Abstract
The Wyoming Department of Health investigated an outbreak of acute gastroenteritis among persons who dined at a tourist saloon in central Wyoming during October 2001. Human caliciviruses (HuCVs) were suspected as the etiological agent of the outbreak based on the incubation period, duration of illness, and symptoms observed in ill patrons. A retrospective cohort study demonstrated that ill patrons were 4.5 times more likely to have exposure to drinking water and/or ice than nonill patrons. No food items were associated with illness. An environmental investigation gave evidence that the saloon's groundwater was contaminated with sewage. Water from the saloon's only well was processed for viruses. The processed water sample and stool samples collected from three ill patrons were analyzed by reverse transcription-PCR (RT-PCR) for the presence of HuCV. All positive RT-PCR results were confirmed by sequence and phylogenetic analyses of cloned RT-PCR products. A genogroup I, subtype 3, HuCV stain was found to be present in the well water sample and two stool samples. In addition, a genogroup II, subtype 6, strain was detected in one stool sample. The identification of the same HuCV strain in both the well water and stool samples strongly suggests a link between exposure to well water and the outbreak of gastroenteritis. The presence of a genogroup II, subtype 6, strain in one of the stool samples suggests that multiple HuCV strains may have been involved in this outbreak. The laboratory isolation of HuCV strains from outbreak-associated drinking water is relatively novel in the United States. This investigation outlines the procedure for virus isolation and illustrates the utility of RT-PCR for the identification of HuCV in large volumes of water and stool samples obtained during outbreaks of acute nonbacterial gastroenteritis.
Human caliciviruses (HuCVs) have been grouped into two genera: Norovirus and Sapovirus (www.ictvdb.iacr.ac.uk/Ictv). Noroviruses that infect humans, which have been previously termed Norwalk-like viruses or small round structured viruses, have been divided into genogroups I and II (3, 4, 21). Norwalk virus is a prototype strain for genogroup I, while Snow Mountain virus is a prototype strain for genogroup II. These genogroups are further divided into subtypes or genetic clusters (4, 14, 43). Sapporo virus is the prototype sapovirus.
HuCVs are a major cause of acute nonbacterial gastroenteritis worldwide. Illness occurs in people of all ages, is characterized by nausea, vomiting, and diarrhea, and is rarely fatal (20, 22, 42). HuCVs are transmitted predominantly through the fecal-oral route but may also be transmitted though person-to-person contact (8, 17, 37, 41). There have been numerous outbreaks due to HuCV-contaminated foods, such as shellfish, salads, and deli sandwiches (1, 12, 14, 19, 26, 31, 38) and to HuCV contamination of water (5, 9, 23, 27, 29, 30, 34, 40). The presence of HuCV in recreational and drinking water sources is a public health concern due to the potential for widespread outbreaks. Rapid and reliable methods to detect HuCV in contaminated water sources are necessary to limit and/or prevent such outbreaks. Unlike other pathogenic viruses, HuCV cannot be propagated in cell culture, and there are no animal models of infection (39).
Reverse transcription-PCR (RT-PCR) has become a favored diagnostic tool in the investigation of outbreaks of gastroenteritis of suspected viral origin (13, 24-26, 28, 31, 32, 35, 44, 45) because it is rapid and sensitive. Several nonbacterial outbreaks of gastroenteritis, which were classified as of unknown etiology, are now being attributed to noroviruses through the use of RT-PCR (15). In this report, we describe an outbreak of gastroenteritis that occurred at a saloon in central Wyoming. Illness was linked to exposure to fecally contaminated groundwater. To our knowledge, this is one of the few reports (2, 5) where epidemiological and environmental data have been supported by molecular data in the analysis of a waterborne norovirus outbreak in the United States.
MATERIALS AND METHODS
Epidemiologic investigation.
A retrospective cohort study was conducted by telephone interview following a report of an outbreak of gastroenteritis in a saloon in central Wyoming during September and October 2001. The epidemiological data were analyzed with Epi Info 2000 (Centers for Disease Control and Prevention, Atlanta, Ga.), and the risk ratio and 95% confidence level for consumption of drinking water from the saloon were calculated.
Environmental investigation.
An environmental survey of the premises was performed to investigate the construction of the well and the possible sources of well water contamination. The investigation included an examination of the well construction log, current well and chlorinator conditions, monitoring records, and potential sources of contamination.
Coliform analysis.
Six samples each of well water (taken at the well head) and tap water were collected on 24 October and tested for the presence of fecal and total coliforms.
Water sample processing.
A standard filter apparatus (16) containing a 1MDS positively charged, 10-inch cartridge filter was used to concentrate viruses from 2,010 liters of the saloon's well water on 26 October. The filter was shipped by overnight delivery to the U.S. Environmental Protection Agency (EPA) laboratories in Cincinnati, Ohio, where virus was eluted from the filter and concentrated by a modification of the Celite procedure of Dahling (11). Briefly, filters were eluted with 1.6 liters of 1.5% beef extract (Adams Scientific, West Warwick, R.I.), pH 9.0, twice. The second elution was performed by filling the cartridge housing with beef extract and storing it at room temperature overnight before finishing the elution with the remaining portion of beef extract. Viruses were further concentrated from each elution with the addition of 1.6 g of Celite (Ohio Valley Specialty Chemical, Marietta, Ohio) and pH adjustment to 4.0 with 1 M HCl. After 10 min of stirring, Celite was collected on a sterile filter (Millipore, Bedford, Mass.) with a Buchner funnel. Viruses collected on the filter were eluted with 80 ml of 0.15 M sodium phosphate (pH 9.0). The concentrate was then adjusted to pH 7.0 and passed through a 0.2-μm-pore-size Acrodisc filter (Pall Gelman Laboratory, Ann Arbor, Mich.).
The concentrates from the first and second eluates were treated for inhibitor removal and viral concentration according to the method of Fout and colleagues (17). Briefly, viruses present in 32 ml of each concentrate were pelleted by ultracentrifugation through a 30% sucrose layer at 131,500 × g for 4.5 h, and the viral pellets were resuspended in phosphate-buffered saline (PBS) with 0.2% bovine serum albumin (BSA) (crystalline grade; U.S. Biochemicals, Cleveland, Ohio). The resuspended pellets were then treated with an equal volume of 0.01% dithiozone (diphenylthiocarbazone)-0.01 M 8-hydroxyquinoline-chloroform-butanol-methanol-trichloroethane (0.1/0.9/1/0.25/0.25, vol/vol/vol/vol/vol), prepared with stock solutions of 0.01% dithiozone (Fisher Scientific, Pittsburgh, Pa.) and 0.01 M 8-hydroxyquinoline (Fisher Scientific) in chloroform. Samples were mixed by vortexing for 30 s, followed by a 15-s incubation at room temperature, a second 30-s vortex, and a 30-s incubation at room temperature. The samples were centrifuged at 18,000 × g for 10 min at 4°C to separate the aqueous and organic phases. The aqueous layer from each sample was removed and further concentrated on Microcon-100 filter units (Amicon, Inc., Beverly, Mass.). Following the concentration and inhibitor removal, the samples were subjected to analysis by RT-PCR either directly or after RNA extraction.
Stool sample processing.
Outbreak-associated stool samples were mixed 1:1 to 1:1.5 with PBS with 0.2% BSA to bring all samples to a volume of 2 ml and similar consistencies. The samples were extracted with an equal volume of 0.01% dithiozone-0.01 M 8-hydroxyquinoline-chloroform-butanol-methanol-trichloroethane (0.1/0.9/1/0.25/0.25, vol/vol/vol/vol/vol) as described above, except that the organic phase from each extraction was reextracted with 0.5 ml of PBS with 0.2% BSA. The two aqueous layers were combined, centrifuged again at 14,000 rpm for 10 min at 4°C, concentrated on Microcon-100 filter units, and stored at 4°C until analysis by RT-PCR.
TRIzol LS extraction of RNA.
Total RNA was extracted from 30 μl of the concentrated water sample with TRIzol LS reagent (Life Technologies, Gaithersburg, Md.). Briefly, 30 μl of sample was brought to a final volume of 0.25 ml with PBS containing 0.2% BSA and then mixed, by vortexing, with 0.75 ml of TRIzol LS reagent. The samples were processed in accordance with the manufacturer's instructions, and the RNA pellet was dissolved in 30 μl of diethyl pyrocarbonate-treated water.
RT-PCR analysis.
Two primer pair sets that amplify a 213-bp region of the polymerase gene of noroviruses (6, 14) were used for RT-PCR analysis (Table 1). RT was performed in a 30-μl volume containing 5 μl of water or stool sample extract or sample RNA, 10 mM Tris, pH 8.3, 50 mM KCl, 0.67 mM (each) deoxynucleoside triphosphate, 1.5 mM MgCl2, and 50 pmol of one set or both sets of cDNA sense primers (Table 1). The RT reaction mixtures were heated to 99°C for 5 min to release and denature the viral RNA and then quick-cooled on ice for 5 min, followed by the addition of 50 U of murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, Calif.) and 7.5 U of RNAsin (Promega, Madison, Wis.). The RT reaction mixtures were incubated for 1 h at 43°C, followed by 5 min at 95°C and a 4°C soak.
TABLE 1.
Oligonucleotide primers
| Name | Sequence (5′-3′)a | Geno- group | Sense |
|---|---|---|---|
| MON431 | TGG ACI AGR GGI CCY AAY CA | II | RNA |
| MON432 | TGG ACI CGY GGI CCY AAY CA | I | RNA |
| MON433 | GAA YCT CAT CCA YCT GAA CAT | II | cDNA |
| MON434 | GAA SCG CAT CCA RCG GAA CAT | I | cDNA |
Oligonucleotide sequences are from reference 7. International Union of Biochemistry ambiguity codes: I, inosine; R, purine (A/G); S, strong (C/G); Y, pyrimidine (C/T).
The PCRs were performed in a final volume of 100 μl, which included the 30-μl RT reaction mixture, with final concentrations of 10 mM Tris, pH 8.3, 50 mM KCl, 0.2 mM (each) deoxynucleoside triphosphate, 2.25 mM MgCl2, 50 pmol of one or both RNA sense primers, and 5 U of Amplitaq Gold DNA polymerase (Applied Biosystems). The reaction mixtures were incubated at 94°C for 10 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 1 min 30 s, and elongation at 72°C for 1 min, and then a final soak at 72°C for 7 min. The RT-PCR products were analyzed for the presence of a 213-bp fragment on a 3% high-resolution blend agarose (Amresco, Solon, Ohio) gel, stained with SYBR Green I, and photographed with a Kodak digital camera. Norovirus-positive and -negative controls were included in all assays.
Cloning of RT-PCR products.
Prior to cloning, PCR products were reamplified by using the PCR conditions described above, except that 0.1 μl of sample from the previous amplification and 60 pmol of each primer were used and amplification was carried out for 25 cycles at an extension temperature of 60 rather than 72°C. The resulting amplicons were ligated into the TA cloning vector pCRII (Invitrogen, Carlsbad, Calif.) and transformed into competent Escherichia coli strain DH5α (Life Technologies) by using the manufacturer's procedures. Clones were screened by PCR using the conditions described above.
For cloning water sample RT-PCR products amplified with genogroup II primers, the DNA band was eluted from 1% agarose (Amresco) and purified with the QIAquick gel extraction kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. It was then cloned into the pCRII vector.
Sequencing.
Clones containing the correct-size insert were sequenced in both directions with T7 and SP6 primers by using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit on an ABI Prism 3700 DNA analyzer. Sequences derived from the viruses present in the water and stool samples were aligned with known HuCV sequences by using MegAlign, version 5.03 (DNAStar, Madison, Wis.).
RESULTS
Epidemiologic investigation.
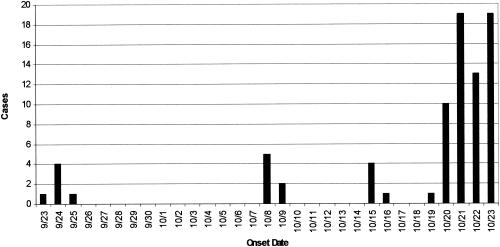
An outbreak of gastroenteritis in patrons of a saloon in central Wyoming occurred during September and October 2001 (Fig. 1). A total of 76% (84 of 111) of the patrons interviewed by telephone developed acute gastroenteritis. Of the ill patrons, 91% reported nausea, 85% reported diarrhea, 82% reported vomiting, and 73% reported muscle aches. The average duration of illness was 2 days. Ill patrons were 4.5 times more likely to have exposure to drinking water and/or ice than nonill patrons (relative risk = 4.5; 95% confidence interval, 1.3 to 15.9). An epidemiologic analysis of the relative risk of 41 food items from the menu indicated that none were statistically associated with illness.
FIG. 1.
Dates of onset of cases of gastroenteritis among saloon patrons.
Environmental investigation.
The Wyoming Department of Environmental Quality construction specifications require a minimum separation distance of 50 ft between a septic tank and a well and 100 ft between the septic tank leach field and a well for facilities with less than 2,000 gallons per day of domestic sewage flow. An environmental survey of the premises revealed that the saloon's only well was 50 ft away from its septic tank. However, the septic tank showed signs of being damaged and the leach field from another septic tank was within 50 ft of the well. The well was also 100 ft away from the effluent disposal of an recreational vehicle park located uphill of the saloon. The well was drilled through fractured basalt in 1977 to a depth of 80 ft. The casing of the well was fitted with a terminal screen and had perforations located between 65 and 75 ft below grade.
Water quality monitoring records of routine quarterly samples indicated that the well had been positive for fecal coliforms in January 1995 and September 2001. Nitrate concentrations ranged from 1.5 to 5.1 mg/liter from 1994 to 2001. A pellet chlorinator was installed on the wellhead following the positive coliform test in 1995. An examination of the chlorinator following this outbreak indicated that it had failed due to pellet dust blockage of the “drop hole.”
Coliform analysis.
Five out of six well samples tested positive for fecal coliforms, and six out six tap water samples were positive for total coliforms.
HuCV analysis.
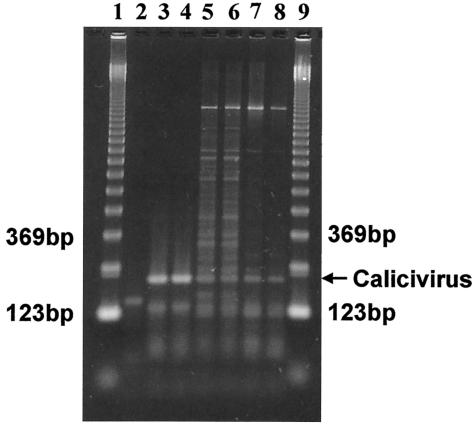
RT-PCRs were performed on total RNA extracted from water sample eluate with a mixture of the genogroup I and II primers. The results of these reactions showed amplification of a 213-bp fragment (data not shown). To give an indication of what types of noroviruses might be present in the water sample, additional reactions were performed with the genogroup I and II primers separately. The results of a representative assay using genogroup I primers are shown in Fig. 2.
FIG. 2.
Agarose gel electrophoresis of PCR products from groundwater. Norwalk virus and groundwater samples were amplified by RT-PCR using HuCV genogroup I-specific primers. Duplicate samples from the Norwalk virus positive control (lanes 3 and 4) and from eluates derived from the initial (lanes 5 and 6) or overnight (lanes 7 and 8) elutions of the 1MDS cartridge filter were analyzed. Lane 2, negative RT-PCR control; lanes 1 and 9, 123-bp ladders.
Three outbreak-associated stool samples were analyzed by RT-PCR using either a mixture of genogroup I and II primers or each primer set separately. All stool samples showed a 213-bp fragment following gel electrophoresis with at least one of the three primer sets used (data not shown).
Since a positive RT-PCR product could represent more than one norovirus strain, products were cloned prior to sequencing. Norovirus-specific clones were obtained from RT-PCRs using mixed genogroup I and genogroup II primers with the well water sample and two stool samples. Norovirus-specific clones could be obtained from a third stool sample only following amplification with genogroup I primers. Eleven clones from the water sample were sequenced, and 2 or 3 from each stool sample were sequenced.
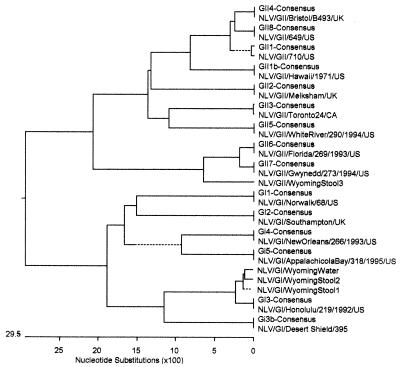
Sequence comparisons showed that the norovirus strain found in water was identical to that found in one stool sample in the 173-bp region between the primers (Fig. 3) and differed by three point mutations from the strain in a second stool sample. Phylogenetic analysis showed that these outbreak isolates had 96% homology to genogroup I, genetic cluster 3 (Fig. 4). The third stool sample showed 89 and 87% homologies to genogroup II, genetic clusters 6 and 7, respectively. Sequences of norovirus isolates from water and the three stool samples were also compared with Norwalk virus (genogroup I positive control) and P99 (genogroup II positive control) to rule out contamination from controls. Results indicated that the control sequences were sufficiently different from those of the water and stool isolates (Fig. 1). Although the RT-PCR results using genogroup II primers were positive based on gel electrophoresis, sequencing of clones obtained from these reactions failed to reveal any homology to genogroup II noroviruses in the water sample.
FIG.3.
Nucleotide sequence alignment for norovirus outbreak isolates. The sequence alignment was performed using MegAlign, version 5.03. Dots indicate identity to the water sample isolate. Dashes indicate sequence deletions to one or more strains. Norwalk virus was used as the genogroup I control, and P99-006 was used as the genogroup II control. P99-006 is most closely related to genogroup II, subtype 4.
FIG. 4.
Phylogenetic tree of norovirus sequences. The analysis was performed using MegAlign, version 5.03. The consensus sequences for the amplified RNA polymerase region were obtained from Stephan S. Monroe of the Centers for Disease Control and Prevention's Viral Gastroenteritis Unit. GenBank accession numbers for the remaining sequences are as follows: NLV/GI/Norwalk/68/US, M87661; NLV/GI/Southampton/UK, I07418; NLV/GI/Honolulu/219/1992/US, AF414403; NLV/GI/Desert Shield/395, U04469; NLV/GI/NewOrleans/266/1993/US, AF414402; NLV/GI/AppalachicolaBay/318/1995/US, AF414406; NLV/GII/710/US, AF493224; NLV/GII/Hawaii/1971/US, U07611; NLV/GII/Melksham/UK, X81879; NLV/GII/Toronto24/CA, U02030; NLV/GII/Bristol/B493/UK, X76716; NLV/GII/WhiteRiver/290/1994/US, AF414423; NLV/GII/Florida/269/1993/US, AF414407; NLV/GII/Gwynedd/273/1994/US, AF414409. GenBank accession numbers for the outbreak isolates are AY210317, AY210318, and AY210319. The abbreviations that precede the consensus strains represent the virus genogroup and subtype. For example, GII4 indicates genogroup II, subtype 4.
DISCUSSION
This report illustrates that noroviruses were associated with an outbreak of gastroenteritis that affected at least 84 patrons of a saloon in central Wyoming. An epidemiological investigation linked illness to the saloon's well water and ice. The outbreak began during the last week of September, but most cases occurred during the fourth week of October (Fig. 1). The State of Wyoming was notified of the outbreak and closed the saloon on 22 October.
An environmental survey of the saloon's public water system indicated that it was vulnerable to fecal contamination. The system is classified as a noncommunity, transient groundwater system. This classification mandates analysis of water samples for coliform bacteria quarterly and nitrate and nitrite analysis annually (40 CFR part 141) and requires that the state be notified following violations. Required monitoring was performed by the facility, but it is not clear whether the two coliform bacteria violations were reported. Monitoring records indicated that the measured nitrate levels of 1.5 to 5.1 mg/liter were below reporting requirements. The environmental survey of the premises indicated that the well was in close proximity to two effluent disposal tanks and that both the well and septic systems were installed in fractured basalt, which is conducive to cross-contamination. In addition, a chlorinator installed after a coliform violation in 1995 was malfunctioning.
Noroviruses are important agents of waterborne nonbacterial gastroenteritis. Outbreaks from these viruses often show a cold weather seasonality (10, 36) although summertime outbreaks are also reported. An interesting finding was the presence in this outbreak of a genogroup I norovirus in the well water and stool samples. Genogroup II strains have been more commonly found in recent outbreaks (18, 33). The presence of a genogroup II strain in one of the stool samples and its absence in well water samples imply that this strain may have been circulating in the water but either was not present at the time of sampling or was present at titers below the detection limit of the assay. Alternatively, it might not be water related.
Currently, surrogates such as total and fecal coliforms are used as indicators of fecal contamination. These surrogates provide great public health benefits but may not always be reliable indicators of the presence of viruses (33), especially for small public systems. Noroviruses have rarely been detected during outbreaks of waterborne diseases, in part due to the lack of suitably sensitive methods. The EPA molecular method used in this investigation has a major sensitivity advantage over methods used in other outbreaks, where less than 1 liter of groundwater could be analyzed per RT-PCR assay. EPA's method provided a degree of concentration such that each RT-PCR assay received the equivalent of 50 liters out of the 2,010 liters of water passed through the 1MDS cartridge filter. This study shows the usefulness of molecular methods as a supplement to environmental and epidemiological data in outbreak investigations. The method, which can be performed in most state regulatory laboratories with the proper training of personnel and adequate equipment and reagent support, provides an additional tool for virus occurrence studies, for studies on indicator-virus relationships, and for testing the criteria for vulnerability under EPA's proposed Groundwater Rule (http://www.epa.gov/ogwdw000/smallsys/ndwac/gwater.html).
Acknowledgments
Sandhya U. Parshionikar and Sandra Willian-True are recipients of fellowships from the Oak Ridge Institute for Science and Education Internship Program for the EPA, National Exposure Research Laboratory, Cincinnati, Ohio.
We thank Stephan S. Monroe of CDC's Viral Gastroenteritis Unit for supplying the primer and consensus sequences used in this study. We also thank David Kingsley of the USDA's Agricultural Research Service for supplying the genogroup II control virus used in this study.
REFERENCES
- 1.Anderson, A. D., V. D. Garrett, J. Sobel, S. S. Monroe, R. L. Fankhauser, K. J. Schwab, J. S. Bresee, P. S. Mead, C. Higgins, J. Campana, and R. I. Glass. 2001. Multistate outbreak of Norwalk-like virus gastroenteritis associated with a common caterer. Am. J. Epidemiol. 154:1013-1019. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, A. D., A. G. Heryford, J. P. Sarisky, C. Higgins, S. S. Monroe, S. R. Beard, C. M. Newport, J. L. Cashdollar, G. S. Fout, D. E. Robbins, S. A. Seys, K. J. Musgrave, C. Medus, J. Vinje, J. S. Bresee, H. M. Mainzer, and R. I. Glass. 2003. A waterborne outbreak of Norwalk-like virus among snowmobilers—Wyoming, 2001. J. Infect. Dis. 187:303-306. [DOI] [PubMed] [Google Scholar]
- 3.Ando, T., M. N. Mulders, D. C. Lewis, M. K. Estes, S. S. Monroe, and R. I. Glass. 1994. Comparison of the polymerase region of small round structured virus strains previously classified in three antigenic types by solid-phase immune electron microscopy. Arch. Virol. 135:217-226. [DOI] [PubMed] [Google Scholar]
- 4.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. [DOI] [PubMed] [Google Scholar]
- 5.Beller, M., A. Ellis, S. H. Lee, M. A. Drebot, S. A. Jenkerson, E. Funk, M. D. Sobsey, O. D. Simmons, S. S. Monroe, T. Ando, J. Noel, M. Petric, J. P. Middaugh, and J. S. Spika. 1997. Outbreak of viral gastroenteritis due to a contaminated well. International consequences. JAMA 278:563-568. [PubMed] [Google Scholar]
- 6.Beuret, C., D. Kohler, A. Baumgartner, and T. M. Luthi. 2002. Norwalk-like virus sequences in mineral waters: one-year monitoring of three brands. Appl. Environ. Microbiol. 68:1925-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beuret, C., D. Kohler, and T. Luthi. 2000. Norwalk-like virus sequences detected by reverse transcription-polymerase chain reaction in mineral waters imported into or bottled in Switzerland. J. Food Prot. 63:1576-1582. [DOI] [PubMed] [Google Scholar]
- 8.Caceres, V. M., D. K. Kim, J. S. Bresee, J. Horan, J. S. Noel, T. Ando, C. J. Steed, J. J. Weems, S. S. Monroe, and J. J. Gibson. 1998. A viral gastroenteritis outbreak associated with person-to-person spread among hospital staff. Infect. Control Hosp. Epidemiol. 19:162-167. [DOI] [PubMed] [Google Scholar]
- 9.Cannon, R. O., J. R. Poliner, R. B. Hirschhorn, D. C. Rodeheaver, P. R. Silverman, E. A. Brown, G. H. Talbot, S. E. Stine, S. S. Monroe, D. T. Dennis, et al. 1991. A multistate outbreak of Norwalk virus gastroenteritis associated with consumption of commercial ice. J. Infect. Dis. 164:860-863. [DOI] [PubMed] [Google Scholar]
- 10.Cubitt, W. D., D. A. McSwiggan, and W. Moore. 1979. Winter vomiting disease caused by calicivirus. J. Clin. Pathol. 32:786-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahling, D. R. 2002. An improved filter elution and cell culture assay procedure for evaluating public groundwater systems for cultural enteroviruses. Water Environ. Res. 74:564-568. [DOI] [PubMed] [Google Scholar]
- 12.Daniels, N. A., D. A. Bergmire-Sweat, K. J. Schwab, K. A. Hendricks, S. Reddy, S. M. Rowe, R. L. Fankhauser, S. S. Monroe, R. L. Atmar, R. I. Glass, and P. Mead. 2000. A foodborne outbreak of gastroenteritis associated with Norwalk-like viruses: first molecular traceback to deli sandwiches contaminated during preparation. J. Infect. Dis. 181:1467-1470. [DOI] [PubMed] [Google Scholar]
- 13.de Leon, R., S. M. Matsui, R. S. Baric, J. E. Herrmann, N. R. Blacklow, H. B. Greenberg, and M. D. Sobsey. 1992. Detection of Norwalk virus in stool specimens by reverse transcriptase-polymerase chain reaction and nonradioactive oligoprobes. J. Clin. Microbiol. 30:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 15.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 16.Fout, G. S., F. W. Schaefer III, J. W. Messer, D. R. Dahling, and R. E. Stetler. 1996. ICR microbial laboratory manual. U.S. Environmental Protection Agency, Washington, D.C.
- 17.Fout, G. S., B. C. Martinson, M. Moyer, and D. R. Dahling. 2003. A multiplex reverse transcription-PCR method for the detection of human enteric viruses in groundwater. Appl. Environ. Microbiol. 69:3158-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaulin, C., M. Frigon, D. Poirier, and C. Fournier. 1999. Transmission of calicivirus by a foodhandler in the pre-symptomatic phase of illness. Epidemiol. Infect. 123:475-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181:S254-S261. [DOI] [PubMed] [Google Scholar]
- 20.Gotz, H., B. de Jong, J. Lindback, P. A. Parment, K. O. Hedlund, M. Torven, and K. Ekdahl. 2002. Epidemiological investigation of a food-borne gastroenteritis outbreak caused by Norwalk-like virus in 30 day-care centres. Scand. J. Infect. Dis. 34:115-121. [DOI] [PubMed] [Google Scholar]
- 21.Gray, J. J., T. G. Wreghitt, W. D. Cubitt, and P. R. Elliot. 1987. An outbreak of gastroenteritis in a home for the elderly associated with astrovirus type 1 and human calicivirus. J. Med. Virol. 23:377-381. [DOI] [PubMed] [Google Scholar]
- 22.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181:S322-S330. [DOI] [PubMed] [Google Scholar]
- 23.Greening, G. E., M. Mirams, and T. Berke. 2001. Molecular epidemiology of ′Norwalk-like viruses' associated with gastroenteritis outbreaks in New Zealand. J. Med. Virol. 64:58-66. [DOI] [PubMed] [Google Scholar]
- 24.Hafliger, D., P. Hubner, and J. Luthy. 2000. Outbreak of viral gastroenteritis due to sewage-contaminated drinking water. Int. J. Food Microbiol. 54:123-126. [DOI] [PubMed] [Google Scholar]
- 25.Huang, P. W., D. Laborde, V. R. Land, D. O. Matson, A. W. Smith, and X. Jiang. 2000. Concentration and detection of caliciviruses in water samples by reverse transcription-PCR. Appl. Environ. Microbiol. 66:4383-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang, X., J. Wang, and M. K. Estes. 1995. Characterization of SRSVs using RT-PCR and a new antigen ELISA. Arch. Virol. 140:363-374. [DOI] [PubMed] [Google Scholar]
- 27.Johansson, P. J., M. Torven, A. C. Hammarlund, U. Bjorne, K. O. Hedlund, and L. Svensson. 2002. Food-borne outbreak of gastroenteritis associated with genogroup I calicivirus. J. Clin. Microbiol. 40:794-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan, J. E., R. Feldman, D. S. Campbell, C. Lookabaugh, and G. W. Gary. 1982. The frequency of a Norwalk-like pattern of illness in outbreaks of acute gastroenteritis. Am. J. Public Health 72:1329-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkwood, C. D., and R. F. Bishop. 2001. Molecular detection of human calicivirus in young children hospitalized with acute gastroenteritis in Melbourne, Australia, during 1999. J. Clin. Microbiol. 39:2722-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kukkula, M., P. Arstila, M. L. Klossner, L. Maunula, C. H. Bonsdorff, and P. Jaatinen. 1997. Waterborne outbreak of viral gastroenteritis. Scand. J. Infect. Dis. 29:415-418. [DOI] [PubMed] [Google Scholar]
- 31.Kukkula, M., L. Maunula, E. Silvennoinen, and C. H. von Bonsdorff. 1999. Outbreak of viral gastroenteritis due to drinking water contaminated by Norwalk-like viruses. J. Infect. Dis. 180:1771-1776. [DOI] [PubMed] [Google Scholar]
- 32.Le Guyader, F., F. H. Neill, M. K. Estes, S. S. Monroe, T. Ando, and R. L. Atmar.1996. Detection and analysis of a small round-structured virus strain in oysters implicated in an outbreak of acute gastroenteritis. Appl. Environ. Microbiol. 62:4268-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieberman, R. J., L. C. Shadix, B. S. Newport, S. R. Crout, S. E. Buescher, R. S. Safferman, R. E. Stetler, D. Lye, G. S. Fout, and D. R. Dahling. 1995. Source water microbial quality of some vulnerable public ground water supplies, p. 1423-1436. In Proceedings of the 1994 American Water Works Association Water Quality Technology Conference. American Water Works Association, Denver, Colo.
- 34.Lodder, W. J., J. Vinje, H. R. van De, A. M. R. Husman, E. J. Leenen, and M. P. Koopmans. 1999. Molecular detection of Norwalk-like caliciviruses in sewage. Appl. Environ. Microbiol. 65:5624-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maguire, A. J., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 1999. Molecular epidemiology of outbreaks of gastroenteritis associated with small round-structured viruses in East Anglia, United Kingdom, during the 1996-1997 season. J. Clin. Microbiol. 37:81-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurer, A. M., and D. Sturchler. 2000. A waterborne outbreak of small round structured virus, campylobacter and shigella co-infections in La Neuveville, Switzerland, 1998. Epidemiol. Infect. 125:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moe, C. L., J. Gentsch, T. Ando, G. Grohmann, S. S. Monroe, X. Jiang, J. Wang, M. K. Estes, Y. Seto, and C. Humphrey. 1994. Application of PCR to detect Norwalk virus in fecal specimens from outbreaks of gastroenteritis. J. Clin. Microbiol. 32:642-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mounts, A. W., T. Ando, M. Koopmans, J. S. Bresee, J. Noel, and R. I. Glass. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181(Suppl. 2):S284-S287. [DOI] [PubMed] [Google Scholar]
- 39.Parashar, U. D., L. Dow, R. L. Fankhauser, C. D. Humphrey, J. Miller, T. Ando, K. S. Williams, C. R. Eddy, J. S. Noel, T. Ingram, J. S. Bresee, S. S. Monroe, and R. I. Glass. 1998. An outbreak of viral gastroenteritis associated with consumption of sandwiches: implications for the control of transmission by food handlers. Epidemiol. Infect. 121:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponka, A., L. Maunula, C. H. von Bonsdorff, and O. Lyytikainen. 1999. An outbreak of calicivirus associated with consumption of frozen raspberries. Epidemiol. Infect. 123:469-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura, M., K. Natori, M. Kobayashi, T. Miyamura, and N. Takeda. 2000. Interaction of recombinant Norwalk virus particles with the 105-kilodalton cellular binding protein, a candidate receptor molecule for virus attachment. J. Virol. 74:11589-11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor, J. W., G. W. Gary, Jr., and H. B. Greenberg. 1981. Norwalk-related viral gastroenteritis due to contaminated drinking water. Am. J. Epidemiol. 114:584-592. [DOI] [PubMed] [Google Scholar]
- 43.Traore, O., G. Belliot, C. Mollat, H. Piloquet, C. Chamoux, H. Laveran, S. S. Monroe, and S. Billaudel. 2000. RT-PCR identification and typing of astroviruses and Norwalk-like viruses in hospitalized patients with gastroenteritis: evidence of nosocomial infections. J. Clin. Virol. 17:151-158. [DOI] [PubMed] [Google Scholar]
- 44.Vainio, K., K. Stene-Johansen, J. T. Oystein, A. L. Bruu, and B. Grinde. 2001. Molecular epidemiology of calicivirus infections in Norway. J. Med. Virol. 65:309-314. [DOI] [PubMed] [Google Scholar]
- 45.Vinje, J., J. Green, D. C. Lewis, C. I. Gallimore, D. W. Brown, and M. P. Koopmans. 2000. Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses.” Arch. Virol. 145:223-241. [DOI] [PubMed] [Google Scholar]