Abstract
The detailed mechanism of how the ribosome decodes protein sequence information with an abnormally high accuracy, after 40 years of study, remains elusive. A critical element in selecting correct transfer RNA (tRNA) transferring correct amino acid is “induced fit” between the ribosome and tRNA. By using single-molecule methods, the induced fit mechanism is shown to position favorably the correct tRNA after initial codon recognition. We provide evidence that this difference in positioning and thermal fluctuations constitutes the primary mechanism for the initial selection of tRNA. This work demonstrates thermal fluctuations playing a critical role in the substrate selection by an enzyme.
Keywords: ribosome dynamics, ribosome selection, single-molecule FRET
Translation, protein synthesis by the ribosome, is a vital cellular process involving the two-subunit ribosomal particle, multiple RNAs, and protein cofactors (1, 2). To synthesize a protein molecule with correct sequence, the ribosome has to select correct transfer RNA (tRNA) as dictated by messenger RNA (mRNA). In the decoding site of the ribosome, the proper matching of the 3-nt codon sequence of the mRNA to the anticodon sequence of tRNA occurs at a rate of 20–30 per second with an error rate of 10−4 (3).
The difference in base-pairing energy between codon and anticodon cannot explain the low error rate of translation (4). In the past decades, considerable progress has been made in understanding the ingenious mechanisms used by the ribosome to achieve this low error rate (2, 4–6). The overall selection process consists of initial selection of the ternary complex [aminoacyl-tRNA (aa-tRNA), GTP, and elongation factor Tu (EF-Tu)], GTP hydrolysis in EF-Tu, and then proofreading (6). The GTP hydrolysis ensures that initial selection is separated from the proofreading by an irreversible reaction. This two-step selection yields overall probability of an error to be the product of the error probabilities of the two selection steps, thus dramatically lowering the overall chance of error (7).
The initial selection of aa-tRNA consists of initial binding of the ternary complex on the ribosome, interaction between tRNA and mRNA (the codon recognition), and stabilization of the ternary complex onto the ribosome. GTPase activation takes place after additional stabilizing contacts are formed between the ribosome and the ternary complex (6, 8). In the initial selection of aa-tRNA, the ternary complex binding induces structural changes in the ribosome (“induced-fit”) (5, 9) in which the 30S subunit of the ribosome changes its structure and forms additional stabilizing contacts with the incoming tRNA after the codon recognition (9). In case of one base-mismatch out of the 3 bp (near cognate), additional binding contacts are weaker, presumably because the mismatched bases form a less compact structure for the ribosome to “wrap” around as it forms additional induced fit contacts. In other words, the tighter binding of cognate tRNA relative to near-cognate tRNA mainly is attributable to shape discrimination at the decoding site of the ribosome (6, 9).
The formation of additional stabilizing contacts between the ternary complex and the 50S subunit of the ribosome (5, 10) requires additional movement beyond the positioning of the ternary complex immediately after the codon recognition. Recent kinetics studies have shown that the rate at which these stabilizing contacts are formed favors cognate ternary complexes over near-cognate ternary complexes (6). Cryo-electron microscopic structures of these stabilizing contacts of the ternary complex with the 50S subunit of the ribosome, particularly with the sarcin–ricin loop (SRL) and the GTPase-associated center (GAC) are shown in supporting information (SI) Fig. 5 (5, 10–12).
Here we present single-molecule fluorescence resonance energy transfer (FRET) studies (13, 14) of the initial selection of aa-tRNA by the ribosome that show how the induced fit plays a dual role in initial selection. In addition to causing more stable binding of the cognate ternary complex, the formation of the additional contacts with the 30S portion of the ribosome positions the ternary complex so that thermal fluctuations are much more likely to form the additional stabilizing contacts that mark the end of initial selection. Thus, large and rare thermal fluctuations are observed to play a critical role in the high substrate selectivity of this enzymatic reaction.
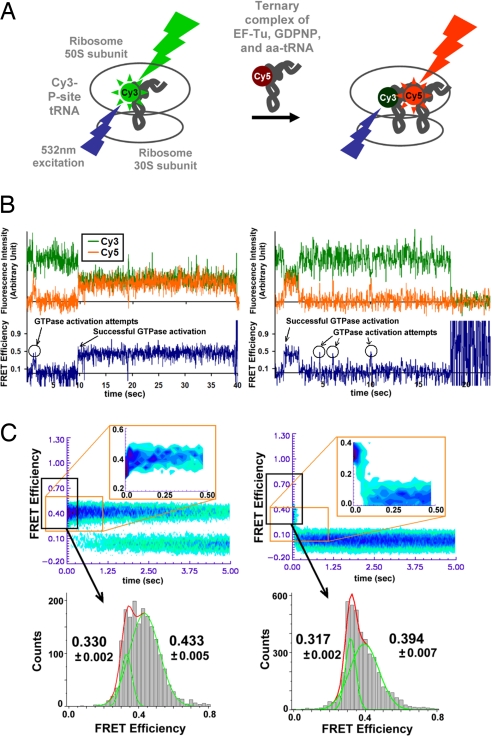
This work followed the single-molecule FRET approach in our earlier work (8, 15). A total internal reflection (TIR) microscope/camera system with single-photon counting sensitivity was used. The ribosome preassembled with biotinylated mRNA and Cy3-labeled peptidyl-tRNA (P-site tRNA) (16, 17) was attached on a quartz surface through streptavidin–biotin interaction. Either cognate or near-cognate ternary complex with Cy5-labeled aa-tRNA was introduced in our stopped-flow, single-molecule apparatus (Fig. 1A). The Cy3-labeled tRNA was excited with a 532-nm laser beam, and the FRET signal was observed as the ternary complex was introduced to the immobilized ribosome complex. A critical improvement in our experiment relative to previous work was that we now were able to record a single-molecule image every 25 ms, a factor of 4 faster than our previous capability. With this higher time resolution, it is possible to observe directly that aa-tRNA is selected by and accommodated into the ribosome based on the codon–anticodon interaction.
Fig. 1.
Schematic of single-molecule FRET experimental setup, examples of resulting FRET traces, and analysis results of cognate (Left) and near-cognate (Right) ternary complex delivery to the ribosome. (A) Single-molecule FRET experimental setup to observe the initial selection of aa-tRNA by the ribosome (8). (B) Typical FRET traces of initial tRNA selection process with the nonhydrolyzable GTP analogue, GDPNP. Materials and methods are explained elsewhere (8). Briefly, the green time trace shows the Cy3 fluorescence intensity, and the orange trace is the Cy5 fluorescence intensity. FRET efficiency is defined as ICy5 /(ICy3 + ICy5). In the presence of GDPNP, aa-tRNA selection process is stalled after the initial selection is complete, thus the system cannot proceed past the mid-FRET state. (C) Postsynchronized histograms and FRET efficiency histograms of cognate and near-cognate delivery of GDPNP stalled ternary complexes (see SI Text). The individual FRET trajectories for the cognate (56 particles) and near-cognate (72 particles) ribosome complexes are superimposed after synchronizing to the first observation of FRET ≥0.27. (Upper) The interaction between the cognate ternary complex and the ribosome results in rapid progression from low- to mid-FRET state, and the majority of near-cognate ribosome complexes retreats from low-FRET state to “0” FRET. (Lower) The mean FRET efficiencies for low- and mid-FRET states in the GDPNP stalled cases. At 15 mM [Mg2+], the mid-FRET efficiencies are 0.433 ± 0.005 and 0.394 ± 0.007 for cognate and near-cognate cases, respectively. The low-FRET efficiencies are 0.330 ± 0.002 and 0.317 ± 0.002 for cognate and near-cognate cases, respectively, showing statistically significant difference in the codon-recognition state. At 5 mM [Mg2+], the mid-FRET efficiencies are 0.438 ± 0.027 and 0.442 ± 0.021 for cognate (59 particles) and near-cognate (55 particles), respectively, and the low-FRET efficiencies are 0.348 ± 0.008 and 0.345 ± 0.004 for cognate and near-cognate cases, respectively (data not shown).
Results
Individual binding events of a cognate ternary complex bearing Cy5-labeled phe-tRNAphe (phenylalanine codon) into the ribosome is shown in Fig. 1B. The FRET efficiency increases in a step-wise fashion as aa-tRNA moves toward the P-site in the ribosome. Changing phenylalanine codon (UUU) to a leucine codon (CUU), thereby introducing one base-mismatch, dramatically reduces the efficiency of initial selection. The real-time, direct observation of the codon-recognition state and the GTPase-activated states allows us to quantify the initial selection efficiency (8, 18). The absolute FRET efficiencies differ from those reported previously because of a difference in quantum yield in the Cy5 spectral region of the newly used charge-coupled device (CCD).
To examine initial selection steps only, experiments were performed in the presence of the nonhydrolyzable GTP analogue 5′-guanylyl-imidodiphosphate (GDPNP). GDPNP stalls the ternary complex on the ribosome at the GTPase-activated state before GTP hydrolysis but has been shown not to affect earlier steps of aa-tRNA selection (19, 20). In the presence of GDPNP, the majority of cognate ternary complex stalls on the ribosome in a stable, GTPase-activated configuration (8, 19, 20).
Both cognate and near-cognate ribosome complexes reach the mid-FRET state (FRET efficiency ≈0.4, required for GTPase activation) from the low-FRET state (FRET efficiency ≈0.3, the codon-recognition state) as shown Fig. 1B (8). To synchronize the behavior of individual ribosome complexes after the codon recognition, individual FRET time traces are superimposed at the first observation point of FRET efficiency ≥0.27. From these “postsynchronized” FRET time traces (8), histograms are compiled (Fig. 1C) to show the majority of cognate ribosome complex progress quickly (<50 ms) from the codon-recognition state to the GTPase-activated state. Near-cognate ribosome complexes, on the other hand, tend to dissociate in <100 ms from the codon-recognition state.
To analyze the detailed kinetics, it is important to properly set the boundary FRET levels among background (0 FRET), the codon-recognition (low-FRET), and the stabilized/GTPase-activated (mid-FRET) states. We initially chose the boundary values to be 1 standard deviation from the mean low- and mid-FRET efficiencies. We then varied the values to achieve the least sensitivity to change of transition rates between the low- and mid-FRET states (see SI Text). This algorithm minimizes the number of false transitions attributable to technical noise of the experimental apparatus. Once the threshold is determined, the lifetime histogram of each state is determined.
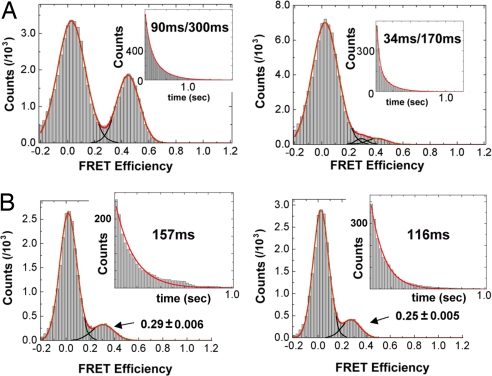
The histogram of the mid-FRET state shows double exponential decay (Fig. 2). These results suggest that the mid-FRET state previously defined as the GTPase-activated state in fact is composed of two different states: a short-lived state (state 3′ in Fig. 3) and a stably bound state (state 3) (8). The detection of the 3′ state suggests that the ribosome complex fluctuates reversibly between the codon-recognition and the GTPase-activated states rather than making a single, discrete transition. The suggested kinetic scheme in Fig. 3 is an analytical solution of what we observed. As an alternative scheme, state 3′ may be an intermediate state between state 2 and state 3. Under this scenario, 3′ is connected directly to 3. It is still possible to get lifetimes that can be fitted with double exponentials under certain conditions (e.g., lifetime of state 3′ is negligibly short compared with that of state 3, and the rate forward from state 3′ is negligibly low compared with the rate backward). Nonetheless, the existence of fluctuations to/from state 3′ is not affected by adopting a different kinetic scheme.
Fig. 2.
FRET histogram and lifetime analysis of the GTPase-activated state and the codon-recognition states for cognate (Left) and near-cognate (Right) cases. (A) At 15 mM [Mg2+], the mid-FRET (0.34–0.6 FRET) state lifetime histograms are well fit as double exponential decay by the equations: 526 exp(−t/0.094) + 561 exp(−t/0.31) and 641 exp(−t/0.034) + 207 exp(−t/0.17) for cognate and near-cognate cases, respectively, where time is given in seconds. At 5 mM [Mg2+] (histograms not shown), the fits are 463 exp(−t/0.085) + 483 exp(−t/0.23) and 192 exp(−t/0.051) + 2.17 exp(−t/0.46) for cognate and near-cognate cases, respectively. (B) FRET efficiency and lifetime histograms of the codon-recognition state in the presence of 100 μM tetracycline. Mean FRET efficiency and lifetime of codon-recognition state in cognate cases are 0.29 ± 0.006 and 157 ms whereas those in near-cognate cases are 0.25 ± 0.005 and 116 ms.
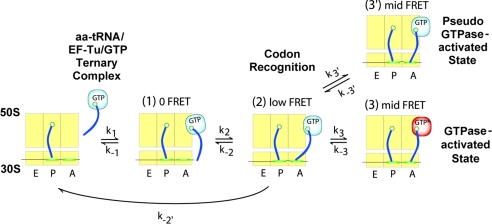
Fig. 3.
A schematic representation of the kinetic model for the initial selection of aa-tRNA. The 30S subunit (lower rectangle) bound to an mRNA (line across the 30S subunit) and the 50S subunit (upper rectangle) are shown divided into small rectangles corresponding to the A, P, and E sites (23). The selection process begins upon the binding of the ternary complex to the ribosome through the interaction between the EF-Tu and the ribosome. We do not observe initial binding with FRET codon-recognition positions with the ternary complex closer to the P site yielding the low-FRET state (state 2). In its attempt to reach the GTPase-activated state (mid-FRET, state 3), the ribosome complex is observed to reversibly sample the pseudo-GTPase-activated state (mid-FRET, state 3′).
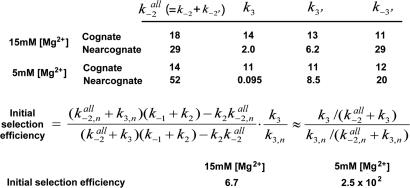
Here, we explicitly assume that the formation of a long-lived mid-FRET state is required for GTPase activation and hydrolysis (5, 20). Even though GTPase activation and hydrolysis are very fast compared with the initial selection process, the fact that cognate tRNA is much more likely to enter into the stably bound mid-FRET state and undergo GTP hydrolysis supports this assumption (6, 8). From the detailed kinetic data, as summarized in Fig. 4, a cognate ribosome complex on average (at 15 mM [Mg2+]) is found to try twice to progress to the GTPase-activated state whereas a near-cognate ribosome complex should try four times per every one successful advancement. Our kinetic analysis also reveals that cognate ribosome complexes not only make fewer unsuccessful trials, but they also try more often. On average, at 15 mM [Mg2+], cognate ribosome complexes try 27 times per second, whereas near-cognate ribosome complexes try only 8 times per second. As a result of such differences, the initial selection efficiency is calculated to 6.7 and 2.5 × 102 for 15 mM and 5 mM [Mg2+] cases, respectively (Fig. 4). In the case of 5 mM [Mg2+], the near-cognate rates we report here have large uncertainties because of our limited data set (see SI Text).
Fig. 4.
Summary of kinetics data and calculated selection efficiencies. Selection efficiency is defined as the ratio of d[state 3]cognate /dt to d[state 3]near-cognate /dt. Both the cognate and near-cognate ribosome complexes fluctuate rapidly and reversibly to the pseudo-GTPase-activated state (state 3′) before reaching successfully the bona fide GTPase-activated state (state 3). Preferential selection of the cognate ternary complex by the ribosome is achieved by the product of the higher frequency and the higher success rate of the attempts to reach the GTPase-activated state. Rate constants (units are s−1) are calculated by counting FRET transitions between states as described in the text. Missed events attributable to too short lifetimes were added (see SI Text). By assuming that the initial binding of the ternary complex to the ribosome (formation of state 1 in Fig. 3) is not rate-determining and making steady state approximations about state 2 and state 3′ in Fig. 3, the initial selection efficiencies are calculated to be 6.7 and 2.5 × 102 at 15 mM [Mg2+] and 5 mM [Mg2+], respectively.
Discussion
Based on these findings, we propose that the codon–anticodon interaction and the domain closure, i.e., the formation of additional ribosome-tRNA-mRNA contacts at the decoding site of the 30S subunit (9, 21), cause the cognate ternary complex to be positioned slightly closer to the P-site tRNA. We observe differences in the average FRET efficiencies of the codon-recognition state for cognate and near-cognate ribosome complexes (Fig. 1C). Cognate ribosome complexes are found to yield significantly higher FRET efficiency than near-cognate complexes for the 15 mM [Mg2+] data. We also performed experiments in which the progression of the aa-tRNA to the position where GTPase activation of EF-Tu also would occur under normal conditions was blocked by adding the antibiotic tetracycline (22). These experiments were performed at 15 mM [Mg2+]. Tetracycline inhibits the tRNA selection process by hindering the stable codon–anticodon interaction and thereby blocking further incorporation of aa-tRNA from the initial codon recognition. Therefore, the position of the aa-tRNA monitored in these experiments would represent the dynamics of the codon-recognition state. In the data shown in Fig. 2B, we filtered out a subpopulation of the ribosome complexes that show normal delivery of aa-tRNA to the full accommodation. Only the first 5–10 s after the ternary complex was introduced was used to build histograms. Within this time window, the percentage of complexes that showed full accommodation was below 10%. The full accommodation percentage increased to 55% over 8 min after the ternary complex delivery. As discussed in the legend of Fig. 2B, cognate ribosome complexes in the codon-recognition state have a longer lifetime and a higher FRET efficiency than do near cognates, further supporting the view that cognate ternary complexes are positioned closer than near-cognate complexes are to the stably docked mid-FRET state.
GTP hydrolysis requires the formation of ternary complex contacts with SRL and GAC as shown in SI Fig. 5 (5, 10). It is the movement of the tRNA to this mid-FRET state that provides the allosteric linkage between action at the decoding site and GTPase activation of EF-Tu at the SRL/GAC (5, 19).
Because of (i) the tens of milliseconds needed for the ternary complex to form stabilizing contacts (k3 in Fig. 4) and (ii) the inferred distance change in going from the low- to mid-FRET states, we conclude that a large and rare thermal fluctuation is required to pivot the tRNA into a position where the contacts between SRL/GAC and the ternary complex can be formed. We now argue that ribosome uses these rare fluctuations as an effective selection mechanism. Assume, for simplicity, that the thermal distribution of distances between the SRL/GAC and the binding sites on the ternary complex is described by a Gaussian distribution exp[−x2/2σ2], where x is the deviation from the equilibrium position and when x is comparable or less than σ. For x ≥2–3 σ, it is plausible that the tail of the distribution of positions might be better described by exp[−x/λ], analogous with thermal activation out of a bound state exp[−ΔE/kT]. The rate of forming stabilizing contacts would be proportional to ν exp[−x/λ], where ν describes a “characteristic frequency” of the thermal fluctuations, most likely in the nano- to microsecond range.
If xc is the critical distance that the near-cognate ternary complex has to move to form contacts with SRL/GAC, our data indicate that the cognate requires a less rare fluctuation xc − ∣δ∣. Thus, the cognate ternary complex is more likely to form stabilizing contacts than near-cognate complex by the ratio exp[−(xc − ∣δ∣)/λ]/exp[−xc/λ] = exp[∣δ∣/λ], provided the attempt frequencies ν of cognate and near-cognate tRNA fluctuations are similar. Note that the rate of forming the stabilizing contacts is made exponentially important in the distance ∣δ∣ because docking must be preceded by a rare thermal fluctuation. The long distance (>7 nm) between the decoding site and the SRL/GAC (5, 11, 12, 20, 23) suggests that the tRNA also may act as a long lever arm to increase ∣δ∣ by magnifying a slight difference in the pivot angle between cognate and near-cognate tRNA at the decoding site.
The simple model presented above does not take into account the fact that the ternary complex can form the bona fide GTPase-activated state with the SRL/GAC or a transient pseudo-GTPase-activated state. However, because states 3 and 3′ are both mid-FRET states, the formation of either state requires a large thermal fluctuation. The shorter lifetime (<100 ms) components of the mid-FRET state plotted in Fig. 2A may represent unsuccessful long-distance fluctuations of the ternary complex to stably dock at the SRL/GAC. Instead, the ternary complex may form only one of the two identified contacts. (SI Fig. 5 shows the structure of the two contacts between ternary complex and SRL/GAC.)
This hypothesis was tested by our recent results from thiostrepton-inhibited ternary complex delivery using the same experimental setup as reported here. Thiostrepton is known to interfere with the ribosome function by binding to GAC (24). The lifetime of inhibited GTPase-activated state attributable to the presence of thiostrepton is found to be <100 ms, and essentially no Phe-tRNAPhe achieves the stabilized mid-FRET state or full accommodation (25). These observations suggest that the shorter lifetime component of mid-FRET state (or pseudo-GTPase-activated state) represents a state where the ternary complex forms a partial contact with SRL only, instead of both SRL and GAC. The sum of the transition rates to this pseudo-GTPase-activated state (k3′) and the long-lived bona fide GTPase-activated state (k3) are found to be strongly dependent on codon–anticodon interaction (Fig. 4) and may be the result of the exponential discrimination exp[∣δ∣/λ] suggested in our model (SI Fig. 6).
In this model, elements of the ribosome that restrict the ternary complex motion would impinge on fidelity of the translation during the initial selection of aa-tRNA, explaining how mutations in such elements (for example, L7, S12, and the SRL) alter fidelity (SI Fig. 5) (26–28). The observation that mutations in tRNA itself influence fidelity (29) can be explained if those mutations affect the structure or rigidity of the molecule or its ability to make proper contacts with the ribosome.
The sensitivity and time resolution of our single-molecule data allow direct single-molecule observation of some of the initial selection steps of aa-tRNA by the ribosome. This work provides evidence that the difference in the ribosome-ternary complex structure at the decoding site uses induced fit in combination with thermal fluctuations for the initial selection of aa-tRNA. Our data indicate that the induced fit interaction serves two purposes. (i) It is responsible for the preferential dissociation of near-cognate tRNAs relative to cognate tRNAs. However, there is only a factor of 4 difference in the off-rates of cognate and near-cognate ternary complexes at 5 mM [Mg2+] (Fig. 4). (ii) Induced fit interactions move the cognate ternary complex into a position where it is more likely to form the stabilizing contacts with the ribosome. Once in this position, large thermal fluctuations in the motion of the ternary complex are needed to form additional contacts with SRL/GAC. We observed unsuccessful attempts to form these contacts as well as successful ones. Because of the slightly favorable position of the cognate ternary complex, this complex is seen to fluctuate into the mid-FRET state more often than the near-cognate complex and have a factor of 100 higher overall rate of forming bona fide contacts. The dynamics of the initial selection of aa-tRNA by the ribosome provides a valuable insight into the critical role of fluctuations in the substrate selection by this enzyme.
Materials and Methods
Materials and detailed methods are explained elsewhere (8). Cy3 attached to the P-site initiator tRNA was excited with a 532-nm laser in the prism-coupled total internal reflection geometry. Cy5 attached to aa-tRNA started fluorescence after the ternary complex bound to the ribosome as the excitation energy was transferred to the Cy5 through resonant energy transfer from the directly excited Cy3. Fluorescence was collected with a 1.2 N.A. ×60 water immersion objective (PlanApo; Nikon, Tokyo, Japan) on a commercial inverted microscope (Eclipse TE2000; Nikon) with a 550-nm long-pass filter (Chroma, Rockingham, VT) and a 650-nm dichroic mirror (Chroma). Fluorescence intensities from Cy3 and Cy5 were recorded simultaneously with an electron multiplying charge-coupled device (CCD) camera (Cascade; Roper Scientific, Tuscon, AZ) to get FRET efficiencies for 9-pixel binned, 25-ms integrated fluorescence until Cy3 bleached.
Supplementary Material
Acknowledgments
S.C.B. was supported by the Giannini Family Foundation. This work was supported by National Institutes of Health Grant GM51266 and funds from the David and Lucille Packard Foundation (to J.D.P.); by grants from the National Science Foundation, National Aeronautics and Space Administration, and Air Force Office of Scientific Research (to S.C.); and by David and Lucille Packard Foundation Interdisciplinary Science Program Grant 2000-01671 (to S.C. and J.D.P.).
Abbreviations
- aa-tRNA
aminoacyl-tRNA
- EF-Tu
elongation factor Tu
- SRL
sarcin–ricin loop
- GAC
GTPase-associated center
- GDPNP
5′-guanylyl-imidodiphosphate.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705988104/DC1.
References
- 1.Ramakrishnan V. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 2.Green R, Noller HF. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 3.Jelenc PC, Kurland CG. Proc Natl Acad Sci USA. 1979;76:3174–3178. doi: 10.1073/pnas.76.7.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogle JM, Carter AP, Ramakrishnan V. Trends Biochem Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 5.Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Nat Struct Mol Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 6.Rodnina MV, Wintermeyer W. Trends Biochem Sci. 2001;26:124–130. doi: 10.1016/s0968-0004(00)01737-0. [DOI] [PubMed] [Google Scholar]
- 7.Hopfield JJ. Proc Natl Acad Sci USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 9.Ogle JM, Murphy I, Frank V, Tarry MJ, Ramakrishnan V. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 10.Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J. EMBO J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 12.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 13.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Proc Natl Acad Sci USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster T. Discuss Faraday Soc. 1959;27:7–17. [Google Scholar]
- 15.Blanchard SC, Kim HD, Gonzalez RL, Puglisi JD, Chu S. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodcock J, Moazed D, Cannon M, Davies J, Noller HF. EMBO J. 1991;10:3099–3103. doi: 10.1002/j.1460-2075.1991.tb07863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moazed D, Noller HF. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 18.Pape T, Wintermeyer W, Rodnina MV. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M. FEBS Lett. 2005;579:959–962. doi: 10.1016/j.febslet.2004.10.105. [DOI] [PubMed] [Google Scholar]
- 20.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BFC, Nyborg J. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 21.Stark H, Rodnina MV, Wieden H-J, Zemlin F, Wintermeyer W, van Heel M. Nat Struct Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- 22.Moazed D, Noller HF. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 23.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J, Musters W, Cundliffe E, Dahlberg AE. EMBO J. 1993;12:1499–1504. doi: 10.1002/j.1460-2075.1993.tb05793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez RL, Chu S, Puglisi JD. RNA. 2007 doi: 10.1261/rna.499407. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powers T, Noller HF. J Mol Biol. 1994;235:156–172. doi: 10.1016/s0022-2836(05)80023-3. [DOI] [PubMed] [Google Scholar]
- 27.Pettersson I, Kurland CG. Proc Natl Acad Sci USA. 1980;77:4007–4010. doi: 10.1073/pnas.77.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tapprich WE, Dahlberg AE. EMBO J. 1990;9:2649–2655. doi: 10.1002/j.1460-2075.1990.tb07447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cochella L, Green R. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.