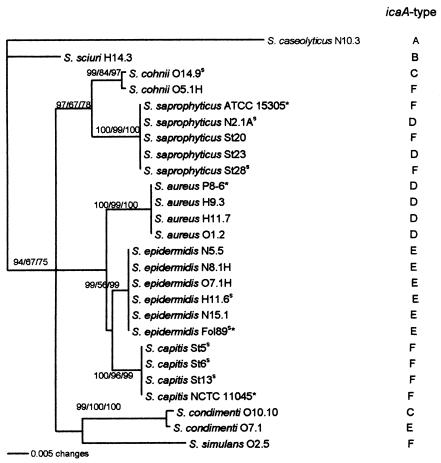
Phylogenetic reconstructions were made for the 26 strains from which both 16S rDNA and icaA sequences were obtained. The 16S rDNA tree is shown in Fig. 3, while the icaA data have been coded as icaA types A to F. The icaA sequence types were determined for the group of sequences with more than 98% internal homology and 100% bootstrap support in the phylogenetic tree (results not shown). The homologies between the different icaA types were in the range of 55 to 82%. For S. saprophyticus, S. cohnii, and S. condimenti, strains from the same species differed considerably in their icaA sequences. S. saprophyticus contained sequences of icaA types F and D and S. cohnii contained icaA types C and F, while S. condimenti contained icaA types C and E. Among strains of the same species, the icaA sequences of S. epidermidis and S. aureus were almost identical.
FIG. 3.
Evolutionary neighbor joining tree for 16S rDNA. The tree was built using the neighbor-joining, maximum-parsimony, and maximum-likelihood methods. The tree shown was constructed on the basis of 479 aligned positions. The genetic distances between two strains are shown as Kimura distances expressed in substitutions per nucleotide in the neighbor-joining tree. The number at each node (maximum likelihood-maximum parsimony-neighbor joining) indicates the percentage of the bootstrap tree in which the cluster descending from the node was found. Strains of nonfood origin are indicated by asterisks, and strong biofilm formers are indicated by a superscript “s.” All strains had >99.8% homology to the type strain of the species to which they were allocated. The type of the icaA sequence for each individual strain (determined on the basis of differences between the strains in the sequenced part of icaA [372 bp]) is indicated in the right column by letters. Strains with identical letters had >98.2% icaA homology. Strains with different letters had 54.8 to 81.9% icaA homology.
Hybridization with atlE probe.
DNAs from the 67 strains tested for hybridization to icaA were also hybridized at 65°C to a probe specific for the atlE gene of S. epidermidis RP62A. All S. epidermidis and S. capitis strains hybridized with the atlE probe. For the species S. aureus, S. piscifermentans, S. hominis, S. cohnii, S. condimenti, S. saprophyticus, and S. schleiferi, weak hybridization signals against the atlE probe were observed (data not shown).
Expression studies.
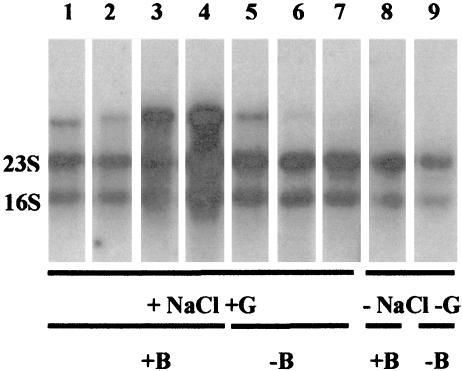
Expression of the ica genes by the strong biofilm formers S. capitis St13, S. saprophyticus St28, S. aureus RN4220, and S. epidermidis RP62A and the non-biofilm formers S. epidermidis N15.1, S. epidermidis N5.5, and S. capitis NCTC11045 (cultivated in BHI broth with a final concentration of 2.5% glucose and 2.5% sodium chloride) was investigated (Fig. 4). BHI broth was used because aggregation and clumping of some strains grown in TSB resulted in difficulties in isolating RNA. Cross-hybridization between the different probes used was observed. Three of the strong biofilm formers (S. capitis St13, S. aureus RN4220, and S. epidermidis RP62A) gave strong or intermediate transcription signals, while the fourth (S. saprophyticus St28) gave a weak transcript. Of the strains not forming biofilm, an intermediate transcript was present for S. epidermidis N15.1 and a very weak transcript was present for S. epidermidis N5.5 while no transcript was observed for S. capitis NCTC 11045. When S. capitis St13 and S. epidermidis N15.1 were cultivated in BHI broth without additional glucose and NaCl (i.e., in BHI broth with 0.2% glucose and 0.5% sodium chloride), no transcripts were detected. The transcripts observed had the same lengths as that of S. epidermidis RP62A (apparently approximately 5.0 kb long), except for S. capitis St13, which gave an approximately 4.8-kb signal.
FIG. 4.
Expression of ica genes in different staphylococci. Hybridization results for an icaA probe specific for S. epidermidis are shown. Lanes 1 and 8, S. capitis St13; lane 2, S. saprophyticus St28; lane 3, S. aureus RN4220; 4, S. epidermidis RP62A; lanes 5 and 9, S. epidermidis N15.1; lane 6, S. epidermidis N5.5; lane 7, S. capitis NCTC11045. Cells were grown in BHI broth with additional sodium chloride and glucose (+NaCl +G) or without additional sodium chloride and glucose (−NaCl −G) as indicated. +B, biofilm formation capabilities; −B, no biofilm formation capabilities. Unspecific hybridization signals were from 16S and 23S rRNA.
DISCUSSION
In this study, strongly biofilm-forming strains were determined among staphylococci from the food industry. Bacteria in biofilms are generally more resistant to stress such as disinfection and mechanical shear than their free-living counterparts. Thus, it is likely that biofilm formation protects bacteria in a food-processing environment. Polystyrene is in use as a packaging material but is not much used in equipment in the food industry. However, other hydrophobic polymers are widely used in the food industry (e.g., in gaskets, conveyor belts, trays, and cutting boards). Since the staphylococci investigated in the present study were isolated from a range of different materials, we chose polystyrene and stainless steel as model systems for biofilm formation on hydrophobic and hydrophilic materials, respectively. Thus, the formation of biofilms of food-related staphylococci on polystyrene and stainless steel may be of relevance for the food industry. It has been shown that in contrast to other isolates, isolates in poultry-processing plants in which S. aureus is endemic had a clumping (aggregation) phenotype (16) and that isolates exhibiting the clumping phenotype are more resistant to disinfection by chlorine than isolates with other phenotypes (6, 38). Thus, it is possible that biofilm formation is of importance for survival of staphylococci in food-processing environments, although to what extent is not known.
Of the food-related strains tested, 132 strains have previously been investigated for presence of qac genes encoding efflux pumps that confer resistance to quaternary ammonium compounds (QAC) (a group of disinfectants) and 19 strains were found to harbor qac genes (26, 51). This information about the prevalence of qac genes can be compared to data on biofilm formation and the presence of icaA for the same strains. Statistical analysis (Fisher's exact test) showed a significantly higher prevalence of qac genes among biofilm-forming isolates (A492 > 0.20) than among biofilm-negative isolates (P = 0.0005). Data on the presence of icaA and qac genes were also positively correlated (P = 0.007). For L. monocytogenes, both QAC resistance and biofilm formation have been proposed to be important for survival in a food-processing environment (1, 36, 41). It may be speculated that there is a synergistic effect between biofilm formation and QAC resistance and that Staphylococcus spp. and L. monocytogenes strains, both being strong biofilm formers and resistant to QAC, are especially well suited to survival in food-processing environments. This should be further investigated by assessing the sensitivity to disinfectants for Staphylococcus spp. in biofilms.
In this study, the ica locus was found in four of eight biofilm-negative S. epidermidis strains (Table 3). It has previously been shown that ica-containing S. epidermidis strains can be biofilm negative, and this has been explained by the insertion of IS256 in the ica locus or by the strict regulation of expression of the ica genes (37, 55). In the present study, no insertion elements were present in the ica locus of the biofilm-negative S. epidermidis strains. Also, all of the tested S. epidermidis strains contained the atlE gene shown to be involved in adhesion to polystyrene (24). For two of the biofilm-negative S. epidermidis strains, it was found that in one strain the ica locus was weakly transcribed while the other strain had an intermediate transcription signal (Fig. 4). Thus, this may partly explain the low biofilm formation of these strains but it is likely that additional mechanisms are involved in biofilm formation. The possibility of point mutations in the ica genes of biofilm-negative strains could not be ruled out.
It has been suggested that the presence of the ica locus can be used to discriminate between virulent and harmless strains of S. epidermidis (4, 5, 20, 21). As shown in the present study and also reported elsewhere, individual strains that are both ica positive and biofilm negative are often found. Since biofilm formation per se has been linked to virulence (12, 46), it seems more appropriate to use biofilm formation and not the presence or absence of the ica locus as one of the criteria to discriminate between potential harmful and harmless strains of S. epidermidis.
Strong biofilm formation and the presence of icaA was positively correlated; thus, the ica locus seemed to be important for strong biofilm formation in the strains tested. Apparently, the ica locus was expressed to a higher degree in strong biofilm formers than in biofilm-negative strains, although the number of strains tested was too low to draw a final conclusion about this. It is not clear why the observed length of the transcripts in the present work (approximately 5.0 kb) was longer than previously reported for transcripts of icaADBC (approximately 3.5 kb) (43). The finding in this study of a high prevalence of the ica locus among biofilm-negative strains might be explainable by low level of expression of the ica locus in those strains, but it may also indicate that additional mechanisms are involved in biofilm formation. Several strains formed weak biofilms (A492 > 0.20), but the ica locus was not detected (Table 3) and biofilm formation in these strains was possibly ica independent. However, the presence of an ica locus with low homology to the S. epidermidis icaA-specific probe used for the hybridization cannot be ruled out. None of the three food-related S. aureus strains formed biofilms, while all were icaA positive. This result is in accordance with previous work showing that the presence of the ica locus has been found in all S. aureus strains investigated; however, only a minor fraction of them are biofilm formers (11).
In the present study, icaA was partially sequenced for 26 strains belonging to nine different Staphylococcus spp. (Fig. 3). With the exception of those of S. aureus and S. epidermidis, the sequences of the icaA genes of these species have not been reported before. The ica locus seemed to be widely distributed among members of the genus Staphylococcus and was found in several of the major 16S rDNA clusters of the genus (Fig. 3) (52). The similarity between the icaA sequences of the different strains was not always consistent with the relatedness of their 16S rDNAs (Fig. 3), which may indicate that the icaA gene has been horizontally transferred.
Previously it has been shown for S. epidermidis and S. aureus that expression of the ica genes and subsequent PIA synthesis and biofilm formation are induced under stress conditions and that the alternative sigma factor SigB, IcaR, and other regulatory genetic loci are involved in the regulation of the ica locus (9, 32, 37, 43, 44, 53). Rachid and coworkers have previously shown increased biofilm formation and induced expression of the ica locus of S. aureus and S. epidermidis in the presence of various antibiotics, SDS, ethanol, sodium chloride, and glucose (42-44). In the present study, no induction of biofilm formation was found in presence of SDS and ethanol; however, positive effects of the presence of sodium chloride and glucose on biofilm formation of staphylococci were found (Fig. 1). For S. capitis St13, the expression of the ica locus in the exponential phase was induced when the strain was cultivated with additional sodium chloride and glucose (Fig. 4). Recently, Dobinsky and coworkers (15) showed that ica expression in S. epidermidis is stimulated by the presence of glucose in the exponential phase but is inhibited by the presence of glucose in the stationary phase. In conclusion, the regulation of expression of the ica genes is complex and relies on many factors.
Biofilm formation has been linked to virulence among clinical S. epidermidis strains (47). Only one biofilm-positive isolate of S. epidermidis was found in the present study. However, human infections have also been reported to be caused by other CNS, e.g., S. capitis, S. cohnii, S. saprophyticus, and S. hominis (40, 45). For S. saprophyticus, which causes urinary tract infections, biofilm formation is recognized to be involved in infection (27, 28). In the present study, isolates among CNS from the food industry were found to be strong biofilm formers belonging to the species of Staphylococcus able to cause infections; however, further investigations are necessary to evaluate whether CNS from food-processing environments have a potential to cause clinical infections.
Acknowledgments
We thank Elisabeth Borch for providing bacterial strains and Tove Maugesten and Birgitta Baardsen for excellent technical assistance.
REFERENCES
- 1.Aase, B., G. Sundheim, S. Langsrud, and L. M. Rørvik. 2000. Occurrence of and a possible mechanism for resistance to a quaternary ammonium compound in Listeria monocytogenes. Int. J. Food Microbiol. 62:57-63. [DOI] [PubMed] [Google Scholar]
- 2.Allignet, J., S. Aubert, K. G. H. Dyke, and N. El Solh. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arciola, C. R., L. Baldassarri, and L. Montanaro. 2001. In catheter infections by Staphylococcus epidermidis the intercellular adhesion (ica) locus is a molecular marker of virulent slime-producing strains. J. Biomed. Mater. Res. 59:557-562. [DOI] [PubMed] [Google Scholar]
- 5.Arciola, C. R., L. Baldassarri, and L. Montanaro. 2001. Presence of icaA and icaD genes in slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 39:2151-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolton, K. J., C. E. R. Dodd, G. C. Mead, and W. M. Waites. 1988. Chlorine resistance of strains of Staphylococcus aureus isolated from poultry processing plants. Lett. Appl. Microbiol. 6:31-34. [Google Scholar]
- 7.Christensen, G. 1982. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, G. D., W. A. Simson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachy. 1985. Adherence of coagulase-negative staphylococci to plastic culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infection. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 11.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deighton, M. A., R. Borland, and J. A. Capstick. 1996. Virulence of Staphylococcus epidermidis in a mouse model: significance of extracellular slime. Epidemiol. Infect. 117:267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deighton, M. A., J. Capstick, E. Domalewski, and T. van Nguyen. 2001. Methods for studying biofilms produced by Staphylococcus epidermidis. Methods Enzymol. 336:177-195. [DOI] [PubMed] [Google Scholar]
- 14.de Silva, G. D. I., A. Justice, A. R. Wilkinson, J. Buttery, M. Herbert, N. P. J. Day, and S. J. Peacock. 2001. Genetic population structure of coagulase-negative staphylococci associated with carriage and disease in preterm infants. Clin. Infect. Dis. 33:1520-1528. [DOI] [PubMed] [Google Scholar]
- 15.Dobinsky, S., K. Kiel, H. Rohde, K. Bartscht, J. K.-M. Knobloch, M. A. Hortskotte, and D. Mack. 2003. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: Evidence for an additional factor required for polysaccharide adhesin synthesis. J. Bacteriol. 185:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodd, C. E. R., B. J. Chaffey, and W. M. Waites. 1988. Plasmid profiles as indicators of the source of contamination of Staphylococcus aureus endemic within poultry processing plants. Appl. Environ. Microbiol. 54:1541-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 19.Fitch, W. M. 1977. On the problem of discovering the most parsimonious tree. Am. Nat. 111:223-257. [Google Scholar]
- 20.Frerbourg, N. B., S. Lefebvre, S. Baert, and J.-F. Lemeland. 2000. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galdbart, J.-O., J. Allignet, H.-S. Tung, C. Rydèn, and N. El Sohl. 2000. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J. Infect. Dis. 182:351-355. [DOI] [PubMed] [Google Scholar]
- 22.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa, M. 1985. Phylogenetic relationships among eukaryotic kingdoms inferred from ribosomal RNA sequences. J. Mol. Evol. 22:32-38. [DOI] [PubMed] [Google Scholar]
- 24.Heilmann, C., M. Hussain, G. Peters, and F. Götz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1025. [DOI] [PubMed] [Google Scholar]
- 25.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 26.Heir, E., G. Sundheim, and A. Holck. 1999. Identification and characterization of quaternary ammonium compound resistant staphylococci from the food industry. Int. J. Food Microbiol. 48:211-219. [DOI] [PubMed] [Google Scholar]
- 27.Hell, W., H.-G. W. Meyer, and S. G. Gatermann. 1998. Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol. 29:871-881. [DOI] [PubMed] [Google Scholar]
- 28.Hjelm, E., and I. Lundell-Etherden. 1991. Slime production by Staphylococcus saprophyticus. Infect. Immun. 59:445-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holah, J., and H. Gibson. 2000. Food industry biofilms, p. 211-235. In L. V. Evans (ed.), Biofilms: recent advances in their study and control. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 30.Kloos, W. E., and T. L. Bannerman. 1994. Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev. 7:117-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kloos, W. E., K.-H. Schleifer, and F. Götz. 1992. The genus Staphylococcus, p. 1369-1420. In A. Balows (ed.), The prokaryotes. Springer Verlag, New York, N.Y.
- 32.Knobloch, J. K.-M., K. Bartscht, A. Sabottke, H. Rohde, H.-H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreiswirth, B. N., S. Löfdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 34.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acids techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 35.Leegaard, T. M., E. Vik, D. A. Caugant, L. O. Frøholm, and E. A. Høiby. 1999. Low occurrence of antibiotic resistance in Escherichia coli and staphylococci isolated from blood cultures in two Norwegian hospitals in 1991-92 and 1995-96. APMIS 107:1060-1068. [PubMed] [Google Scholar]
- 36.Lunden, J. M., M. K. Miettinen, T. J. Autio, and H. J. Korkeala. 2000. Persistent Listeria monocytogenes strains show enhanced adherence to food contact surface after short contact times. J. Food Prot. 63:1204-1207. [DOI] [PubMed] [Google Scholar]
- 37.Mack, D., H. Rohde, S. Dobinsky, J. Riedewald, M. Nedelmann, J. K.-M. Knobloch, H.-A. Elsner, and H. H. Feucht. 2000. Identification of three essential regulatory gene loci governing expression of Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect. Immun. 68:3799-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mead, G. C., and B. W. Adams. 1986. Chlorine resistance of Staphylococcus aureus isolated from turkeys and turkey products. Lett. Appl. Microbiol. 3:131-133. [Google Scholar]
- 39.Mettler, E., and B. Carpentier. 1998. Variations over time of microbial load and physicochemical properties of floor materials after cleaning in food industry premises. J. Food Prot. 61:57-65. [DOI] [PubMed] [Google Scholar]
- 40.Molnàr, C., Z. Hevessy, F. Rozgonyi, and C. G. Gemmell. 1994. Pathogenicity and virulence of coagulase negative staphylococci in relation to adherence, hydrophobicity, and toxin production in vitro. J. Clin. Pathol. 47:743-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norwood, D. E., and A. Gilmour. 1999. Adherence of Listeria monocytogenes strains to stainless steel coupons. J. Appl. Microbiol. 86:576-582. [DOI] [PubMed] [Google Scholar]
- 42.Rachid, S., S. Cho, K. Ohlsen, J. Hacker, and W. Ziebuhr. 2000. Induction of Staphylococcus epidermidis biofilm formation by environmental factors: the possible involvement of the alternative transcription factor SigB. Adv. Exp. Med. Biol. 485:159-166. [DOI] [PubMed] [Google Scholar]
- 43.Rachid, S., K. Ohlsen, W. Witte, J. Hacker, and W. Ziebuhr. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:3357-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rupp, M. E., and G. L. Archer. 1994. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19:231-245. [DOI] [PubMed] [Google Scholar]
- 46.Rupp, M. E., J. S. Ulphani, P. D. Fey, K. Bartscht, and D. Mack. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rupp, M. E., J. S. Ulphani, P. D. Fey, and D. Mack. 1999. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect. Immun. 67:2656-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 50.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 51.Sundheim, G., T. Hagtvedt, and R. Dainty. 1992. Resistance of meat associated staphylococci to a quaternary ammonium compound. Food Microbiol. 9:161-167. [Google Scholar]
- 52.Takahashi, T., I. Satoh, and N. Kikuchi. 1999. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int. J. Syst. Bacteriol. 49:725-728. [DOI] [PubMed] [Google Scholar]
- 53.Valle, J., A. Toledo-Arana, C. Berasain, J.-M. Ghigo, B. Amorena, J. R. Penadès, and I. Lasa. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 54.Ziebuhr, W., C. Heilmann, F. Götz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesin gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lössner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]