Abstract
Efficient enrichment of staphylococcal cells displaying specific heterologous affinity ligands on their cell surfaces was demonstrated by using fluorescence-activated cell sorting. Using bacterial surface display of peptide or protein libraries for the purpose of combinatorial protein engineering has previously been investigated by using gram-negative bacteria. Here, the potential for using a gram-positive bacterium was evaluated by employing the well-established surface expression system for Staphylococcus carnosus. Staphylococcus aureus protein A domains with binding specificity to immunoglobulin G or engineered specificity for the G protein of human respiratory syncytial virus were expressed as surface display on S. carnosus cells. The surface accessibility and retained binding specificity of expressed proteins were demonstrated in whole-cell enzyme and flow cytometry assays. Also, affibody-expressing target cells could be sorted essentially quantitatively from a moderate excess of background cells in a single step by using a high-stringency sorting mode. Furthermore, in a simulated library selection experiment, a more-than-25,000-fold enrichment of target cells could be achieved through only two rounds of cell sorting and regrowth. The results obtained indicate that staphylococcal surface display of affibody libraries combined with fluoresence-activated cell sorting might indeed constitute an attractive alternative to existing technology platforms for affinity-based selections.
Recent advances within the field of combinatorial protein engineering have led to the development of several complementary technologies for the selection of novel protein variants from large libraries. So far, phage display has been the preferred format for directed evolution efforts (39), but more recently, other techniques, such as ribosomal display (29), covalent display, and different formats of cell display, have become attractive alternatives (46). Cell surface display combined with fluorescence-activated cell sorting (FACS) constitutes a powerful strategy for isolation of novel ligands with improved affinity, stability, or enzymatic activity (4, 28, 37). The high throughput and quantitative multiparameter population analysis of modern flow cytometers makes FACS ideal for protein engineering applications (7, 46). However, FACS sorting would only be applicable with cell display systems, since the phage particles are too small to be sorted with the present state-of-the-art flow cytometers (6, 43).
Traditionally, engineering of antibody fragments has been the dominating strategy for generating novel proteins with specific ligand-binding properties (17). More recently, other protein scaffolds have also been investigated as sources for novel affinity ligands (26, 38). One such novel class of affinity proteins, called affibody ligands, i.e., engineered Staphylococcus aureus protein A (SpA) domains, has recently been described (23). Combinatorial libraries were created through simultaneous randomization of 13 amino acid residues. Through the genetic fusion of these libraries to the coat protein III of filamentous phage M13, phage libraries adapted for the selection of novel affinity variants were created. This strategy has been successfully used to select affibody ligands to diverse targets, including Taq DNA polymerase, human recombinant factor VIII, a human apolipoprotein variant, and the G protein of human respiratory syncytial virus (RSV) (15, 23, 25). The same strategy has also been employed to select affibody ligands to immunoglobulins (Igs) for which SpA has no inherent affinity, e.g., human IgA and IgE (12, 30). So far, phage display has been used for selection of affibody ligands, but a number of bacterial expression systems would also be available (3, 18, 33). Regarding bacterial display for protein engineering purposes, combinatorial libraries have so far been displayed only on gram-negative bacteria but inherent morphological properties of gram-positive bacteria would make them an attractive alternative in this context (40). These properties include the observed high viability and robustness of staphylococcal cells in high-speed flow cytometry cell sorting (40, 42) and the convenience of C-terminal anchoring of proteins on the cell surface that is characteristic of gram-positive bacteria (33), which allows insertion of extended sequences without interference with the translocation machinery.
Two previously described systems for cell surface display on the nonpathogenic food-grade bacteria Staphylococcus carnosus (34) and Staphylococcus xylosus (16) have been extensively investigated in various biotechnological applications (43), most frequently for development of live bacterial vaccine delivery vehicles through the display of heterologous immunogens at the cell surface (6, 40). Furthermore, single-chain Fv antibody fragments, as well as IgE- and IgA-specific affibody ligands, have been displayed, creating potential whole-cell diagnostic devices (12, 13). In addition, staphylococci with increased metal-binding capacity have been created through display of polyhistidyl peptides (35), and novel metal-binding proteins have been generated through combinatorial protein engineering approaches (19, 44).
An improved S. carnosus display vector exhibiting enhanced growth characteristics and DNA stability was recently developed (45) for intended use in future display of peptide and protein libraries and subsequent affinity-based selections of fluorescence-labeled target cells by flow cytometry. The observed high viability and robustness of the staphylococcal cells in the flow cytometer might be of considerable importance for this type of application (45). Here, we report the use of a model system employing surface-displayed engineered SpA domains to test the functionality of this S. carnosus display system for the specific enrichment of target cells from a large excess of background cells by flow cytometry. The accessibility and binding specificity of affibody ligands exposed on the surface were analyzed in whole-cell enzyme assays, and target cells were enriched by using single or multiple rounds of sorting and reamplification by growth. Implications for the display of whole affibody libraries and subsequent affinity-based selections of novel binding proteins will be discussed.
MATERIALS AND METHODS
Plasmid constructions.
The Escherichia coli strain RRIΔM15 (31) was used as the host strain during plasmid constructions. Gene fragments corresponding to an engineered IgG-binding SpA domain, Zwt (22), or to an SpA-based affibody binding to the G protein of human respiratory syncytial virus (RSV), ZRSV1 (15), were amplified by PCR. The 5′ PCR primer SAPA27 introduced a BamHI restriction enzyme site in the upstream region of the affibody sequence. Similarly, the 3′ primer SAPA28 added a SalI restriction enzyme site in the downstream region. The generated PCR fragments were ligated into the surface display vector pSCXm, which had previously been digested with BamHI and SalI (45), thus generating the plasmids pSCXZwt and pSCXZRSV1, respectively. Positive clones were identified by PCR screening and verified by DNA sequencing performed on the Mega-BACE 1000 DNA sequencing system (Amersham Biosciences, Uppsala, Sweden) by using MegaBACE terminator chemistry (Amersham Biosciences) in a cycle-sequencing protocol.
Preparation of biotinylated human IgG.
Human IgG was biotinylated by using the EZ-Link Sulfo-NHS-LC-Biotin labeling kit (Pierce, Rockford, Ill.) according to the supplier's recommendations. The unreacted free biotin was removed by Microcon centrifugal filter devices (Millipore Corp., Bedford, Mass.). The final concentration of IgG-biotin was determined by a standard Bradford analysis using bovine serum albumin as a standard.
Evaluation of surface display efficiency and IgG binding.
Overnight cultures of recombinant and wild-type staphylococci were diluted 1:200 in growth medium (containing chloramphenicol when appropriate) and grown at 37°C to an A578 of 1. The cells were harvested and washed twice with phosphate-buffered saline (PBS) supplemented with 0.05% Tween 20 (PBST) (pH 7.5) before being resuspended in PBST to an A578 of 1. One-milliliter aliquots from these suspensions were incubated at room temperature with either biotinylated human serum albumin (HSA) (1.75 μl of biotinylated HSA [2.39 mg/ml] in 1 ml of PBST) or biotinylated IgG (4 μl of biotinylated IgG [11.5 μM] in 1 ml of PBST) for surface expression and binding analysis. The cells were washed twice in PBST prior to resuspension in 1 ml of PBST containing 0.5 units of streptavidin-alkaline phosphatase conjugate (Boehringer, Mannheim, Germany) and then incubated for another 30 min at room temperature. After two additional washings, the cells were diluted 1:10 in substrate buffer (1 M diethanolamine-HCl [pH 9.8], 0.5 mM MgCl2), and five 100-μl aliquots of each cell type were loaded into a microtiter plate before addition of 100 μl of the substrate solution (p-nitrophenylphosphate; Sigma, St. Louis, Mo.). The change in A405 nm was measured for 20 min in an enzyme-linked immunosorbent assay plate reader (SUNRISE, Tecan, Grödingen, Austria).
FACS.
Overnight cultures grown at 37°C were harvested and washed twice in PBS before being resuspended at an optical density at 600 nm of 1 in PBS. Positive (pSCXZwt) and negative (pSCXZRSV1) cell populations were mixed at the desired ratios, and 100-μl aliquots were withdrawn, pelleted, and resuspended in a solution of 14 nM biotin-IgG in PBS. After incubation for 30 min on ice, the cells were washed once with ice-cold PBS before being resuspended in Alexa-Fluor488-streptavidin (Molecular Probes, Eugene, Oreg.) conjugate diluted 1,600× in PBS. Following another incubation for 30 min on ice, the cells were washed and resuspended in PBS at approximately 107 cells per ml and either counted or sorted on the basis of fluorescence intensity with a FACS Vantage SE flow cytometer (Becton Dickinson, Sunnyvale, Calif.). The cytometer was set to enhanced normal-R mode for the single-step enrichments and to enrich for the library simulations. The cells were sorted directly into tryptic soy broth medium and either plated onto chloramphenicol plates or reamplified by incubation overnight and subjected to another round of cell sorting and analysis. After incubation at 37°C for 48 h, the sorted cells were screened by PCR by using primer pairs specific for the genes encoding the Zwt and ZRSV1 variants. The fraction of sorted cells expressing the desired variant could thus be determined.
RESULTS
Background.
In this study, we have explored the possibility of using staphylococcal surface display and flow cytometry as an alternative to filamentous phages for affinity-based selection of engineered SpA domains, the so-called affibody ligands. A previous study demonstrated that affibody ligands were efficiently displayed with retained binding specificity on the surfaces of S. carnosus cells (12). Recently, the plasmid vector for surface display on S. carnosus was significantly improved with respect to genetic stability with retained surface expression efficiency (45) in order to be better suited for library selection applications. This increased stability should potentially enable the display of whole affibody ligand libraries at the cell surface. In this study, a model system consisting of the IgG-binding affibody scaffold, Zwt (22, 24), and a previously described affibody, ZRSV1 (15), selected using phage display and specific for the G protein of RSV, was used for simulated library selections.
Display vectors.
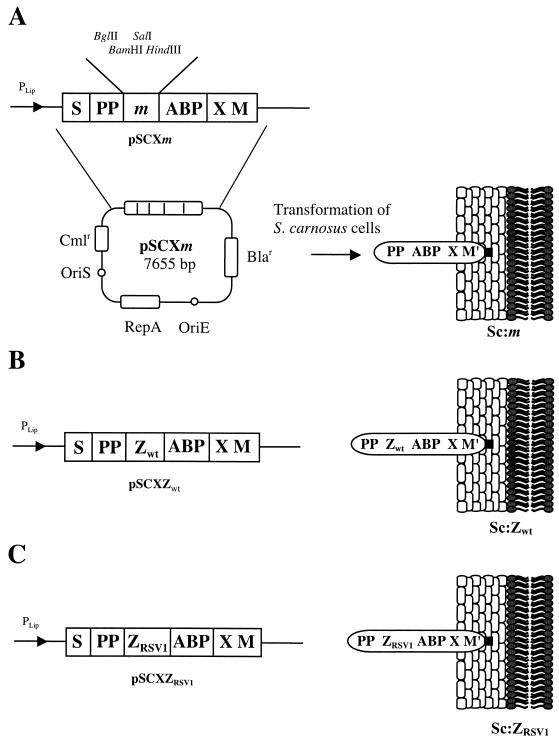
Two novel plasmid vectors were constructed for display of chimeric proteins containing the Zwt or ZRSV1 affibody ligands (15, 24) on the S. carnosus cell surface. In Figure 1, the parental vector, pSCXm (45), as well as the two novel constructs, pSCXZwt and pSCXZRSV1, are schematically depicted together with their encoded gene products as anchored to the staphylococcal cell wall. For simplicity, the recombinant S. carnosus strains were designated Sc:m, Sc:Zwt and Sc:ZRSV1. The pSCXm display vector utilizes the promoter, signal sequence, and propeptide from a Staphyloccocus hyicus lipase for optimized expression and secretion in S. carnosus (20). The lipase propeptide, which is cleaved off in its homologous host, S. hyicus (2), but not in S. carnosus (9), has been extensively studied and proved to be essential for efficient secretion of heterologous gene fusion products in the lipase expression system (8, 32). The vector system also contains the gene fragments X and M from the C-terminal cell wall anchoring part of the SpA gene for covalent anchoring of expressed proteins to the peptidoglycan cell wall (21, 36, 42). In addition, the gene encoding an albumin binding protein (ABP) (27), derived from streptococcal protein G, is also present in the expression vector. The ABP region is expressed as the part of the chimeric surface protein closest to the cell wall-anchoring motifs functioning as a spacer molecule, increasing the accessibility of expressed chimeric proteins at the bacterial surface (41). Also, ABP-containing surface proteins can be extracted from the cell wall by lysostaphin treatment and can subsequently be purified by affinity chromatography on HSA resins (31), utilizing the ABP moiety as an affinity handle. Using this method is a convenient strategy for verifying that the chimeric surface proteins are expressed as full-length proteins and not subjected to degradation. Finally, the ABP tag functions as a reporter molecule in enzyme- and flow cytometry-based whole-cell assays (34, 45) for monitoring expression levels and density of surface-displayed receptors.
FIG. 1.
The expression cassettes of the different expression vectors suitable for surface display on S. carnosus together with a schematic picture of the processed gene fusion products, illustrated as anchored to the cell surface. The names of the constructed expression vectors are given below the expression cassettes, and the abbreviated names of the recombinant staphylococci are shown to the right. When a target gene is introduced into the multiple cloning sites of the vector, the encoded target protein is exposed, as illustrated, anchored to the staphylococcal cell surface. M′ represents the processed and covalently anchored form of the M sequence of SpA. Note that the propeptide (PP) from S. hyicus is not processed in S. carnosus. Abbreviations: S, secretion signal from S. hyicus lipase; Bla, gene encoding β-lactamase; Cml, gene encoding chloramphenicol acetyltransferase; OriE, origin of replication from Escherichia coli; OriS, origin of replication from S. aureus; PLip, S. hyicus promoter region designed for S. hyicus lipase production in S. carnosus; X, charged repetitive region postulated to interact with the peptidoglycan cell wall.
Surface expression and IgG-binding analysis.
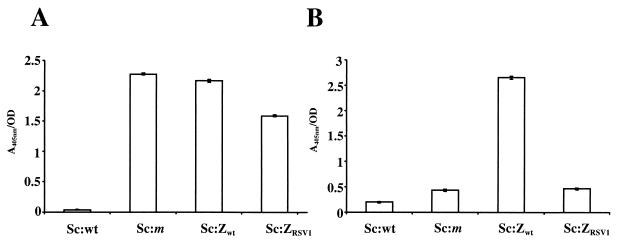
Before fluorescence-activated cell sorting could be performed, it was necessary to determine whether the Zwt and ZRSV1 domains were efficiently expressed in a functional form at the cell surface. In order to analyze the expression levels and accessibility of the displayed affibody ligands, the ABP region was used as a reporter molecule in a previously described whole-cell enzyme assay (34). Briefly, recombinant and wild-type staphylococcal cells were grown to early logarithmic phase and incubated with biotinylated HSA, followed by incubation with a streptavidin-alkaline-phosphatase conjugate. The presence of surface proteins containing the ABP domain could then be detected through the addition of a chromogenic substrate. The results, shown in Fig. 2A, indicate that cells harboring the pSCXZwt or pSCXZRSV1 plasmid express ABP-containing surface proteins at a level similar to that of cells containing the parental pSCXm plasmid. In analogy with previous studies, this would mean that there are approximately 10,000 ABP-containing proteins displayed per cell, enabling the use of flow cytometry for sorting for affinity-based selection (1). The wild-type S. carnosus cells, not expressing any albumin-binding domains, were, as expected, negative in this assay (Fig. 2A). The IgG-binding capacity of the recombinant strains was analyzed in a similar assay by replacing the biotinylated HSA with biotinylated IgG in the probing step. The Sc:Zwt cells were positive in the assay, while Sc:wt, Sc:m, and Sc:ZRSV1 cells demonstrated only background reactivity (Fig. 2B), which shows that the Zwt domain may be efficiently expressed in a functional form on the cell surface with retained binding specificity. The results also demonstrated very low levels of unspecific interactions with wild-type S. carnosus cells and recombinant strains expressing non-IgG-binding affibody ligands.
FIG. 2.
Histogram representation of the results of colorimetric assays for determining the accessibility and IgG-binding capacity of affibodies displayed on cell surfaces. (A) Results for detection of the accessibility of receptors displayed on the cell surface and containing regions with albumin-binding capacity. Wild-type and recombinant S. carnosus cells were incubated with biotinylated HSA for binding to successfully exposed receptors containing ABP. Streptavidin-alkaline phosphatase conjugate was added, and absorbance at 405 nm was monitored after addition of substrate. (B) Results for detection of IgG-binding capacity by using biotinylated IgG in an assay similar to that used for panel A. Wild-type cells (Sc:wt) were used together with pSCXm-transformed S. carnosus cells (Sc:m) as controls in all experiments. OD, optical density.
Single-step FACS enrichment of Zwt-expressing cells.
Whether working with the screening of combinatorial libraries or classical protein purification applications, making a calculated choice between yield and purity is necessary. For library screening applications, the goal is to obtain the purest possible product without losing any valuable target cells. Thus, when working with library selections, the maximum attainable single-step enrichment factor becomes an important parameter. In an initial study, we therefore investigated the possibility of quantitative sorting of Sc:Zwt cells from a moderate excess of control cells. Cells expressing a previously selected affibody ligand, ZRSV1, capable of specific recognition of the G protein of RSV served as a relevant nontarget background population. Specifically, overnight cell cultures of Sc:Zwt target cells were mixed with Sc:ZRSV1 control cells at ratios of 1:1, 1:100 and 1:1,000. The cell mixtures were incubated with biotinylated IgG and an Alexa-Fluor488-streptavidin conjugate before being analyzed and sorted on the basis of fluorescence. Since these initial sortings aimed at obtaining a high degree of purity in a single sorting round, a relatively narrow sorting gate was set to minimize the number of nontarget background cells being sorted. The fraction of the total cell population that was sorted corresponded to approximately 50, 1, and 0.1%, respectively, for the 1:1, 1:100, and 1:1,000 mixtures (data not shown). The cells were collected and filtered through a sterile filter and applied to chloramphenicol plates. To verify that the sorted cells were indeed Sc:Zwt, PCR primer pairs specific for either Zwt or ZRSV1 were used for clone-specific PCR amplification and screening purposes directly on staphylococcal colonies. The results, presented in Table 1, clearly indicate that essentially quantitative sorting of target cells may be obtained from a moderate excess of background cells in a single sorting round. For the 1:1 mixture, 100% of the analyzed cells were identified as Sc:Zwt, while the numbers for the 1:100 and 1:1,000 mixtures were 95 and 80%, respectively. This means there was a 571-fold enrichment of Zwt-expressing target cells in a single sorting round for the 1:1,000 mixture. Furthermore, the high viability observed for the sorted staphylococcal cells suggests a possible advantage of using gram-positive bacteria for cell-sorting applications. More than 95% of the analyzed and sorted cells were capable of growth on chloramphenicol plates (Table 1), indicating that the inherent structure of the staphylococcal cell wall, i.e., the thick peptidoglycan layer, indeed seems to improve ability to withstand the elevated shear forces acting on the individual cells in the flow cytometer.
TABLE 1.
Single-step enrichment of Zwt-expressing staphylococci at different Zwt:ZRSV1 ratios
| Zwt:ZRSV1 ratioa | Cell sorting outcome
|
||
|---|---|---|---|
| % Sc:Zwt cellsb | % Viabilityc | Fold enrichment (theoretical)d | |
| 1:1 | 100 | >95 | 1.6 (1.6) |
| 1:100 | 95 | >95 | 84 (87) |
| 1:1,000 | 80 | >95 | 571 (595) |
Based on optical density measurements of overnight cultures.
As determined by PCR screening of sorted cells by using Zwt-specific primer pairs.
Defined as the percentage of sorted cells capable of growth on chloramphenicol agar plates.
Theoretical enrichment factors are based on the actual Zwt:ZRSV ratio calculated from FACS analysis of the initial cell populations.
Sorting of very rare target cells from a large background.
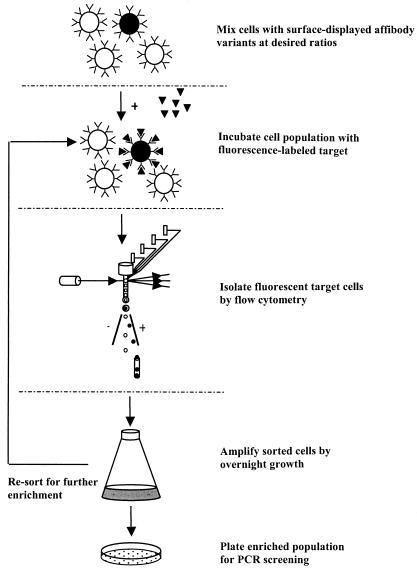
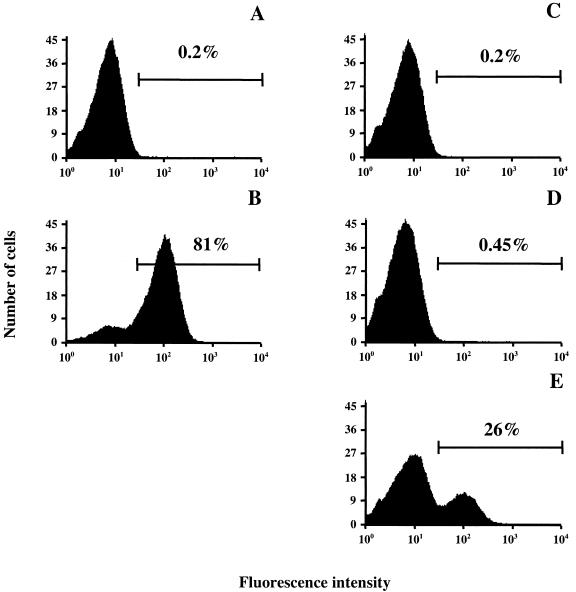
The possibility of also using FACS for the isolation of very rare target cells from a large excess of background cells was evaluated. S. carnosus cells expressing surface displays of Zwt or ZRSV1 affibody ligands were mixed at a ratio of 1:100,000 and labeled with biotinylated IgG and Alexa-Fluor488-streptavidin conjugate to create a simulated library situation. In this experiment, the starting mixture was incubated with the fluorescence-labeled probes, sorted on the basis of fluorescence, and enriched through multiple rounds of cell sorting and amplification by overnight growth (Fig. 3). Since the initial number of positive cells in this experiment was very small, a wide sorting gate was set so as not to lose any valuable target cells. Initially, approximately 0.2% of the control Sc:ZRSV cells fell within the sorting window (Fig. 4C), thus ensuring that most of the target cells were effectively sorted in the first sorting round. One million cells from the input mixture were run through the flow cytometer, and Zwt-expressing target cells that fell within the sorting window were collected and grown overnight in fresh medium and re-sorted. The cell fluorescence distribution after two rounds of sorting and regrowth are shown in Fig. 4C through E, and that of the positive and negative control populations is shown in Fig. 4A and B. The bar in each graph represents the sorting gate setting, i.e., the minimum fluorescence intensity defined as a positive event, with the results for nonstained Sc:ZRSV1 bacterial cells displayed to the left in the histograms and the results for cells exposing Zwt-containing proteins on their surfaces shifted to the right. Approximately 26% of the cells fell within the positive window (Fig. 4E), which corresponds to a more-than-25,000-fold enrichment of target cells in only two rounds of cell sorting. The identities of the sorted cells were further analyzed by PCR screening as before, confirming the identity of selected cells (data not shown). These data demonstrate that specific enrichment of rare target cells can be efficiently achieved by FACS sorting by using the above staphylococcal display system. Importantly, the observed enrichment is not due to a growth advantage of the Zwt-expressing S. carnosus cells, since repeated regrowth of the starting mixture without in-between cell sorting did not result in any significant bias when analyzed by PCR screening using Zwt-specific primers (data not shown).
FIG. 3.
General procedure for the enrichment of fluorescence-labeled, Zwt-expressing target cells by flow cytometry. Staphylococcal cells with surface display of affibodies are mixed at the desired ratios and incubated with fluorescence-labeled probes. Target cells are selected on the basis of fluorescence and enriched through multiple rounds of cell sorting and amplification by growth. The cells are plated, and individual clones are screened by PCR for identification.
FIG. 4.
Histograms of results from cell-sorting experiments. Wild-type and recombinant S. carnosus cells were probed with biotinylated IgG and stained with an Alexa-Fluor488-streptavidin conjugate. The bar in each graph represents the sorting gate setting, i.e., the minimum fluorescence intensity defined as a positive event. (A) Sc:ZRSV1 sample used as a negative control; (B) Sc:Zwt sample used as a positive control; (C) a 1:100,000 mixture of Sc:Zwt to Sc:ZRSV1 cells analyzed prior to the first cell-sorting round; (D) a 1:100,000 mixture of Sc:Zwt to Sc:ZRSV1 cells recovered from the first cell-sorting round; (E) a 1:100,000 mixture of Sc:Zwt to Sc:ZRSV1 cells recovered from the second cell-sorting round. The histograms show results for nonstained Sc:ZRSV1 bacterial cells to the left and results for cells exposing Zwt-containing proteins on their surfaces shifted to the right. Fluorescence intensity is shown on the x axis, and the number of cells is shown on the y axis.
DISCUSSION
In this study, we have demonstrated the possibility of using staphylococcal surface display and flow cytometry as a potential alternative to filamentous phages for the specific and quantitative isolation of novel affinity pairs from surface-displayed affibody libraries. Recombinant S. carnosus strains with engineered SpA domains specific for IgG or the G protein of human RSV exposed on the surface were created. The chimeric surface proteins were shown to be expressed in an accessible form with retained binding specificity at the staphylococcal surface by an enzyme assay based on the ABP domain. Also, affibody-expressing target cells could be sorted essentially quantitatively from a moderate excess of nontarget staphylococci in a single sorting round by using a high-stringency sorting mode. Furthermore, by using a simulated library situation, a more-than-25,000-fold enrichment of target cells was achieved through only two successive rounds of cell sorting and regrowth. This indicates that real affinity-based selections from surface-displayed affibody libraries would be feasible. In addition, the staphylococcal cells seem to be highly suited for flow cytometry applications, since they also demonstrate a high degree of viability after high-speed sorting. More than 95% of the sorted cells grow well on chloramphenicol plates after sorting. This capability is probably due to their high resistance to shear forces related to the thick peptidoglycan cell wall.
One important advantage of cell-based display systems compared to monovalent phage display is the apparent lack of avidity effects. While phage display systems based on pIII fusions rely on single molecular binding events, a so-called monovalent display, the staphylococcal display system described here typically expresses around 10,000 copies of the displayed protein per cell. This means that random variation in expression levels or stability should not interfere with library selections. Furthermore, the presence of the ABP reporter molecule in the surface chimeras would make it possible to normalize ligand binding by taking into account the number of surface proteins per cell, avoiding artifacts related to expression biases when working with small differences in affinity. This would be of utmost importance if working with real library selections involving fine affinity discrimination of library members.
One obvious challenge when using cell-based systems, like bacterial or yeast display (46), rather than in vitro-based ribosomal display or oil-water emulsion techniques (14) is the limited library size that can be obtained due to restrictions in the transformation step. For the staphylococcal display system described here, the routinely obtained transformation frequency using present transformation methods would be around 105 to 106 transformants per μg of DNA (10), comparable to the numbers reported for yeast and mammalian cell-based display systems (5). Nevertheless, one important application that obviates the need for very large libraries is the use of the S. carnosus system in affinity maturation strategies, where small biased libraries can be constructed on the basis of previously selected binders, thus eliminating the need for very large libraries (11, 25). In the future, it would therefore be interesting to test the staphylococcal display system described here for the affinity maturation of previously selected affibody ligands.
Acknowledgments
We are grateful to George Georgiou for letting us conduct initial model experiments in his lab.
This work was financially supported by Cell Factory for Functional Genomics, a program funded by the Swedish Foundation for Strategic Research. P.S. was supported by a postdoctoral fellowship from the Wenner-Gren Foundation.
REFERENCES
- 1.Andréoni, C., L. Goetsch, C. Libon, P. Samuelson, T. N. Nguyen, A. Robert, M. Uhlén, H. Binz, and S. Ståhl. 1997. Flow cytometric quantification of surface-displayed recombinant receptors on staphylococci. BioTechniques 23:696-702, 704. [PubMed] [Google Scholar]
- 2.Ayora, S., P. E. Lindgren, and F. Götz. 1994. Biochemical properties of a novel metalloprotease from Staphylococcus hyicus subsp. hyicus involved in extracellular lipase processing. J. Bacteriol. 176:3218-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benhar, I. 2001. Biotechnological applications of phage and cell display. Biotechnol. Adv. 19:1-33. [DOI] [PubMed] [Google Scholar]
- 4.Boder, E. T., K. S. Midelfort, and K. D. Wittrup. 2000. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl. Acad. Sci. USA 97:10701-10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boder, E. T., and K. D. Wittrup. 2000. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 328:430-444. [DOI] [PubMed] [Google Scholar]
- 6.Cano, F., H. Plotnicky-Gilquin, T. N. Nguyen, S. Liljeqvist, P. Samuelson, J. Bonnefoy, S. Ståhl, and A. Robert. 2000. Partial protection to respiratory syncytial virus (RSV) elicited in mice by intranasal immunization using live staphylococci with surface-displayed RSV-peptides. Vaccine 18:2743-2752. [DOI] [PubMed] [Google Scholar]
- 7.Daugherty, P. S., B. L. Iverson, and G. Georgiou. 2000. Flow cytometric screening of cell-based libraries. J. Immunol. Methods 243:211-227. [DOI] [PubMed] [Google Scholar]
- 8.Demleitner, G., and F. Götz. 1994. Evidence for importance of the Staphylococcus hyicus lipase pro-peptide in lipase secretion, stability and activity. FEMS Microbiol. Lett. 121:189-197. [DOI] [PubMed] [Google Scholar]
- 9.Götz, F. 1990. Staphylococcus carnosus: a new host organism for gene cloning and protein production. Soc. Appl. Bacteriol. Symp. Ser. 19:49S-53S. [DOI] [PubMed]
- 10.Götz, F., and B. Schumacher. 1987. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40:285-288. [Google Scholar]
- 11.Gunneriusson, E., K. Nord, M. Uhlén, and P. Å. Nygren. 1999. Affinity maturation of a TaqDNA polymerase specific affibody by helix shuffling. Protein Eng. 12:873-878. [DOI] [PubMed] [Google Scholar]
- 12.Gunneriusson, E., P. Samuelson, J. Ringdahl, H. Grönlund, P.-Å. Nygren, and S. Ståhl. 1999. Staphylococcal surface display of immunoglobulin A (IgA)- and IgE-specific in vitro-selected binding proteins (affibodies) based on Staphylococcus aureus protein A. Appl. Environ. Microbiol. 65:4134-4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunneriusson, E., P. Samuelson, M. Uhlén, P.-Å. Nygren, and S. Ståhl. 1996. Surface display of a functional single-chain Fv antibody on staphylococci. J. Bacteriol. 178:1341-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanes, J., and A. Plückthun. 1997. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 94:4937-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansson, M., J. Ringdahl, A. Robert, U. Power, L. Goetsch, T. N. Nguyen, M. Uhlén, S. Ståhl, and P. Å. Nygren. 1999. An in vitro selected binding protein (affibody) shows conformation- dependent recognition of the respiratory syncytial virus (RSV) G protein. Immunotechnology 4:237-252. [DOI] [PubMed] [Google Scholar]
- 16.Hansson, M., S. Ståhl, T. N. Nguyen, T. Bachi, A. Robert, H. Binz, A. Sjölander, and M. Uhlén. 1992. Expression of recombinant proteins on the surface of the coagulase-negative bacterium Staphylococcus xylosus. J. Bacteriol. 174:4239-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt, L. J., C. Enever, R. M. de Wildt, and I. M. Tomlinson. 2000. The use of recombinant antibodies in proteomics. Curr. Opin. Biotechnol. 11:445-449. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. Y., J. H. Choi, and Z. Xu. 2003. Microbial cell-surface display. Trends Biotechnol. 21:45-52. [DOI] [PubMed] [Google Scholar]
- 19.Lehtiö, J., T. T. Teeri, and P. Nygren. 2000. Alpha-amylase inhibitors selected from a combinatorial library of a cellulose binding domain scaffold. Proteins 41:316-322. [DOI] [PubMed] [Google Scholar]
- 20.Liebl, W., and F. Götz. 1986. Studies on lipase directed export of Escherichia coli beta-lactamase in Staphylococcus carnosus. Mol. Gen. Genet. 204:166-173. [DOI] [PubMed] [Google Scholar]
- 21.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson, B., T. Moks, B. Jansson, L. Abrahmsén, A. Elmblad, E. Holmgren, C. Henrichson, T. A. Jones, and M. Uhlén. 1987. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1:107-113. [DOI] [PubMed] [Google Scholar]
- 23.Nord, K., E. Gunneriusson, J. Ringdahl, S. Ståhl, M. Uhlén, and P. Å. Nygren. 1997. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat. Biotechnol. 15:772-777. [DOI] [PubMed] [Google Scholar]
- 24.Nord, K., J. Nilsson, B. Nilsson, M. Uhlén, and P.-Å. Nygren. 1995. A combinatorial library of an alpha-helical bacterial receptor domain. Protein Eng. 8:601-608. [DOI] [PubMed] [Google Scholar]
- 25.Nord, K., O. Nord, M. Uhlén, B. Kelley, C. Ljungqvist, and P.-Å. Nygren. 2001. Recombinant human factor VIII-specific affinity ligands selected from phage-displayed combinatorial libraries of protein A. Eur. J. Biochem. 268:4269-4277. [DOI] [PubMed] [Google Scholar]
- 26.Nygren, P.-Å., and M. Uhlén. 1997. Scaffolds for engineering novel binding sites in proteins. Curr. Opin. Struct. Biol. 7:463-469. [DOI] [PubMed] [Google Scholar]
- 27.Nygren, P.-Å., M. Eliasson, L. Abrahmsén, M. Uhlén, and E. Palmcrantz. 1988. Analysis and use of the serum albumin binding domains of streptococcal protein G. J. Mol. Recognit. 1:69-74. [DOI] [PubMed] [Google Scholar]
- 28.Olsen, M., B. Iverson, and G. Georgiou. 2000. High-throughput screening of enzyme libraries. Curr. Opin. Biotechnol. 11:331-337. [DOI] [PubMed] [Google Scholar]
- 29.Roberts, R. W. 1999. Totally in vitro protein selection using mRNA-protein fusions and ribosome display. Curr. Opin. Chem. Biol. 3:268-273. [DOI] [PubMed] [Google Scholar]
- 30.Rönnmark, J., H. Grönlund, M. Uhlén, and P.-Å. Nygren. 2002. Human immunoglobulin A (IgA)-specific ligands from combinatorial engineering of protein A. Eur. J. Biochem. 269:2647-2655. [DOI] [PubMed] [Google Scholar]
- 31.Rüther, U. 1982. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 10:5765-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samuelson, P., F. Cano, A. Robert, and S. Ståhl. 1999. Engineering of a Staphylococcus carnosus surface display system by substitution or deletion of a Staphylococcus hyicus lipase propeptide. FEMS Microbiol. Lett. 179:131-139. [DOI] [PubMed] [Google Scholar]
- 33.Samuelson, P., E. Gunneriusson, P.-Å. Nygren, and S. Ståhl. 2002. Display of proteins on bacteria. J. Biotechnol. 96:129-154. [DOI] [PubMed] [Google Scholar]
- 34.Samuelson, P., M. Hansson, N. Ahlborg, C. Andréoni, F. Götz, T. Bachi, T. N. Nguyen, H. Binz, M. Uhlén, and S. Ståhl. 1995. Cell surface display of recombinant proteins on Staphylococcus carnosus. J. Bacteriol. 177:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samuelson, P., H. Wernérus, M. Svedberg, and S. Ståhl. 2000. Staphylococcal surface display of metal-binding polyhistidyl peptides. Appl. Environ. Microbiol. 66:1243-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 37.Shusta, E. V., P. D. Holler, M. C. Kieke, D. M. Kranz, and K. D. Wittrup. 2000. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat. Biotechnol. 18:754-759. [DOI] [PubMed] [Google Scholar]
- 38.Skerra, A. 2000. Engineered protein scaffolds for molecular recognition. J. Mol. Recognit. 13:167-187. [DOI] [PubMed] [Google Scholar]
- 39.Smith, G. P., and V. A. Petrenko. 1997. Phage display. Chem. Rev. 97:391-410. [DOI] [PubMed] [Google Scholar]
- 40.Ståhl, S., A. Robert, E. Gunneriusson, H. Wernérus, F. Cano, S. Liljeqvist, M. Hansson, T. N. Nguyen, and P. Samuelson. 2000. Staphylococcal surface display and its applications. Int. J. Med. Microbiol. 290:571-577. [DOI] [PubMed] [Google Scholar]
- 41.Ståhl, S., P. Samuelson, M. Hansson, C. Andréoni, L. Goetsch, C. Libon, S. Liljeqvist, E. Gunneriusson, H. Binz, N. Nguyen, and M. Uhlén. 1997. Development of non-pathogenic staphylococci as vaccine delivery vehicles, p. 62-81. In G. Pozzi and J. Wells (ed.), Gram-positive bacteria: vaccine vehicles for mucosal immunization. Landes Bioscience, Georgetown, Tex.
- 42.Ton-That, H., S. K. Mazmanian, K. F. Faull, and O. Schneewind. 2000. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J. Biol. Chem. 275:9876-9881. [DOI] [PubMed] [Google Scholar]
- 43.Wernérus, H., J. Lehtiö, P. Samuelson, and S. Ståhl. 2002. Engineering of staphylococcal surfaces for biotechnological applications. J. Biotechnol. 96:67-78. [DOI] [PubMed] [Google Scholar]
- 44.Wernérus, H., J. Lehtiö, T. Teeri, P.-Å. Nygren, and S. Ståhl. 2001. Generation of metal-binding staphylococci through surface display of combinatorially engineered cellulose-binding domains. Appl. Environ. Microbiol. 67:4678-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wernérus, H., and S. Ståhl. 2002. Vector engineering to improve a staphylococcal surface display system. FEMS Microbiol. Lett. 212:47-54. [DOI] [PubMed] [Google Scholar]
- 46.Wittrup, K. D. 2001. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 12:395-399. [DOI] [PubMed] [Google Scholar]