Abstract
Many bacteria possess 2 or more genes for the chaperonin GroEL and the cochaperonin GroES. In particular, rhizobial species often have multiple groEL and groES genes, with a high degree of amino-acid similarity, in their genomes. The Rhizobium leguminosarum strain A34 has 3 complete groE operons, which we have named cpn.1, cpn.2 and cpn.3. Previously we have shown the cpn.1 operon to be essential for growth, but the two other cpn operons to be dispensable. Here, we have investigated the extent to which loss of the essential GroEL homologue Cpn60.1 can be compensated for by expression of the other two GroEL homologues (Cnp60.2 and Cpn60.3). Cpn60.2 could not be overexpressed to high levels in R. leguminosarum, and was unable to replace Cpn60.1. A strain that overexpressed Cpn60.3 grew in the absence of Cpn60.1, but the complemented strain displayed a temperature-sensitive phenotype. Cpn60.1 and Cpn60.3, when coexpressed in Escherichia coli, preferentially selfassembled rather than forming mixed heteroligomers. We conclude that, despite their high amino acid similarity, the GroEL homologues of R. leguminosarum are not functionally equivalent in vivo.
INTRODUCTION
Molecular chaperones are defined broadly as proteins that enable the correct folding of other proteins into their final active state, without themselves being a part of that state (Ellis 1993). Many chaperones are also heat shock proteins and are important in both normal growth and survival of stresses that cause the unfolding of cytoplasmic proteins. Only 1 chaperone is known to be essential at all temperatures in Escherichia coli. This is the chaperone GroEL (also referred to as chaperonin), a complex of 14 identical subunits in 2 rings that assists the folding of proteins by temporarily encapsulating them in the central cavities of the rings. This enables them to fold in an environment where they are protected from aggregating and which favors the folded state (Ranson et al 1998; Sigler et al 1998; Saibil 2000). Loss of groEL, or of groES (the gene for GroES that is required for function of GroEL), is lethal to E. coli at all temperatures (Fayet et al 1989). GroEL and GroES homologues exist in nearly all bacteria for which complete genome sequences are available, the exceptions being a few of the mycoplasmas.
The substrates for GroEL have been defined (Ewalt et al 1997; Houry et al 1999). Although many proteins can bind to and be folded by the GroEL complex, the number that absolutely requires this complex for folding is relatively small. Among the obligate substrates are several essential proteins, which explains why groEL and groES are essential (Kerner et al 2005). These proteins show no similarity at the amino acid sequence level, but there is good evidence for an enrichment among the substrates for proteins with (βα)8 TIM-barrel domains. The chaperonins thus may have evolved in part specifically to fold this class of proteins.
Given that the number of obligate substrates in E. coli is low and the structural specificity of GroEL for its substrates is somewhat restricted, it is surprising that around 20% of sequenced bacterial genomes contain more than 1 groEL gene. What is the reason for these multiple genes? Do they encode GroEL proteins with a different range of substrate specificities, have some of them evolved to have novel functions, or do they simply represent a mechanism for increasing cellular levels of GroEL? Evidence that would enable us to distinguish these hypotheses is limited. It is known that, in all cases tested where multiple genes are present, the presence of at least 1 groEL gene is essential (eg, Servant et al 1993; Lee at al 1997; Rodríguez-Quiñones et al 2005). It is also known that expression levels of groE operons in the same organism can vary widely (Fischer et al 1993; Lehel et al 1993; Glatz et al 1997; Lee et al 1997; Karunakaran et al 2003; Rodríguez-Quiñones et al 2005). Multiple groEL genes in some organisms are quite closely related, whereas in others they are diverged significantly, suggesting that they may have different functions (Hughes 1993; Karlin and Brocchieri 2000).
Rhizobia have unusually large numbers of groEL and groES homologues. In examples with sequenced genomes, Bradyrhizobium japonicum has 7 groEL homologues (Kaneko et al 2002), Mesorhizobium loti and Sinorhizobium meliloti both have 5 (Kaneko et al 2000; Galibert 2001), and Rhizobium leguminosarum and Rhizobium etli both have 4 (Young et al 2006; Gonzalez et al 2006). The reasons for these high numbers are not clear; other heat shock proteins (for example DnaK and ClpB) have only 1 or 2 copies per rhizobial genome. However, there does appear to be a role for GroEL proteins in nitrogen fixation. Thus, 1 of the B. japonicum groE operons is regulated by NifA, and a reduction in the overall level of chaperonin expressed in this organism leads to a defect in nitrogen fixation caused by severely reduced levels of some nitrogenase enzymes (Fischer et al 1993, 1999). In S. meliloti, mutations in 1 of the groE operons leads to the formation of ineffective nodules, and it has been shown that 1 of the GroEL proteins interacts with the NodD activator proteins as well as having a role in quorum sensing (Ogawa and Long 1995; Yeh et al 2002; Marketon and Gonzalez 2002).
We have been studying the groEL genes of R. leguminosarum A34 to understand the roles of multiple GroEL proteins in more detail. A34 has 3 complete groE operons (1 less than the genome strain, R. leguminosarum biovar viciae 3841; Wallington and Lund 1994). The GroEL proteins produced by these operons all have been purified and show distinct properties in vitro (George et al 2004). The 3 operons are expressed at very different levels, and both of the less well-expressed operons can be deleted with no effect on strain growth, whereas the most highly expressed operon is essential (Rodríguez-Quiñones et al 2005). Thus this constitutes a good system to test whether the different operons encode proteins with distinct functions, by testing to see whether overexpression of either of the nonessential operons can compensate for loss of the essential operon. We report here the results of this analysis.
MATERIALS AND METHODS
Strains and plasmids
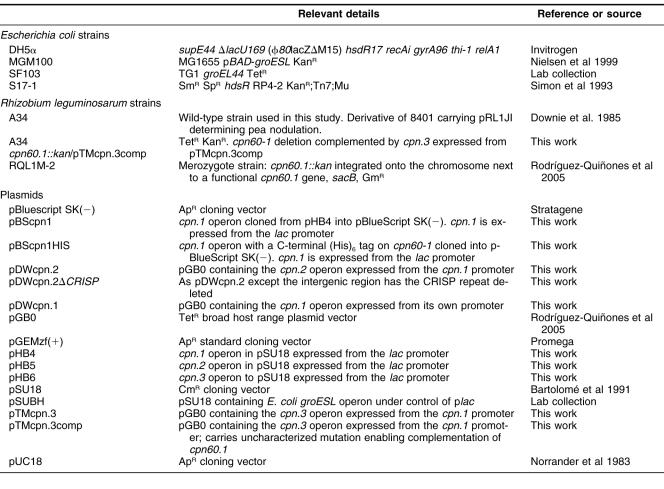
All strains and plasmids are listed in Table 1. E. coli strains were grown in Luria broth (LB) at 37°C. R. leguminosarum strains were grown in TY (Beringer 1974) or GGM (Rodríguez-Quiñones et al 1989) at 28°C, unless otherwise stated. Antibiotics were used at the following concentrations: ampicillin (200 μg/mL), gentamicin (50 μg/mL), kanamycin (30 μg/mL), and tetracycline (10 μg/mL). Where required, arabinose or glucose was added to 0.2%. Five percent sucrose was added to counterselect against sacB expression when required.
Table 1.
Strains and plasmids used in this study
Strain manipulation (transformations, conjugations, and disruption of chromosomal genes) were done as described (Rodríguez-Quiñones et al 2005). Insertion of a kan cassette into the chromosomal cpn60.1 gene was confirmed by polymerase chain reaction (PCR) using primers cpn60.1F (AAGCAGGTCGGTCTCGAC) and cpn60.1R (GACGGTGATCTTCGTGGA), which amplify across the insert site within the cpn60.1 gene.
All plasmid constructions were done using standard molecular biology methods, with enzymes used according to the manufacturers' instructions. The DNA sequences of all PCR products were confirmed after cloning. The plasmids pHB4, pHB5, and pHB6 contain the entire cpn.1, cpn.2, and cpn.3 operons, respectively, in each case with a unique NdeI site introduced at the initiator ATG of the respective cpn10 genes, cloned downstream of the plac promoter in the expression plasmid pSU18 (Bartolomé et al 1991). Subsequently, each operon was cloned downstream of a 780 base pairs (bp) fragment containing the promoter and regulatory regions of the cpn.1 operon, using the NdeI site so that each operon was at the same position relative to the cpn.1 promoter, in the shuttle plasmid pGB0 (Rodríguez-Quiñones et al 2005). This yielded the plasmids pDWcpn.1 (containing cpn.1), pDWcpn.2 (containing cpn.2), and pTMcpn.3 (containing cpn.3).
To reduce the size of the intergenic region and delete the conserved Rhizobial intergenic sequence palindrome (CRISP) repeat from the cpn.2 operon, the cpn.2 operon was cloned into pSU18 and the cpn10.2 gene and the 5′-end of cpn60.2 were amplified by PCR as 2 separate fragments, using primers GCAGGGAATTCATGCGGCCTC and CTTTTCCTGGAATTCCTCTGG that both contained a novel EcoRI site, paired with an universal M13 reverse and forward primer, respectively. The primers were designed such that, when the PCR products were ligated at the EcoRI site, most of the intergenic region was lost. The new intergenic region generated was 32 bases long. The entire cpn.2 operon, under the control of the cpn.1 promoter and with the shorter intergenic region, was cloned into pGB0 to give the plasmid pDWcpn.2ΔCRISP.
To add DNA encoding a (His)6 tag to the 3′ end of the cpn60.1 gene, the 5′-end of the cpn60.1 gene was amplified using the primers GGCGACATGATCGCCATGGGT and GCGCGGATCCTCAATGATGATGATGATGATGCATCATGTCCATACCGCCCATGCCGCCCA. The PCR product was digested with NcoI and BamHI and ligated to the remainder of the cpn.1 operon excised from pHB4 with XbaI and NcoI. The completed cpn.1(his)6 operon was cloned into the vector pBlueScript SK(-) digested with XbaI and BamHI generating the plasmid pBScpn.1HIS. As a control, a copy of the cpn.1 operon with no his-tag was excised from pHB4 with XbaI and HindIII and ligated into pBlueScript SK(-), to generate pBScpn.1.
Protein analysis
Western blots, 35S-labelling, and protein quantification were done as described previously (Rodríguez-Quiñones et al 2005; Gould et al 2006). All quantifications using 35S were based on 3 independent repeats. (His)6-tagged Cpn60.1 protein was purified using Ni-NTA resin (Qiagen), as recommended by the manufacturer. Proteins were eluted with 500 mM imidazole (dissolved in 20 mM Tris, 0.5 M NaCl pH 8.0) and precipitated using trichloro-acetic acid (TCA) for further analysis.
RESULTS
Comparative analysis of Cpn60 sequences
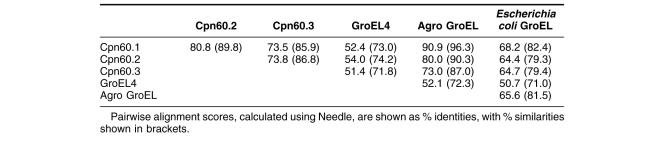
We used the program NEEDLE in Emboss-align (www.ebi.ac.uk) to make all possible pairwise comparisons between the 3 Cpn60 proteins from R. leguminosarum A34, the fourth GroEL from the genome strain (which is on a plasmid not present in A34), and GroEL from A. tumefacienes, which is closely related to R. leguminosarum but which cannot fix nitrogen or nodulate roots and has only a single groE operon. As can be seen in Table 2, the 3 Cpn60 proteins are much more closely related to each other than any of them are to the fourth protein found in the genome strain, and the major chaperonin protein Cpn60.1 shows the highest similarity to the GroEL protein from A. tumefaciens, which is closely related to R. leguminosarum.
Table 2.
Pairwise identity and similarity scores between different GroEL homologues
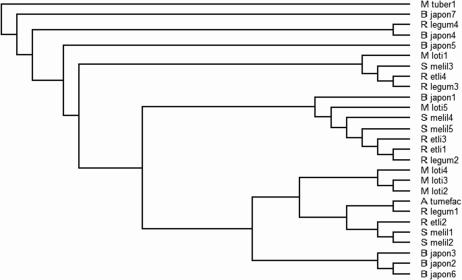
Phylogenetic analysis of all GroEL multiples from Rhizobia with completely sequenced genomes confirmed this result. We used several different methods to build phylogenetic trees from a ClustalW alignment of these proteins and, although there were small differences in the resulting trees, the main features were fully conserved. An example is shown in Figure 1. Essentially the same tree was obtained whether the R. leguminosarum chaperonin sequences from the sequenced genome, or from strain A34, were used (data not shown). A degree of clustering by species is observed, indicating that some homologues are likely to have arisen by recent gene duplications, but the clustering is not universal, and indeed the 4 R. leguminosarum GroEL homologues are all more similar to GroEL homologues from other Rhizobia than they are to each other. It is likely therefore that the multiple groEL homologues in Rhizobia have arisen by a mixture of gene duplication, speciation, and horizontal gene transfer. Because of the high similarity between the genes, the details of this are unlikely to be decipherable, a conclusion also reached in a recent detailed analysis of GroEL phylogeny (Goyal et al 2006).
Fig 1.
Cladogram showing relationships between different rhizobial Cpn60 proteins. Mycobacterium tuberculosis cpn60 was used as an outgroup, and the Cpn60 protein from Agrobacterium tumefaciens also was included. Sequences were aligned by ClustalW, and analysis was done using the programs seqboot and protpars in the phylip package (Felsentein 1989), using the default settings. The tree was displayed using treeview (Page 1996)
We next looked at alignments between the 3 Cpn60 proteins from R. leguminosarum A34 and GroEL, to see whether any residues known to be of particular importance from analysis of GroEL were altered in any of these proteins. Thirty-three positions, which have been identified as being important in genetic studies on GroEL, were compared. In all 3 Cpn60 proteins, the corresponding residues were either identical or similar to GroEL (data not shown).
One intriguing point of difference between the 3 Cpn60 homologues is at their extreme C-termini. Most GroEL homologues have long runs of glycines and methionines at their C-terminus (usually referred to as the GGM tail). The role of this region in E. coli is apparently fairly minor (Burnett et al 1994; McLennan et al 1994), but nevertheless it shows good conservation across species. However, of all the homologues present in R. leguminosarum, only Cpn60.1 possesses a GGM tail similar to that of E. coli GroEL, with 4 GGM repeats compared to 3 in E. coli. Cpn60.3 contains a single GGM motif and perhaps the remains of a second one, and Cpn60.2 does not have any GGM motifs at all.
Attempted overexpression of and complementation with Cpn60.2
The system used to check for the ability of different Cpn60 homologues to replace Cpn60.1 was as follows. We had previously constructed a copy of the cpn.1 operon with the cpn60.1 gene disrupted with a kanamycin resistance cassette and ligated this into a suicide plasmid, which then was integrated into the R. leguminosarum A34 chromosome by homologous recombination between the cpn.1 operon on the chromosome and the disrupted copy on the plasmid. This strain was called RQL1-M2 (Rodrí guez-Quiñones et al 2005). The plasmid also contained a gentamicin resistance cassette and a sacB gene. Expression of sacB is lethal on cells plated on media containing sucrose, so the ability of any other chaperonin to functionally replace Cpn60.1 can be tested by introducing it on a plasmid and selecting for a second round of recombination by growth of the cells on 5% sucrose. Recombination either will restore the original chromosome or remove the intact cpn60.1 gene from the chromosome. These 2 possibilities can be distinguished by the ability of the cells to grow on kanamycin. Strains carrying point mutations in the sacB gene can be detected as they remain gentamicin resistant.
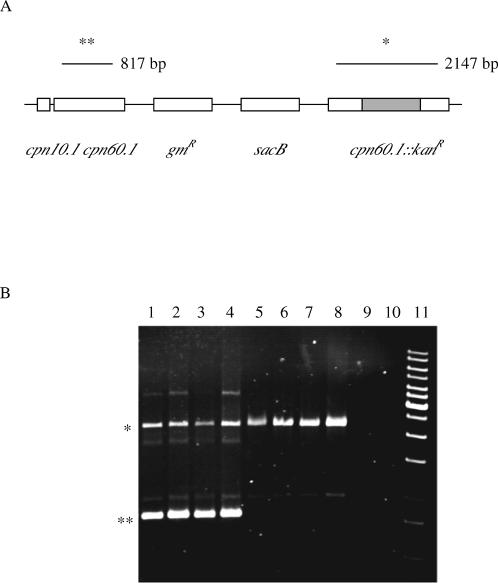
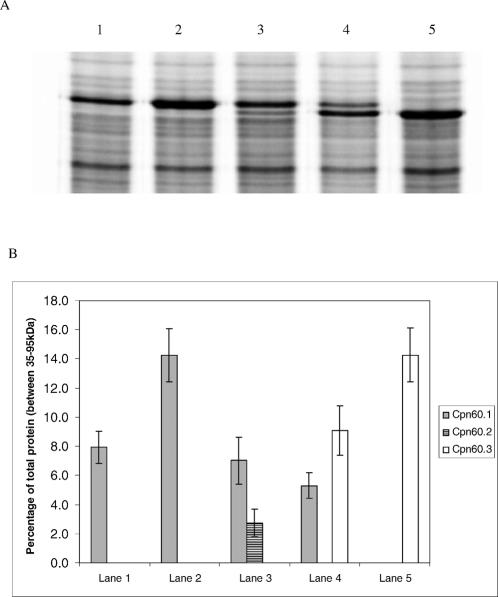
In the presence of the plasmid pDWcpn.1, which contains the cpn.1 operon under the control of its own promoter, a high frequency of sucrose-resistant colonies was seen, and those that were kanamycin resistant also were gentamicin sensitive, showing that the chromosomal copy of the cpn60.1 gene can be disrupted if a complementing source of Cpn60 is present. To determine whether Cpn60.2 functionally could replace Cpn60.1, the plasmid pDWcpn.2 that contains the cpn.2 operon under the control of the cpn.1 operon promoter, was introduced into RQL1-M2, and cells were plated out on sucrose or sucrose plus kanamycin. In repeated experiments, the frequency of sucrose-resistant colonies was reproducibly 10- to 100-fold lower than when pDWcpn.1 was present, and no kanamycin-resistant, gentamicin-sensitive colonies ever were found when pDWcpn.2 was used. PCR analysis of 4 of the sucrose-resistant colonies is shown in Figure 2 (tracks 1–4), confirming that the wild-type copy of cpn60.1 still is present in these strains.
Fig 2.
Polymerase chain reaction (PCR) analysis of potential cpn60-1 knockouts. (A) Genetic map of the merozygote region of strain RQL1M-2 showing position of primers cpn60.1F (▸) and cpn60.1R (◂) and sizes of PCR products. (B) PCR analysis. Lanes 1–4: RQL1M-2/pDWcpn.2 KanR SucR GmR; lanes 5–8: RQL1M-2/pTMcpn.3comp KanR SucR GmS; lane 9: DH5α; lane 10: blank (no DNA); lane 11: 1 kb ladder
Lack of ability of cpn60.2 to complement for loss of cpn60.1 could be due to poor expression of the Cpn60.2 protein, or inability of the Cpn60.2 protein to function in folding essential substrates, or both. We measured the level of Cpn60.2 protein expressed using Western blotting (data not shown) and quantitative labeling (Fig 3). The level of Cpn60.2 produced by pDWcpn.2 was only about a third of that seen for Cpn60.1 and significantly lower than the amount of Cpn60.1 produced from pDWcpn.1 (compare lanes 2 and 3). The cpn.2 operon is the presence of an unusually long spacer region of 159 bases between the cpn.10.2 and cpn60.2 genes (the average distance in chaperonin operons is around 70 bases). This spacer contains a sequence that is very abundant in all rhizobial genomes, but absent from other genomes examined so far. We call this element CRISP, and it is probably a repetitive extragenic palindromic (REP) element of some kind (Gould 2003). We therefore deleted all but 32 bases of this intergenic region in the plasmid pDWcpn.2 to produce the plasmid pDWcpn.2ΔCRISP, and transformed this plasmid into the merodiploid strain RQL1-M2. However, the amount of Cpn60.2 produced by this plasmid was unchanged relative to pDWcpn.2 (data not shown) and, when the strain was plated onto sucrose plus kanamycin and the resulting sucrose-resistant colonies were screened, all of them were still gentamicin resistant, showing that they had arisen from mutation of the sacB gene and not by loss of the cpn60.1 gene.
Fig 3.
Levels of expression of Cpn60 proteins in RQL1-M2. (A) 35S-labelled protein extracts from RQL1-M2 containing the following chaperonin expression plasmids: pGB0 (lane 1), pDWcpn.1 (lane 2), pDWcpn.2 (lane 3), pTMcpn.3 (lane 4). Lane 5 is an extract from A34 cpn60.1::kan/pTMcpn.3comp. Cpn60.1 levels are indicated by the single arrowhead; Cpn60.2 and Cpn60.3 levels are indicated by the double arrowhead. (B) Mean levels of chaperonins shown as a percentage of total proteins between 35–95 kDa
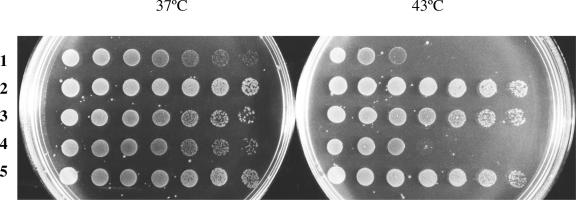
We compared expression of Cpn60.1 and Cpn60.2 in E. coli from the plasmids pHB4 and pHB5, which express the whole cpn.1 and cpn.2 operons respectively under the control of the lac promoter. The results showed that, even when induced by isopropyl β-D-1-thiogalactopyranoside (IPTG), Cpn60.2 was less well expressed than Cpn60.1 under these conditions, although expression was higher than that of the endogenous GroEL (data not shown). We compared strains of E. coli containing pHB4 or pHB5 for their ability to complement for mutation or loss of the endogenous groEL gene. Cpn60.1, but not Cpn60.2, was able to rescue growth of SF103 (which carries a groELts mutation) at the nonpermissive temperature (Fig 4). We thus conclude that, in both R. leguminosarum and E. coli, the Cpn60.2 protein is poorly expressed and, for this reason at least, unable to replace the function of the major endogenous chaperonin.
Fig 4.
Complementation analysis of cpn60.1, cpn60.2, and cpn60.3 in Escherichia coli. Spots are of 10-fold dilutions of log phase cultures of SF103. Plates were incubated at 37°C or 43°C. All plates contained 0.1 mM IPTG. Strains contain the following plasmids: (1) pSU18 (negative control); (2) pSUBH (expresses E. coli GroES and GroEL); (3) pHB4 (expresses Cpn10.1 and Cpn60.1), (4) pHB5 (expresses Cpn10.2 and Cpn60.2), (5) pHB6 (expresses Cpn10.3 and Cpn60.3)
Attempted overexpression of and complementation with Cpn60.3
We next investigated whether overexpression of Cpn60.3 could compensate for loss of expression of Cpn60.1, using the same approach. The plasmid pTMcpn.3, which contains the cpn.3 operon under the control of the cpn.1 promoter, was introduced into the merodiploid strain RQL1-M2, and colonies were selected on sucrose and kanamycin. This experiment was repeated 3 times. On 2 occasions, all of the colonies that grew on kanamycin and sucrose were still gentamicin resistant, showing they were likely to have arisen by mutations in the sacB gene and not by homologous recombination and loss of the wild-type cpn.1 operon. On a third occasion, however, we obtained several colonies that were gentamicin-sensitive. Four of these colonies were checked using PCR with oligonucleotides internal to the cpn60.1 gene, and this showed that, in each of these colonies, the wild-type cpn60.1 gene had been lost (Fig 2). This was confirmed by analysis of the protein profiles of the different strains, which showed loss of Cpn60.1 expression in one of the putative cpn60.1 knockout strain (Fig 3). We observed that the level of expression of Cpn60.3 in this strain was significantly higher than in the merozygote from which it was derived (Fig 3, compare lanes 4 and 5). Because we had only obtained successful complementation of cpn60.1 in 1 out of 3 experiments, this raised the possibility that a mutation had arisen in the pTMcpn.3 plasmid that was increasing Cpn60.3 expression, which in turn was improving its ability to replace Cpn60.1. To test this hypothesis, we isolated plasmid DNA from the strain in which plasmid-expressed Cpn60.3 was able to complement Cpn60.1 and reintroduced it into the original merozygote strain RQL1-M1. The complementing plasmid was named pTMcpn.3comp. Approximately 50% of the sucrose-resistant, kanamycin-resistant colonies derived from this strain were now gentamicin sensitive, compared to 0% of the original strain, showing that the ability to complement for Cpn60.1 segregated with the plasmid pTMcpn.3comp. We determined that the amount of Cpn60.3 produced by pTMcpn.3comp was about 2.5 times greater than that produced by pTMcpn.3, equivalent to nearly 4 times the endogenous level of Cpn60.1 (data not shown). We thus conclude that a mutation in the plasmid has increased the level of Cpn60.3 protein and that this is needed for effective complementation to occur. We resequenced the entire cpn.3 operon and cpn.1 promoter in pTMcpn.3comp, but no mutations were found. It is thus likely that the mutation has arisen elsewhere in the plasmid and alters its copy number.
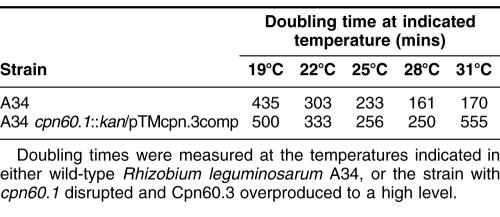
How effective is Cpn60.3 at replacing the function of Cpn60.1 when expressed at this higher level? To investigate this, we compared the doubling time during log phase of A34 and A34 cpn60.1::kan/pTMcpn3.comp over a range of temperatures from 19°C to 31°C (Table 3). As can be seen, the strain overexpressing Cpn60.3 and lacking Cpn60.1 grew less well at all temperatures, and growth was particularly poor at high temperatures, with cells at this temperature failing to reach a final optical density at 600 nm (OD600) of 0.1 under our growth conditions. Overexpression of Cpn60.3 from this plasmid in a wild-type background had no effect on growth (data not shown). It is thus the case that, even when overexpressed, Cpn60.3 cannot fully replace Cpn60.1 in vivo.
Table 3.
Ability of Cpn60.3 to replace Cpn60.1
We also tested to see whether or not overexpression of Cpn60.3, together with its cognate cochaperonin Cpn10.3, could replace E. coli GroEL. The results of this experiment, shown in Figure 4, confirm that Cpn60.3 can replace GroEL even at 43°C, showing the temperature-sensitive phenotype of A34 cpn60.1::kan/pTMcpn.3comp is not due to inherent lack of activity of Cpn60.3 at higher temperatures.
Can the different Cpn60 proteins form mixed oligomers?
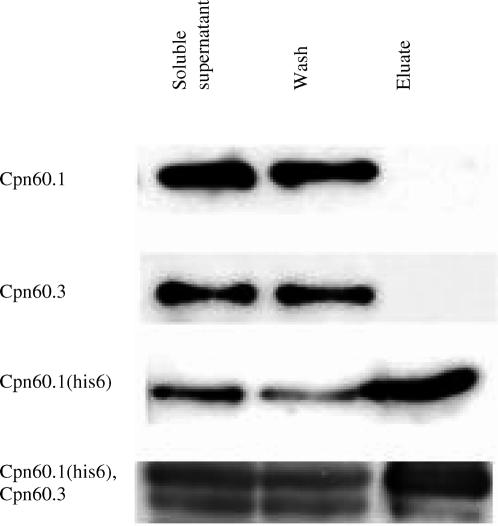
We wished to determine whether or not different Cpn60 proteins assemble into the same complex, or whether they form unique, homo-oligomeric complexes when coexpressed. To do this, we first constructed a version of the cpn.1 operon that included codons for a histidine tail on the Cpn60.1 protein, as described in the Materials and Methods section. The tagged Cpn60.1 still was able to functionally replace GroEL in E. coli, showing that the presence of the tag has no significant effect on the in vivo function of Cpn60.1 (data not shown). The tagged protein was strongly retained on a Ni-NTA resin, whereas neither Cpn60.1 nor Cpn60.3 were retained (Fig 5). We cotransformed MGM100 with the plasmid expressing the his-tagged Cpn60.1 and pHB6, which overexpresses Cpn60.3. When the extract from this strain (grown on glucose to eliminate expression of the endogenous GroEL) was passed through Ni-NTA resin and bound proteins eluted with imidazole, a small amount of Cpn60.3 was detected in addition to the strong Cpn60.1 band, but the proportion of Cpn60.3 to Cpn60.1 was considerably lower than in the initial protein extract (Fig 5). We conclude from this that these two proteins can form mixed complexes, but that they preferentially form homo-oligomers. Because of the poor expression of Cpn60.2 in E. coli, we could not test for the formation of mixed rings with Cpn60.1.
Fig 5.
Formation of mixed oligomers between Cpn60.1 and Cpn60.3 in Escherichia coli. Crude extracts from strains expressing either Cpn60.1 alone, Cpn60.3 alone, Cpn60.1(his)6, or Cpn60.1(his)6 and Cpn60.3, were passed through a Ni-NTA column and subsequently eluted with imidazole. Protein in the soluble supernatant, the initial wash through the column, and the eluted fraction, were detected using an anti-GroEL monoclonal antibody (4-3F) that cross-reacts equally well with Cpn60.1 and Cpn60.3
DISCUSSION
It is not clear why some bacteria have multiple genes encoding chaperonin homologues, but several different hypotheses can be proposed. The different homologues may have different pools of substrates and may be needed under particular growth conditions. Alternatively, the presence of multiple genes may enable the production of higher levels of chaperonins under stress conditions. Finally, different chaperonins may have evolved to have quite different functions, which are not related to protein folding. We have chosen the organism R. leguminosarum to study the phenomenon of multiple chaperonin genes. The chaperonin genes present in this organism show a high degree of sequence similarity. Our previous studies showed that only the most highly expressed of the 3 genes is essential and that it is regulated differently to the second most highly expressed gene (Rodríguez-Quiñones et al 2005; Gould et al 2006). All 3 chaperonins have been purified and analyzed, and they show distinct biochemical properties and different degrees of ability to act as chaperones in vitro (George et al 2004). The current study extends these findings to show that 1 of the nonessential chaperonins (Cpn60.2) cannot be expressed at high levels and the other (Cpn60.3) cannot completely replace the function of the essential Cpn60.1 protein in vivo even when overexpressed. The data in this paper, combined with our earlier analyses, demonstrate that the multiple chaperonins in R. leguminosarum are not functionally equivalent.
The fact that we were unable to obtain good overexpression of Cpn60.2 in either R. leguminosarum or E. coli, under conditions where the other chaperonins were well expressed, means that the in vivo activity of this protein could not be assessed. In vitro, we have found previously that it was able to chaperone the folding of denatured lactate dehydrogenase and that it had approximately half the adenosine triphosphatase (ATPase) activity of Cpn60.1 and a third that of E. coli GroEL. Thus there were no compelling reasons to suppose that it would not act as a chaperone in vivo. We also found it to be the least stable in vitro of the chaperonin proteins in R. leguminosarum, and it is possible that this leads to more rapid turnover of the protein in vivo.
The Cpn60.3 chaperonin is interesting because it is only expressed normally in vivo when cells are incubated anaerobically, and its expression requires a functional NifA protein (Rodríguez-Quiñones et al 2005). This implies it may have some role in nitrogen fixation, and strains that do not express this protein show a 50% to 60% reduction in nitrogenase activity in root nodules (P.A. Lund and J.A. Downie, unpublished results). However, it is clearly not specialized solely for such function, because it can complement well for loss of the E. coli GroEL protein. In this study, we only were able to get functional replacement of Cpn60.1 in R. leguminosarum when an uncharacterized plasmid mutation led to an increase in the level of expression of Cpn60.3 to a level 4-fold greater than that of Cpn60.1 in a wild-type strain. Even then, Cpn60.3 was not able to fully substitute for Cpn60.1 function, as shown by the temperature sensitivity of the complemented strains. Thus the reason for the presence of multiple chaperonin genes in some strain is clearly not simply to provide an additional means of increasing chaperonin levels at times of stress. Interestingly, Cpn60.3 has an intrinsically low ATPase activity (about 20% that of Cpn60.1). Although it can assemble into a complex with Cpn60.1, it appears to assemble more preferentially into homo-oligomeric complexes. The fact that this protein can function well in E. coli, but not in R. leguminosarum, implies that Cpn60.1 may be required for the folding of 1 or more proteins that do not require GroEL for folding in E. coli, and that Cpn60.3 may be poor at chaperoning these proteins.
Sequence alignments provide some suggestions for future work to explain the differences in properties of these 3 chaperonin proteins. In particular, the lack of the conserved GGM motif at the C-terminal tail of the nonessential chaperonins is intriguing. It has been suggested recently that the alternations in the C-terminal tail can affect the kinetics of folding of some GroEL substrate proteins (Tang et al 2006); whether this also might lead to some substrate selectivity in organisms that express multiple GroEL homologues remains to be tested. Future studies swapping domains between the different homologues should enable us to determine which regions of the protein are critical for effective function in the host organism.
Acknowledgments
The plasmids pDWcpn.2, pDWcpn.1, and pTMcpn.3 were constructed by David Winwood and Tristan Magnay as part of their undergraduate projects. One of us (P.S.G.) was funded by a Biotechnology and Biological Sciences Research Council studentship.
REFERENCES
- Bartolomé B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i.0378-1119(1991)102[0075:CAPOAF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Beringer JE. R factor transfer in Rhizobium. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188.0022-1287(1974)084[0188:RFTIR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Burnett BP, Horwich AL, Low KB. A carboxy-terminal deletion impairs the assembly of GroEL and confers a pleiotropic phenotype in Escherichia coli K-12. J Bacteriol. 1994;176:6980–6985. doi: 10.1128/jb.176.22.6980-6985.1994.0021-9193(1994)176[6980:ACDITA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie JA, Knight CD, Johnston AWB. Identification of genes and gene products involved in nodulation of peas by Rhizobium leguminosarum. Mol Gen Genet. 1985;198:255–262.0026-8925(1985)198[0255:IOGAGP]2.0.CO;2 [Google Scholar]
- Ellis RJ. The general concept of molecular chaperones. Phil Trans Roy Soc B. 1993;339:257–261. doi: 10.1098/rstb.1993.0023.1471-2970(1993)339[0257:TGCOMC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ewalt KL, Hendrick JP, Houry WA, Hartl FU. In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell. 1997;90:491–500. doi: 10.1016/s0092-8674(00)80509-7.0092-8674(1997)090[0491:IVOOPF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Fayet O, Ziegelhoffer T, Georgopoulos C. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol. 1989;171:1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989.0021-9193(1989)171[1379:TGAGHS]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 32) Cladistics. 1989;5:164–166.1096-0031(1989)005[0164:PIPV]2.0.CO;2 [Google Scholar]
- Fischer HM, Babst M, and Kaspar T. et al. 1993 One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is coregulated with symbiotic nitrogen fixation genes. EMBO Jou. 12:2901–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer HM, Schneider K, Babst M, Hennecke H. GroEL chaperonins are required for the formation of a functional nitrogenase in Bradyrhizobium japonicum. Arch Microbiol. 1999;171:279–289.0302-8933(1999)171[0279:GCARFT]2.0.CO;2 [Google Scholar]
- Galibert F, Finan TM, and Long SR. et al. 2001 The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 293:668–672. [DOI] [PubMed] [Google Scholar]
- George R, Kelly SM, and Price NC. et al. 2004 Three GroEL homologues from Rhizobium leguminosarum have distinct in vitro properties. Biochem Biophys Res Commun. 324:822–828. [DOI] [PubMed] [Google Scholar]
- Glatz A, Horvath I, and Varvasovszki V. et al. 1997 Chaperonin genes of the Synechocystis PCC 6803 are differentially regulated under light–dark transition during heat stress. Biochem Biophys Res Commun. 239:291–297. [DOI] [PubMed] [Google Scholar]
- Gonzalez V, Santamaria RI, and Bustos P. et al. 2006 The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc Natl Acad Sci U S A. 103:3834–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould P 2003 Regulation and role of the three chaperonin operons of Rhizobium leguminosarum. PhD thesis, University of Birmingham, UK. [Google Scholar]
- Gould P, Maguire M, Lund PA. Distinct mechanisms regulate expression of the two major groEL homologues in Rhizobium leguminosarum. Arch Microbiol. 2007;187:1–14. doi: 10.1007/s00203-006-0164-y.0302-8933(2007)187[0001:DMREOT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Goyal K, Qamra R, Mande SC. Multiple gene duplication and rapid evolution in the groEL gene: functional implications. J Mol Evol. 2006;63:781–787. doi: 10.1007/s00239-006-0037-7.0022-2844(2006)063[0781:MGDARE]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Houry WA, Frishman D, and Eckerskorn C. et al. 1999 Identification of in vivo substrates of the chaperonin GroEL. Nature. 402:147–154. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Contrasting evolutionary rates in the duplicate chaperonin genes of Mycobacterium tuberculosis and M. leprae. Mol Biol Evol. 1993;10:1343–1359. doi: 10.1093/oxfordjournals.molbev.a040064.0737-4038(1993)010[1343:CERITD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, and Sato S. et al. 2000 Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331–338. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, and Sato S. et al. 2002 Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:225–256. [DOI] [PubMed] [Google Scholar]
- Karlin S, Brocchieri L. Heat shock protein 60 sequence comparisons: duplications lateral transfer and mitochondrial evolution. Proc Natl Acad Sci U S A. 2000;97:11348–11353. doi: 10.1073/pnas.97.21.11348.1091-6490(2000)097[11348:HSPSCD]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunakaran KP, Noguchi Y, and Read TD. et al. 2003 Molecular analysis of the multiple GroEL proteins of Chlamydiae. J Bacteriol. 185:1958–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, and Ishihama Y. et al. 2005 Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 122:209–220. [DOI] [PubMed] [Google Scholar]
- Lee WT, Terlesky KC, Tabita FR. Cloning and characterisation of two groESL operons of Rhodobacter sphaeroides: transcriptional regulation of the heat induced groESL operon. J Bacteriol. 1997;179:487–495. doi: 10.1128/jb.179.2.487-495.1997.0021-9193(1997)179[0487:CACOTG]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehel C, Los D, and Wada H. et al. 1993 A second groEL-like gene organized in a groESL operon is present in the genome of Synechocystis sp PCC 6803. J Biol Chem. 268:1799–1804. [PubMed] [Google Scholar]
- Marketon MM, Gonzalez JE. Identification of two quorum-sensing systems in Sinorhizobium meliloti. J Bacteriol. 2002;184:3466–3475. doi: 10.1128/JB.184.13.3466-3475.2002.0021-9193(2002)184[3466:IOTQSI]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan NF, McAteer S, Masters M. The tail of a chaperonin: the C-terminal region of Escherichia coli GroEL protein. Mol Microbiol. 1994;14:309–321. doi: 10.1111/j.1365-2958.1994.tb01292.x.0950-382X(1994)014[0309:TTOACT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nielsen KL, McLennan N, Masters M, Cowan NJ. A single-ring mitochondrial chaperonin (Hsp60–Hsp10) can substitute for GroEL–GroES in vivo. J Bacteriol. 1999;181:5871–5875. doi: 10.1128/jb.181.18.5871-5875.1999.0021-9193(1999)181[5871:ASMCHC]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9.0378-1119(1983)026[0101:COIMVU]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ogawa J, Long SR. The Rhizobium meliloti groELc locus is required for regulation of early nod genes by the transcription activator NodD. Genes Dev. 1995;15:714–729. doi: 10.1101/gad.9.6.714.0890-9369(1995)015[0714:TRMGLI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Comp Apps Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357.1460-2059(1996)012[0357:TAATDP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ranson NA, White HE, Saibil HR. Chaperonins. Biochem Jou. 1998;333:233–242. doi: 10.1042/bj3330233.0264-6021(1998)333[0233:C]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Quiñones F, Maguire M, and Wallington EJ. et al. 2005 Two of the three groEL homologues in Rhizobium leguminosarum are dispensable for normal growth. Arch Microbiol. 183:253–265. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Quiñones F, Fernández-Burriel M, and Banzalvi Z. et al. 1989 Identification of a conserved reiterated DNA region that influences the efficiency of nodulation in strain RS1051 of Rhizobium leguminosarum bv trifolii. Mol Plant-Microbe Interact. 2:75–83. [Google Scholar]
- Saibil H. Molecular chaperones: containers and surfaces for folding stabilizing or unfolding proteins. Curr Opin Struct Biol. 2000;10:251–258. doi: 10.1016/s0959-440x(00)00074-9.0959-440X(2000)010[0251:MCCASF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Servant P, Thompson C, Mazodier P. Use of new Escherichia coli/Streptomyces conjugative vectors to probe the functions of the two groEL-like genes of Streptomyces albus G by gene disruption. Gene. 1993;134:25–32. doi: 10.1016/0378-1119(93)90170-8.0378-1119(1993)134[0025:UONESC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sigler PB, Xu ZH, and Rye HS. et al. 1998 Structure and function in GroEL-mediated protein folding. Ann Rev Biochem. 67:581–608. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology. 1993;1:784–791.0168-1656(1993)001[0784:ABHRMS]2.0.CO;2 [Google Scholar]
- Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M. Structural features of the GroEL–GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125:903–14. doi: 10.1016/j.cell.2006.04.027.0092-8674(2006)125[0903:SFOTGN]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Wallington EJ, Lund PA. Rhizobium leguminosarum contains multiple chaperonin (cpn60) genes. Microbiology. 1994;140:113–122. doi: 10.1099/13500872-140-1-113.0026-2617(1994)140[0113:RLCMCC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Yeh KC, Peck MC, Long SR. Luteolin and GroESL modulate in vitro activity of NodD. J Bacteriol. 2002;184:525–530. doi: 10.1128/JB.184.2.525-530.2002.0021-9193(2002)184[0525:LAGMIV]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JP, Crossman LC, and Johnston AW. et al. 2006 The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]