Abstract
A novel S-adenosyl-l-methionine (SAM)-dependent methyltransferase catalyzing the O methylation of several chlorophenols and other halogenated phenols was purified 220-fold to apparent homogeneity from mycelia of Trichoderma longibrachiatum CECT 20431. The enzyme could be identified in partially purified protein preparations by direct photolabeling with [methyl-3H]SAM, and this reaction was prevented by previous incubation with S-adenosylhomocysteine. Gel filtration indicated that the Mr was 112,000, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed that the enzyme was composed of two subunits with molecular weights of approximately 52,500. The enzyme had a pH optimum between 8.2 and 8.5 and an optimum temperature of 28°C, with a pI of 4.9. The Km values for 2,4,6-trichlorophenol and SAM were 135.9 ± 12.8 and 284.1 ± 35.1 μM, respectively. S-Adenosylhomocysteine acted as a competitive inhibitor, with a Ki of 378.9 ± 45.4 μM. The methyltransferase was also strongly inhibited by low concentrations of several metal ions, such as Cu2+, Hg2+, Zn2+, and Ag+, and to a lesser extent by p-chloromercuribenzoic acid, but it was not significantly affected by several thiols or other thiol reagents. The methyltransferase was specifically induced by several chlorophenols, especially if they contained three or more chlorine atoms in their structures. Substrate specificity studies showed that the activity was also specific for halogenated phenols containing fluoro, chloro, or bromo substituents, whereas other hydroxylated compounds, such as hydroxylated benzoic acids, hydroxybenzaldehydes, phenol, 2-metoxyphenol, and dihydroxybenzene, were not methylated.
The cork stopper, which is manufactured from the bark of cork oak (Quercus suber), is currently recognized as the best material to seal bottles of wine (27). Unfortunately, cork stoppers are responsible for cork taint of 2 to 7% of the bottles of wine produced annually (8), causing severe economic losses to the wine industry. Traditionally, cork taint has been related to the transfer of compounds, mainly of microbial origin, from the cork stopper to the wine (8, 27). In fact, cork is a complex ecosystem harboring very large microbial populations (1, 2, 17, 18) directly implicated in the production of such compounds. Although several chemical compounds have been related to this off-odor problem (4, 17, 18), chloroanisoles and especially 2,4,6-TCA have been blamed as the main agents responsible for cork taint. In fact, 2,4,6-TCA has been reported to be responsible for at least 80% of the cases of cork taint (8), and in a recent study Pollnitz and colleagues reported that 2,4,6-TCA could be detected in 100% of tainted wines (23). The role of chloroanisoles as contaminant agents is not restricted to wine, since the presence of these compounds is well documented in other food materials, such as eggs and poultry (12), chickens (11), pulp chips (10), dried fruits (30), Brazilian coffee (29), and drinking water (21).
Information about the O methylation reactions leading to the formation of anisoles and their derivatives is rather scarce. In some Rhodococcus and Acinetobacter strains, a constitutive SAM-dependent methyltransferase is involved in the formation of anisoles and thioanisoles (20), although no extensive characterization of this process has been conducted. Also, a 2,4-disubstitued phenol O-methyltransferase has been purified from the white-rot basidiomycete Phanerochaete chrysosporium. This enzyme preferentially methylates 3-methoxy- and 3,5-dimethoxy-substitued 4-hydroxybenzaldehydes, 4-hydroxybenzoic acids, and 4-hydroxyacetophenones, but it can also O methylate xenobiotic compounds like 2,4-DCP and 2,4-dibromophenol, yielding the corresponding anisoles (9). Although the substrate specificity of the enzyme indicates that it has a role in the methylation of lignin degradation products, the possibility that the enzyme is involved in the formation of 2,4,6-TCA and PCA observed in cultures of this fungus under nonlimiting nitrogen source conditions cannot be eliminated (5, 6).
Recently, it has been reported that a number of filamentous fungi can convert 2,4,6-TCP into 2,4,6-TCA when they are growing directly on cork. This conversion was shown to occur by O methylation of 2,4,6-TCP in a reaction catalyzed by an inducible SAM-dependent O-methyltransferase (1). In this paper we describe the purification and properties of a novel SAM-dependent CPOMT from Trichoderma longibrachiatum that is highly specific for halogenated phenols. Our results suggest that this enzyme is responsible for the formation on cork of several anisoles, including 2,4,6-TCA. Accordingly, CPOMT is an important candidate for mediating cork taint of wines under naturally occurring conditions.
MATERIALS AND METHODS
Abbreviations.
The following abbreviations are used in this paper: CPOMT, chlorophenol O-methyltransferase; CP, chlorophenol; CA, chloroanisole; DBrP, dibromophenol; DCP, dichlorophenol; DCA, dichloroanisole; TCP, trichlorophenol; TCA, trichloroanisole; TeCP, tetrachlorophenol; TeCA, tetrachloroanisole; PCP, pentachlorophenol; PCA, pentachloroanisole; TBrP, tribromophenol; TFP, trifluorophenol; TIA, triiodoanisole; TIP, triiodophenol; DTT, dithiothreitol; PMSF, phenylmethylsulfonyl fluoride; SAHC, S-adenosylhomocysteine; SAM, S-adenosyl-l-methionine; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Chemicals.
2-CP, 3-CP, 4-CP, 2-CA, 3-CA, 4-CA, 2,3-DCP, 2,4-DCP, 2,5-DCP, 2,6-DCP, 3,4-DCP, 3,5-DCP, 2,3-DCA, 2,4-DCA, 2,6-DCA, 3,5-DCA, 2,3,4-TCP, 2,3,6-TCP, 2,4,5-TCP, 2,4,6-TCP, 2,4,6-TCA, 2,3,4,5-TeCA, PCP, 2,4,6-TBrP, 2,4,6-tribromoanisole, 2,4,6-TFP, and 2,4,6-TIP were obtained from Aldrich-Chemie (Steinheim, Germany). 2,3,4-TCA, 2,3,6-TCA, 2,3,4,5-TeCP, 2,3,4,6-TeCP, 2,3,5,6-TeCP, and 2,3,4,6-TeCA were purchased from Fluka Chemie AG (Buchs, Switzerland). 2,4,5-TCA, PCA, and 2,3,5,6-TeCA were obtained from Supelco (Bellefonte, Pa.). 2,5-DCA and 3,4-DCA were purchased from Acros Organics (Geel, Belgium). Phenol, anisole, 2-methoxyphenol (guaiacol), 1,2-dihydroxybenzene (catechol), 3,4-dihydroxybenzoic acid (protocatechuic acid), 4-hydroxy-3-methoxybenzoic acid (vanillic acid), 3-hydroxy-4-methoxybenzoic acid (isovanillic acid), 4-hydroxy-3-methoxybenzaldehyde (vanillin), 3,4,-dihydroxybenzaldehyde, DTT, PMSF, SAM, and SAHC were obtained from Sigma Chemical Co. (St. Louis, Mo.).
Fungal strain and culture conditions.
T. longibrachiatum CECT 20431, a strain isolated from cork, was routinely maintained and grown as described previously (1). After 48 h, 2,4,6-TCP (10 μg ml−1) was added to induce enzyme synthesis. Cultures were harvested 4 h later by filtration through a glass fiber filter. The mycelium was extensively washed with a sterile saline solution to remove any trace of phenolic compounds. Superficial moisture was removed by pressing mycelia gently between pieces of filter paper before they were frozen at −20°C until they were used. The typical mycelium yields obtained by this cultivation process were approximately 18 g (wet weight) liter−1.
Preparation of cell extracts.
The frozen mycelia were thawed slowly in an ice bath, resuspended (2 ml g of mycelium−1) in 50 mM Tris-HCl (pH 8.2) containing 5 mM MgCl2, 100 mM NaCl, 1 mM DTT, 1 mM PMSF, and 20% glycerol (breaking buffer), and disrupted by sonication (150-W MSE ultrasonic disintegrator) by using 15 10-s bursts. Cell debris was removed by centrifugation at 20,000 × g for 20 min at 4°C. The supernatant was desalted by passage through PD-10 columns (Amersham Pharmacia Biotech, Uppsala, Sweden) to remove any trace of phenolic compounds, and the eluate in 50 mM Tris-HCl (pH 8.2) containing 5 mM MgCl2 and 10% glycerol (standard buffer) was used as the crude enzyme preparation.
CPOMT assay.
The CPOMT activity was routinely determined by assaying production of 2,4,6-TCA from 2,4,6-TCP. The reaction mixture (total volume, 0.5 ml) contained, in 50 mM Tris-HCl (pH 8.2), 0.1 to 2 μg of enzyme, 1 mM SAM, 0.25 mM 2,4,6-TCP, 1 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, and 10% glycerol. Optimal SAM and 2,4,6-TCP concentrations had been determined previously in enzymatic assays. Saturation of the system was observed with 1 mM SAM and 0.25 mM 2,4,6-TCP. Also, a noteworthy inhibitory effect was detected when the 2,4,6-TCP concentration in the reaction mixture was higher than 350 μM. The enzyme reaction mixtures were incubated at 28°C for 3 h before the reactions were terminated with 20 μl of 6 N HCl. The reaction mixtures were processed, and the 2,4,6-TCA content was estimated as reported previously (1) by high-performance liquid chromatography (1100 series; Hewlett-Packard, Wilmington, Del.) in a Zorbax SB-C8 column (4.5 by 150 mm; Agilent Technologies, Madrid, Spain) with methanol-water (75:25) as the mobile phase at a flow rate of 1 ml min−1. Eluted peaks were detected at 230 nm. 2,4,6-TCA was quantified by using 2,3,4,6-TeCA as the internal standard. Specific activities were expressed in picomoles of product formed per minute per milligram of protein. The total soluble protein content was determined by the method of Bradford (7) by using the Bio-Rad protein reagent (Bio-Rad, Hercules, Calif.) and bovine serum albumin as the standard.
Specificity of substrates.
To check the specificity of substrates of the CPOMT, reactions were performed and reaction mixtures were processed as described above, except that the corresponding substrate was substituted for 2,4,6-TCP. The reaction mixtures and negative controls without SAM performed in parallel were analyzed by high-performance liquid chromatography in a Zorbax SB-C8 column (4.5 by 150 mm; Agilent Technologies) with a linear 5 mM formic acid-acetonitrile gradient (30 to 85% acetonitrile in 20 min) at a flow rate of 1 ml min−1. The eluted peaks were detected at 230 nm. Products were quantified by using 2,4,6-TCA as an internal standard (when the substrate was any TCP, the internal standard was changed to PCA). The area under each peak was referred to calibration curves obtained with standards. The identity of each product was confirmed by gas chromatography-mass spectrometry as previously described (1).
Induction of CPOMT activity.
Induction of the CPOMT was measured in resting cell systems as reported previously (1), except that the methyltransferase activity was determined as described above. The putative inducers were each added to a final concentration of 10 μg μl−1. Experiments with negative noninduced controls were always performed in parallel.
CPOMT purification.
All purification steps were performed at 4°C with a fast protein liquid chromatography ÄKTA system (Amersham Pharmacia Biotech).
(i) Step 1: preparation of cell extracts.
A 6-g sample of mycelia was used to obtain cell extracts in breaking buffer as described above. After passage through four PD-10 columns, the proteins (14 ml) were recovered in 10 mM Tris-HCl (pH 8.0) containing 2 mM MgCl2, 1 mM DTT, and 3.5 M NaCl (buffer PA).
(ii) Step 2: Resource PHE column chromatography.
The preparation was loaded onto a Resource PHE column (Amersham Pharmacia Biotech) which previously had been equilibrated with 5 bed volumes of buffer PA, and was washed with 5 bed volumes of the same buffer. Then the enzyme was eluted with 20 ml of 10 mM Tris-HCl (pH 8.0) containing 2 mM MgCl2 and 1 mM DTT (buffer PB) at a flow rate of 1 ml min−1 with the following elution gradient: 0 to 60% buffer PB for 5 min and 60 to 100% buffer PB for 15 min. Fractions containing enzyme activity were pooled and subsequently processed.
(iii) Step 3: ultrafiltration.
An 80-fold-concentrated enzyme preparation was obtained by ultrafiltration through a polyethersulfone 50-kDa-cutoff membrane in a Vivaspin concentrator (Vivascience, Göttingen, Germany). The buffer of the enzyme preparation was changed to 50 mM Tris-HCl (pH 8.0) containing 2 mM MgCl2, 1 mM DTT, and 10% glycerol (buffer DA) by using a HiTrap desalting column (Amersham Pharmacia Biotech) according to procedures recommended by the supplier.
(iv) Step 4: HiTrap DEAE column chromatography.
Next, the enzyme solution in buffer DA was applied to a 1-ml-bed-volume HiTrap DEAE column (Amersham Pharmacia Biotech) that had been equilibrated previously in the same buffer. The column was washed, and the enzymatic activity was eluted with buffer DA containing 1 M NaCl (buffer DB) at a flow rate of 1 ml min−1 with the following elution gradient: 0 to 2% buffer DB for 2 min, 2% buffer DB for 6 min, 2 to 10% buffer DB for 12 min, 10 to 15% buffer DB for 4 min, and 15 to 100% buffer DB for 2 min.
(v) Step 5: Resource Q column chromatography.
The active fractions from step 4 were pooled, the buffer was changed to 50 mM Tris-HCl (pH 8.2) containing 2 mM MgCl2, 1 mM DTT, and 10% glycerol (buffer QA), and the preparation was applied to a Resource Q column (Amersham Pharmacia Biotech) that had been equilibrated previously with 5 bed volumes of buffer QA. The adsorbed enzyme was eluted at a flow rate of 1 ml min−1 with buffer QA containing 1 M NaCl (buffer QB) with the following elution gradient: 0 to 2.5% buffer QB for 2 min, 2.5% buffer QB for 6 min, 2.5 to 6.5% buffer QB for 6 min, 6.5 to 10% buffer QB for 5 min, 10 to 15% buffer QB for 3 min, and 15 to 100% buffer QB for 2 min.
(vi) Step 6: Superdex 200 HR 10/30 column chromatography.
The fractions from step 5 containing CPOMT activity were combined, concentrated by using a 7.5-kDa-cutoff Vivapore concentrator (Vivascience), and applied to a Superdex 200 HR 10/30 column (Amersham Pharmacia Biotech) that was equilibrated and eluted at a flow rate of 0.25 ml min−1 in buffer DA. Fractions (0.75 ml) were collected and assayed for CPOMT activity, and the fractions showing activity that contained the purified enzyme were combined and preserved at −20°C for subsequent analysis.
Determination of native molecular mass.
The native molecular mass (Mr) was determined by gel filtration chromatography on a Superdex 200 HR 10/30 column that was eluted with buffer DA at a flow rate of 0.2 ml min−1. The column was calibrated with the following standards: thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), bovine serum albumin (67 kDa), and carbonic anhidrase (29 kDa), all of which were obtained from Amersham Pharmacia Biotech.
pI determination.
The pI of the enzyme was estimated by chromatofocusing on a PBE 94 column (4 by 1 cm; Amersham Pharmacia Biotech). Each sample analyzed was applied to the column in 25 mM imidazole-HCl buffer (pH 7.4) containing 2 mM MgCl2. The column was eluted with 20 ml of Polybuffer 74 (Amersham Pharmacia Biotech) covering a pH range from 7 to 4, and 1-ml fractions were collected and immediately assayed for enzyme activity.
pH and temperature optima.
The optimal pH for enzymatic activity was determined by using phosphate buffer (pH 6.0 and 7.0), Tris-HCl buffer (pH 8.0, 8.2, 8.5, and 9.0), or CAPS (Sigma Chemical Co.) buffer (pH 10.0). All buffers (50 mM) contained 2 mM MgCl2. The optimal temperature was determined in buffer DA by using a reaction mixture incubated for 10 min at each temperature tested before the enzyme preparation was added.
Stability at 4 and 42°C.
The stability of the enzyme was studied by using semipurified enzyme preparations (from step 5 of the purification process), which were extensively dialyzed against 50 mM Tris-HCl (pH 8.2) and maintained at 4 or 42°C for up to 15 days. The effect of 0.1 mM SAM, 1 mM DTT, 2 mM MgCl2, or 10% glycerol on stability was tested by including the compound in an enzyme preparation.
Kinetic analysis.
Highly purified CPOMT (from step 6 of the purification procedure) was used for all the kinetic studies. Kinetic parameters (Vmax and Km) were determined for 2,4,6-TCP and SAM from Lineweaver-Burk double-reciprocal plots under initial velocity conditions. Values for the variable substrate 2,4,6-TCP (0.05 to 0.3 mM) were determined at four different fixed concentrations of SAM (0.1, 0.2, 0.3, and 0.4 mM). In a similar way values for the variable substrate SAM (0.1 to 0.6 mM) were determined at different fixed concentrations of 2,4,6-TCP (0.1, 0.15, 0.2, and 0.3 mM). The Ki for SAHC was determined in reaction mixtures containing 0.25 mM 2,4,6-TCP, 0.1 to 0.4 mM SAM, and 0.4 mM SAHC. The kinetic constants were determined in duplicate in three independent experiments by using Lineweaver-Burk plots and the EZ-fit program developed by Perrella (22).
SDS-PAGE.
SDS-PAGE was performed by the method of Laemmli (16). The protein standards used to estimate subunit molecular masses were broad-range SDS-PAGE standards obtained from Bio-Rad.
Photolabeling of CPOMT with [methyl-3H]SAM.
[3H]SAM (84 Ci mmol−1) was purchased from Amersham Pharmacia Biotech. The labeling reaction mixtures (40 μl) without a methyl acceptor in 50 mM Tris-HCl (pH 8.0) contained 1 to 15 μg of protein, 2 mM MgCl2, 1 mM DTT, and [3H]SAM (0.1 μg μl−1). Each mixture was placed in a round-bottom 96-well microtiter plate and preincubated for 10 min at room temperature. Cross-linking of [3H]SAM was induced by irradiating the preparation at 4°C with a 254-nm UV light source (Spectroline, Westbury, N.Y.) at a distance of 5 cm. The reactions were stopped by mixing the preparations with 10 μl of 5× sample loading buffer (16) and heating them at 100°C for 2 min, and they were subjected to SDS-PAGE (8 to 12% gel). After electrophoresis the gel was fixed by immersion for 20 min at room temperature in a 5% acetic acid-20% methanol fixing solution. Then the gel was soaked in Amplify solution (Amersham Pharmacia Biotech) for 30 min before drying. Exposure with an enhancing screen at −80°C was performed with Kodak X-Omat AR film for up to 60 days.
RESULTS
Induction of CPOMT.
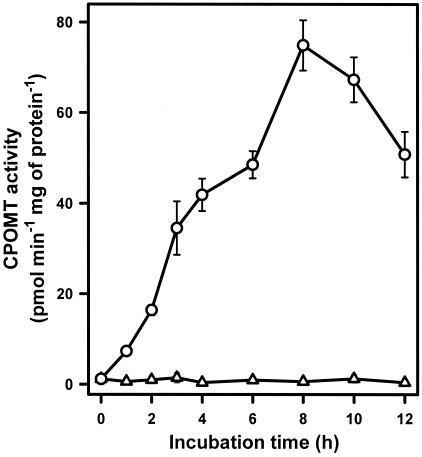
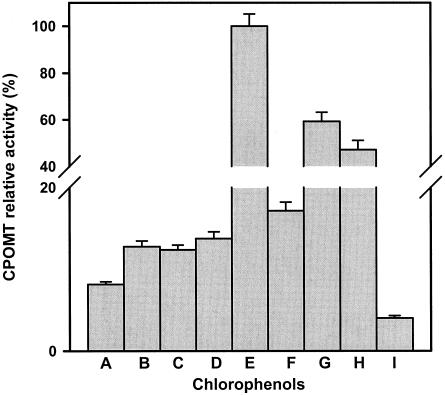
Previously (1), preliminary evidence that CPOMT from T. longibrachiatum is an inducible enzyme was obtained. As shown in Fig. 1 only traces of enzyme activity could be detected when the mycelia were incubated in a resting cell system in the absence of 2,4,6-TCP. By contrast, significant levels of activity were detected as soon as 1 h after addition of the inducer. The activity increased steadily during the next 7 h and then declined (Fig. 1). Under the conditions tested, the lowest concentration of 2,4,6-TCP able to induce CPOMT activity was 7.5 μg ml−1. Maximal induction was observed at concentrations of inducer of 10 to 20 μg ml−1, and higher concentrations were deleterious for induction (data not shown). This was probably due to a toxic effect of 2,4,6-TCP on cell metabolism. The relative rates of induction by a variety of CPs are shown in Fig. 2. All the CPs containing at least two chlorine atoms were able to induce CPOMT activity, but TCPs, TeCPs, and PCP were better inducers than DCPs, whereas 2-CP could not support induction of this activity. In particular 2,4,6-TCP proved to be the best inducer. By contrast, phenol, anisoles (2,4,6-TCA and PCA), halogenated compounds (1,3,5-trichlorobenzene, hexachlorocyclohexane, hexachlorobenzene), and other aromatic nonhalogenated alcohols like vanillic acid, catechol, and guaiacol failed to induce activity.
FIG. 1.
Time course of induction of methyltransferase activity by 2,4,6-TCP (○) in a resting-cell system and activity in noninduced mycelia (▵). The values are estimates resulting from duplicate determinations in three independent experiments. The error bars indicate standard deviations of the means.
FIG. 2.
Induction of CPOMT activity by several CPs, including 2,4-DCP (bar A), 2,6-DCP (bar B), 2,3-DCP (bar C), 3,4-DCP (bar D), 2,4,6-TCP (bar E), 2,4,5-TCP (bar F), 2,3,4,6-TeCP (bar G), and PCP (bar H), and activity of noninduced mycelium (bar I). A value of 100 was arbitrarily assigned to the level of induction obtained with 2,4,6-TCP (bar E). The error bars indicate standard deviations of the means.
Purification of CPOMT.
A six-step purification procedure (Table 1) was used to obtain apparently pure CPOMT from T. longibrachiatum cell extracts. All the purification procedures were conducted at 4°C, and the enzyme was stabilized throughout this process by inclusion of 1 mM DTT and 2 mM MgCl2 in buffers. A cell extract (step 1) suspended in a high-ionic-strength buffer containing 3.5 M NaCl was applied to a Resource PHE column (step 2). Virtually all the activity was retained in the column, and it was subsequently eluted by using a decreasing NaCl gradient. Under the conditions used, the activity eluted at 1.20 to 0 M NaCl (36 to 0% buffer PB). This step was highly efficient since it resulted in 11.3-fold purification with a yield of 81% in terms of enzymatic activity. The active fractions were pooled and concentrated by ultrafiltration. This step resulted in additional purification (1.2-fold), although some activity was lost. The concentrate was applied to a HiTrap DEAE FF column (step 4). The enzyme activity was confined to fractions that eluted at NaCl concentrations between 40 and 120 mM, and maximum elution occurred at about 75 mM NaCl. After this step, the preparation was purified 22.2-fold. The most active fractions were pooled and loaded onto a Resource Q column. The enzyme eluted in a narrow salt range (25 to 65 mM NaCl; maximum at around 45 mM). Finally, the active fractions were combined, and the proteins were resolved by gel filtration through a Superdex 200 column. The enzyme activity was associated with a single peak at an elution volume (Ve) void volume (Vo) ratio of 1.51, whereas the rest of the proteins in the sample eluted in other fractions. Overall, we obtained a 220-fold-purified preparation, with a yield of 0.9%. Analysis of this preparation by SDS-PAGE indicated that it contained a single protein with an apparent molecular mass of 52,500 Da (Fig. 3A, lane 2). Since some SAM-dependent methyltransferases had been identified by specific photolabeling with [3H]SAM (25, 28), we attempted to identify CPOMT by this technique. As shown in Fig. 3B (lane 3), analysis of the protein extract after passage through the DEAE column revealed the presence of a single band that migrated at the same level as the purified protein. Photolabeling was specific since it could be prevented by incubation of the sample with 1 mM SAHC (Fig. 3B, lane 4). Other additional bands, probably corresponding to other SAM-dependent methyltransferases, were detected in samples from previous steps of the purification process (data not shown).
TABLE 1.
Purification of CPOMT from T. longibrachiatum
| Step | Procedure | Total activity (pmol min−1) | Total protein (mg) | Sp act (pmol min−1 mg of protein−1)a | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| 1 | Crude extract | 1,190 | 481 | 2.5 | 100 | 1 |
| 2 | Resource PHE | 963 | 34 | 28.3 | 81 | 11.3 |
| 3 | Ultrafiltration | 667 | 19 | 35.1 | 56 | 14.0 |
| 4 | HiTrap DEAE | 289 | 5.2 | 55.6 | 24 | 22.2 |
| 5 | Resource Q | 75 | 0.8 | 93.7 | 6.3 | 37.5 |
| 6 | Superdex 200 | 11 | 0.02 | 550.0 | 0.9 | 220.0 |
Specific activity at 28°C.
FIG. 3.
Electrophoretic analysis of CPOMT. (A) SDS-PAGE of purified CPOMT: Coomassie blue staining of molecular weight standards (lane 1) and the purified enzyme (lane 2). (B) Detection of CPOMT by photolabeling with [3H]SAM. Lanes 1 and 2, Coomassie blue staining of broad-range SDS-PAGE molecular weight standards (lane 1) and the enzyme preparation after step 4 of the purification process (lane 2); lanes 3 and 4, fluorography of the photolabeling reaction mixture with the protein preparation shown in lane 2 in the absence (lane 3) and in the presence (lane 4) of 1 mM SAHC. The position of CPOMT is indicated by an arrow.
Stability of CPOMT.
Semipurified enzymatic preparations (from step 5 of the purification process) in 50 mM Tris-HCl (pH 8.2) had a half-life of approximately 50 h at 4°C. Addition of 2 mM MgCl2, 0.1 mM SAM, or 1 mM DTT resulted in a significant increase in the half-life of the enzyme, to 72, 170, and 290 h, respectively. Freezing of the preparations resulted in an almost complete loss of activity. However, inclusion of 10% glycerol in the buffer prevented denaturation of the enzyme in such a way that the protein samples could be frozen at −20°C for up to 3 months without a significant loss of activity. Notably, enzyme preparations from the final purification step with Superdex 200 became extremely labile; the half-life at 4°C was about 20 h, and freezing of the samples resulted in a complete lost of activity, even in presence of glycerol. In view of the stability data, preparations from step 5 were routinely used for enzyme characterization. The enzyme exhibited high lability during incubation at 42°C. In fact, the enzyme retained 46 and 11% of its activity after 30 min and 1 h of incubation at 42°C, respectively.
Enzyme properties.
Under standard assay conditions (28°C, 1 mM SAM and 0.25 mM 2,4,6-TCP), the rate of O methylation remained linear for at least 4 h. CPOMT showed maximal activity at a narrow pH range, pH 8.2 to 8.5; at pH 7.0 the activity decreased to 50% of the optimal value, and it decreased to less than 1% of the optimal value at pH 5.0 or 10.0. The pI of the enzyme, as estimated by chromatofocusing on a PBE 94 column, was 4.9. The effect of temperature on activity was determined by using a temperature range of 20 to 42°C. Maximal activity occurred at 28°C when a 3-h incubation period was used in the standard assay, whereas higher temperatures were deleterious for the catalytic process (less than 6% of the activity was detected at 42°C). In fact, rapid denaturation of the enzyme occurred at 42°C or at higher temperatures, as deduced from the stability data described above. The Km values of the enzyme were 135.9 ± 12.8 μM for 2,4,6-TCP and 284.1 ± 35.1 μM for SAM. The reciprocal plot for 2,4,6-TCP was not linear at concentrations above 350 μM, indicating that there was some inhibition at higher concentrations of this substrate. Upon gel filtration, CPOMT emerged as a discrete peak at a relative elution volume corresponding to a native molecular mass of 112,000 Da. On the other hand, in SDS-PAGE the purified enzyme migrated as a single band at 52,500 Da (Fig. 3A, lane 2). These results suggest that there are two subunits in the native enzyme.
Effects of thiol-containing reagents, inhibitors, and metal ions on CPOMT activity.
The effects of several compounds, including thiols, and metal ions on CPOMT activity were tested by using an enzymatic preparation from step 5 of the purification procedure, which had been extensively dialyzed against 50 mM Tris-HCl (pH 8.2) to remove any traces of MgCl2, DTT, and glycerol. The results are shown in Table 2. Addition of 10% glycerol or 1 mM 5-methyltetrahydrofolic acid resulted in weak enhancement of the activity. Several thiol-containing reagents, such as β-mercaptoethanol, thioglycolic acid, and DTT, did not have a significant effect on the activity, whereas l-Cys was slightly inhibitory. However, the Ser and Cys reagent and protease inhibitor PMSF caused a slight reduction in activity. The effect of PMSF was reversed by DTT; moreover, simultaneous addition of both compounds resulted in a notable and reproducible increase in activity. O methylation was almost completely eliminated by several metal ions, including Cu2+, Hg2+, Zn+2, and Ag+, whereas the inhibitory effects of Ca2+, Cs+, Fe2+, and NH4+ were weak. In contrast, Li+ and Mg2+ did not have any effect on activity. The failure of the chelating agent EDTA (1 mM) to inhibit the reaction suggests that there is not a metal ion requirement. The levels of inhibition of several typical inhibitors of SAM-dependent methyltransferases were as follows (means ± standard errors, based on three determinations): SAHC, 87.1% ± 5.6%; iodoacetamide, 18.3% ± 2.9%; N-ethylmaleimide, 16.5% ± 1.9%; and p-chloromercuribenzoate, 45.2% ± 6.6%. A conserved characteristic of the transmethylation reactions involving SAM is that they are strongly inhibited by low concentrations of the demethylated product SAHC. In agreement with this, SAHC behaved as a competitive inhibitor of SAM, with a Ki of 378.9 ± 45.4 μM. The enzyme was partially inhibited by certain thiol-blocking agents, such as p-chloromercuribenzoate, whereas the sensitivity to iodoacetamide and N-ethylmaleimide was lower.
TABLE 2.
Effects of several compounds, thiols, and metal ions on CPOMT activitya
| Compound, thiol, or metal ion addedb | Final concn (mM) | Relative activity (%)c |
|---|---|---|
| None | 100 | |
| MTHFd | 1 | 119 ± 11 |
| Glycerol | 1,085.7e | 122 ± 9 |
| l-Cys | 1 | 88 ± 10 |
| PMSF | 1 | 85 ± 6 |
| PMSF + DTT | 1 + 1 | 147 ± 12 |
| Ca2+ | 1 | 84 ± 4 |
| Cu2+ | 1 | 4 ± 1 |
| Fe2+ | 1 | 82 ± 9 |
| Hg2+ | 0.1 | 4 ± 1 |
| Zn2+ | 0.1 | 18 ± 5 |
| Ag+ | 1 | 6 ± 2 |
| Cs+ | 1 | 85 ± 3 |
| NH4+ | 1 | 84 ± 5 |
Other compounds (1 mM) and ions (1 mM) tested, such as EDTA, β-mercaptoethanol, DTT, thioglycolic acid, Mg2+, and Li+, did not have a significant effect on the activity (the variations observed were always less than 10% compared with the control reaction).
The enzymatic preparation in 50 mM Tris-HCl (pH 8.2) was preincubated with each compound or ion for 10 min before addition of the substrate.
The activity of the enzyme with no addition was assigned a value of 100. The values are means ± standard deviations.
MTHF, 5-methyltetrahydrofolic acid.
10% (vol/vol).
Substrate specificity.
The rates of O methylation of a number of different phenolic substrates by CPOMT are shown in Table 3. The enzyme was highly specific for halogenated phenols. In fact, several nonhalogenated hydroxylated compounds, such as hydroxylated derivatives of benzoic acid (vanillic, isovanillic, and protocatechuic acids), hydroxybenzaldehydes (including vanillin), phenol, 2-methoxyphenol (guaiacol), and dihydroxybenzene, were not methylated at all, an indication that the presence of a halogen atom on the molecule was a requirement for enzyme attack. The only single CP methylated was 2-CP; neither 3-CP nor 4-CP was an effective substrate. The substrate 2,4-DCP supported the highest rate of O methylation, and the activity decreased in the following order: 2,3-DCP, 2,5-DCP, 2,6-DCP, and 3,4-DCP. Activity could not be detected when 3,5-DCP was used as a substrate. All the TCPs tested were methylated; within this group, the maximal activity was observed with 2,3,4-TCP, whereas there was increasingly reduced activity with 2,4,5-TCP, 2,4,6-TCP, and 2,3,6-TCP. The only TeCP that was methylated was 2,3,4,5-TeCP, since no activity was detected with 2,3,4,6-TeCP and 2,3,5,6-TeCP. Finally, it should be noted that CPOMT was also able to methylate 2,4-DBrP, 2,4,6-TBrP, and 2,4,6-TIP, indicating that the specificity is not restricted to CPs but extends to other halogenated phenols. Still, not all such compounds were O methylated, since 2,4,6-TFP was not an effective substrate for the enzyme.
TABLE 3.
Substrate specificity of purified T. longibrachiatum CPOMTa
| Substrate | Rateb |
|---|---|
| 4-Hydroxy-3-methoxybenzoic acid (vanillic acid) | 0 |
| 3-Hydroxy-4-methoxybenzoic acid (isovanillic acid) | 0 |
| 3,4-Dihydroxybenzoic acid (protocatechuic acid) | 0 |
| 2-Methoxyphenol (guaiacol) | 0 |
| 1,2-Dihydroxybenzene (catechol) | 0 |
| 3,4-Dihydroxybenzaldehyde | 0 |
| 4-Hydroxy-3-methoxybenzaldehyde (vanillin) | 0 |
| Phenol | 0 |
| 2-CP | 9.61 |
| 3-CP | 0 |
| 4-CP | 0 |
| 2,3-DCP | 22.51 |
| 2,4-DCP | 100 |
| 2,5-DCP | 19.58 |
| 2,6-DCP | 10.36 |
| 3,4-DCP | 9.26 |
| 3,5-DCP | 0 |
| 2,3,4-TCP | 93.33 |
| 2,3,6-TCP | 55.58 |
| 2,4,5-TCP | 64.19 |
| 2,4,6-TCP | 59.91 |
| 2,3,4,5-TeCP | 10.73 |
| 2,3,4,6-TeCP | 0 |
| 2,3,5,6-TeCP | 0 |
| PCP | 10.07 |
| 2,4-DBrP | 36.80 |
| 2,4,6-TBrP | 12.98 |
| 2,4,6-TFP | 0 |
| 2,4,6-TIP | NQc |
The data were obtained on the basis of a comparison of substrate specificity at a concentration of 0.25 mM.
Rate of O methylation on a molar basis relative to that with 2,4-DCP as the substrate. The values are the averages for duplicate estimates obtained in two independent experiments.
NQ, not quantified. 2,4,6-TIP was methylated, but the reaction could not be quantified because no 2,4,6-TIA was available.
DISCUSSION
Chloroanisoles are thought to be responsible for cork taint of wines (4, 17, 18). The presence of these compounds in cork stoppers is attributed to O methylation of contaminant CPs present on cork (1, 26), and accordingly, this reaction has applied significance. Although production of chloroanisoles from halogenated phenols has been widely detected in nature (1, 3, 5, 6, 9-14, 19-21, 29-31), the characteristics of the methyltranferases involved in such reactions are largely unknown. Here we report for the first time characterization of a T. longibrachiatum methyltransferase which specifically O methylates CPs and other halogenated phenols, yielding the corresponding anisoles.
Several characteristics of CPOMT are worth mentioning. Certain O-methyltransferases of plant and animal origin require the presence of a bivalent ion, such as Mg2+, for maximum activity (24). However, this cation was not essential for CPOMT activity. Consistent with this, the enzyme was not inhibited by EDTA. On the other hand, several classic inhibitors (thiol blocking reagents) of methyltransferases had a weak inhibitory effect on CPOMT. PMSF was also slightly inhibitory, and this inhibition was clearly eliminated by inclusion of DTT in the reaction mixture. These data are consistent with the presence of a thiol group at the active center of the enzyme. Interestingly, O methylation remained linear with time up to 4 h at 28°C.
A bacterial methyltransferase that catalyzes the O methylation of a broad range of CPs, bromophenols, and chlorothiophenols has been detected in cellular extracts of Rhodococcus, Acinetobacter, and Pseudomonas strains (20). Nevertheless, this activity is markedly different from the CPOMT activity of T. longibrachiatum, since it is constitutive, requires MgCl2, has an optimum pH of 7.0, and is also active against other halogenated aromatic compounds, such as chloro- and bromoguaiacols and chloro- and bromocatechols.
O methylation of 2,4-DCP and 2,4,-DBrP by a 2,4-disubstituted phenol methyltransferase has been reported for P. chrysosporium. Although the wide substrate range suggests that the enzyme has a putative role in catabolism or detoxification of lignin degradation products (9), this activity could be also responsible for the formation of 2,4,6-TCA and PCA from 2,4,6-TCP and PCP, respectively, in cultures of this basidiomycete when it is growing in high-carbon—high-nitrogen conditions (5, 6).
It should be noted that the specific activity of purified CPOMT is very low when it is compared with the specific activities of other SAM-dependent fungal phenol methyltransferases reported previously (9, 15). This fact could be explained if halogenated phenols are not the true physiological substrates of the enzyme. In fact, CPs are xenobiotic compounds which in a few decades have become contaminants of ecosystems. Given the sort period that has lapsed since these compounds first appeared in nature, it is difficult to imagine that filamentous fungi could have acquired de novo an enzyme specifically designed to detoxify them. From this point of view, it could be speculated that the CPOMT activity detected represents the nonspecific activity of a methyltransferase involved in the methylation of some other physiological substrates, which remain to be identified.
In contrast to the wide variety of substrates used by the enzymes mentioned above, CPOMT from T. longibrachiatum was highly specific for halogenated phenols. Within this group, it displayed methylating activity with a wide range of compounds, especially CPs. Interestingly, the enzyme was able to methylate several DCPs containing a chlorine atom in position 2, showing the highest activity against 2,4-DCP, whereas it exhibited very low or undetectable activity against 3,4-DCP and 3,5-DCP. Furthermore, the only CP to be methylated was 2-CP. These results suggest that a chlorine (or halogen) substituent in position 2 relative to the hydroxyl group of the substrates is important (although not essential, as deduced from the low level of methylation of 3,4-DCP) for efficient attack of a substrate. All the TCPs tested were readily methylated, but the presence of four or five chlorine atoms resulted in a marked decrease in the activity. In addition, introduction of substituents at position 6 interfered with the catalytic action. Thus, 2,3,6-TCP and 2,4,6-TCP were methylated to a lesser extent than 2,3,4-TCP and 2,4,5-TCP. In a similar way, 2,3,4,6-TeCP and 2,3,5,6-TeCP did not support methylation, whereas 2,3,4,5-TeCP was biomethylated, although at a low rate. Also, 2,4-DCP was methylated at a rate 10 times higher than the rate of methylation of 2,6-DCP. Finally, the nature of the halogen substituents does not appear to be critical once certain steric criteria are satisfied. Remarkably, 2,4,6-TCP, 2,4,6-TBrP, and 2,4,6-TIP were O methylated, unlike 2,4,6-TFP. These data suggest that the methylation activity of CPOMT depends on the structural or steric properties rather than the electronic properties of the substituents, as has been described for the 2,4-disubstitued phenol O-methyltransferase from P. chrysosporium (9).
T. longibrachiatum CPOMT is clearly inducible. The induction is specifically carried out by CPs, especially if they contain three to five chlorine atoms, whereas phenol and other nonhalogenated phenolic compounds are unable to induce the enzyme. Moreover, as described previously, the activity is not repressed by high glucose or high ammonium contents (1). These observations are important since they imply that CPOMT is produced solely in response to the presence of these highly toxic compounds, independent of the metabolic or physiological conditions, and therefore CPOMT can be efficiently induced and synthesized to inactivate the harmful CP inducers. Our substrate specificity data, as well as the inducible character of CPOMT, suggest that the enzyme is physiologically involved in the detoxification of CPs that could cross the plasma membrane. Accordingly, this enzyme might represent a second barrier of resistance to these pesticides, superimposed on the battery of extracellular enzymes like laccases, lignin peroxidases, and manganese peroxidases that are present in a variety of fungi (5, 6), which are thought to mediate the degradation of these compounds outside the cell.
Traditionally, the filamentous fungi have been blamed as the microorganisms responsible of cork taint of wines (1, 8, 17, 18, 27). Accordingly, some authors have proposed that cork stoppers should be manufactured in a clean or microorganism-free environment since the cork stopper should be considered a part of the wine and consequently treated like a food product (8). However, we point out that the real cause of cork taint is not the presence of filamentous fungi growing on cork but the presence of CPs contaminating the cork planks. The development of both biotechnological and mechanical treatments to remove CPs from cork could be a powerful tool to obtain CP-free cork planks and accordingly cork stoppers and wine that are free of chloroanisoles.
Acknowledgments
J.-J.R.C. and M.L.A.-R. contributed similarly to this work.
We gratefully acknowledge Leocadia Franco for technical assistance and Richard Calderone for helpful comments.
This work was supported by the European Community, by the Ministerio de Educación y Cultura of Spain (grant 1FD97-1172), and also by the Junta de Extremadura (grant 2PR01A009).
REFERENCES
- 1.Álvarez-Rodríguez, M. L., L. López-Ocaña, J. M. López-Coronado, E. Rodríguez, M. J. Martínez, G. Larriba, and J. J. R. Coque. 2002. Cork taint of wines: role of the filamentous fungi isolated from cork in the formation of 2,4,6-trichloroanisole by O methylation of 2, 4,6-trichlorophenol. Appl. Environ. Microbiol. 68:5860-5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Álvarez-Rodríguez, M. L., C. Belloch, M. Villa, F. Uruburu, G. Larriba, and J. J. R. Coque. 2003. Degradation of vanillic acid and production of guaiacol by microorganisms isolated from cork samples. FEMS Microbiol. Lett. 220:49-55. [DOI] [PubMed] [Google Scholar]
- 3.Allard, A.-S., M. Remberger, and A. H. Neilson. 1987. Bacterial O-methylation of halogen-substituted phenols. Appl. Environ. Microbiol. 53:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amon, J. M., J. M. Vandepeer, and R. F. Simpson. 1989. Compounds responsible for cork taint in wine. Wine Ind. J. 4:62-69. [Google Scholar]
- 5.Bhasker Reddy, G. V., M. D. Sollewijn Gelpke, and M. H. Gold. 1998. Degradation of 2,4,6-trichlorophenol by Phanerochaete chrysosporium: involvement of reductive dechlorination. J. Bacteriol. 180:5159-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhasker Reddy, G. V., and M. H. Gold. 2000. Degradation of pentachlorophenol by Phanerochaete chrysosporium: intermediates and reactions involved. Microbiology 146:405-413. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Butzke, C. E., T. J. Evans, and S. E. Ebeler. 1999. Detection of cork taint in wine using automated solid-phase microextraction in combination with GC/MS-SIM, p. 208-216. In A. L. Waterhouse and S. E. Ebeler (ed.), Chemistry of wine flavour. ACS Symposium Series. American Chemical Society, Washington, D.C.
- 9.Coulter, C., J. T. Kennedy, W. C. McRoberts, and D. B. Harper. 1993. Purification and properties of an S-adenosylmethionine:2,4-disubstitued phenol O-methyltransferase from Phanerochaete chrysosporium. Appl. Environ. Microbiol. 59:706-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cserjesi, A. J., and E. L. Johnson. 1972. Methylation of pentachlorophenol by Trichoderma virgatum. Can. J. Microbiol. 18:45-49. [DOI] [PubMed] [Google Scholar]
- 11.Curtis, R. F., D. G. Land, N. M. Griffiths, M. G. Gee, D. Robinson, J. L. Peel, C. Dennis and, J. M. Gee. 1974. 2,3,4,6-Tetrachloroanisole: association with musty taint in chickens and microbiological formation. Nature 235:223-224. [DOI] [PubMed] [Google Scholar]
- 12.Engel, C., A. P. DeGroot, and C. Weurman. 1966. Tetrachloroanisole: a source of musty taste in eggs and broilers. Science 154:270-271. [DOI] [PubMed] [Google Scholar]
- 13.Gee, J. M., and J. L. Peel. 1974. Metabolism of 2,3,4,6-tetrachlorophenol by microorganisms from broiler house litter. J. Gen. Microbiol. 85:237-243. [DOI] [PubMed] [Google Scholar]
- 14.Harper, D. B., J. T. G. Hamilton, J. T. Kennedy, and K. J. McNally. 1989. Chloromethane, a novel methyl donor for biosynthesis of esters and anisoles in Phellinus pomaceus. Appl. Environ. Microbiol. 55:1981-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffers, M. R., W. C. McRoberts, and D. B. Harper. 1997. Identification of a phenolic 3-O-methyltransferase in the lignin-degrading fungus Phanerochaete chrysosporium. Microbiology 143:1975-1981. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lee, T. H., and R. F. Simpson. 1993. Microbiology and chemistry of cork taint in wine, p. 353-372. In G. H. Fleet (ed.), Wine microbiology and bio/technology. G. Harwood Academic Publishers, Chur, Switzerland.
- 18.Maujean, A., P. Millery, and H. Lemaresquier. 1985. Explications biochimiques et metaboliques de la confusion entre goût de bouchon et goût de moisi. Rev. Fr. Oenol. 99:55-59. [Google Scholar]
- 19.McNally, K. J., and D. B. Harper. 1991. Methylation of phenol by chloromethane in the fungus Phellinus pomaceus. J. Gen. Microbiol. 137:1029-1032. [Google Scholar]
- 20.Neilson, A. H., C. Lindgren, P.-A. Hynning, and M. Remberger. 1988. Methylation of halogenated phenols and thiophenols by cell extracts of gram-positive and gram-negative bacteria. Appl. Environ. Microbiol. 54:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nystrom, A., A. Grimvall, C. Krantzrulcker, R. Savenhed, and K. Akerstrand. 1992. Drinking-water off-flavor caused by 2,4,6-trichloroanisole. Water Sci. Technol. 25:241-249. [Google Scholar]
- 22.Perrella, F. W. 1988. EZ-FIT: a practical curve fitting microcomputer program for the analysis of enzyme kinetic data on IBM PC compatible computers. Anal. Biochem. 174:437-447. [DOI] [PubMed] [Google Scholar]
- 23.Pollnitz, A. P., K. H. Pardon, D. Liacopoulos, G. K. Skouroumounis, and M. A. Sefton. 1996. The analysis of 2,4,6-trichloroanisole and other chloroanisoles in tainted wines and corks. Aus. J. Grape Wine Res. 2:184-190. [Google Scholar]
- 24.Poulton, J. E. 1981. Transmethylation and demethylation reactions in the metabolism of secondary plant products, p. 667-723. In P. K. Stumpf and E. E. Conn (ed.), The biochemistry of plants. Academic Press, London, United Kingdom.
- 25.Ruiz, O. A., and R. A. Ugalde. 1998. Partial characterization and photolabeling of a Rhizobium meliloti polysaccharide methyltransferase with S-adenosylmethionine. Int. Microbiol. 1:225-230. [PubMed] [Google Scholar]
- 26.Silva Pereira, C., A. Pires, M. J. Valle, L. Vilas Boas, J. J. Figueiredo Marques, and M. V. San Romao. 2000. Role of Chrysonilia sitophila in the quality of cork stoppers for sealing wine bottles. J. Ind. Microbiol. Biotechnol. 24:256-261. [Google Scholar]
- 27.Silva Pereira, C., J. J. Figueiredo Marques, and M. V. San Romao. 2000. Cork taint in wine: scientific knowledge and public perception—a critical review. Crit. Rev. Microbiol. 26:147-162. [DOI] [PubMed] [Google Scholar]
- 28.Som, S., and S. Friedman. 1990. Direct photolabelling of the EcoRII methyltransferase with S-adenosyl-l-methionine. J. Biol. Chem. 265:4278-4283. [PubMed] [Google Scholar]
- 29.Spadone, J.-C., G. Takeoka, and R. Liandron. 1990. Analytical investigation of Rio off-flavor in green coffee. J. Agric. Food Chem. 38:226-233. [Google Scholar]
- 30.Tindale, C. R., F. B. Whitfield, S. D. Levingston, and T. H. L. Nguyen. 1989. Fungi isolated from packaging materials: their role in the production of 2,4,6-trichloroanisole. J. Sci. Food Agric. 49:437-447. [Google Scholar]
- 31.Whitfield, F. B., T. H. L. Nguyen, and C. R. Tindale. 1991. Effect of relative-humidity and incubation-time on the O-methylation of chlorophenols in fiberboard by Paecilomyces variotii. J. Sci. Food Agric. 55:19-26. [Google Scholar]