Abstract
Membrane microdomains (lipid rafts) are now recognized as critical for proper compartmentalization of insulin signaling. We previously demonstrated that, in adipocytes in a state of TNFα-induced insulin resistance, the inhibition of insulin metabolic signaling and the elimination of insulin receptors (IR) from the caveolae microdomains were associated with an accumulation of the ganglioside GM3. To gain insight into molecular mechanisms behind interactions of IR, caveolin-1 (Cav1), and GM3 in adipocytes, we have performed immunoprecipitations, cross-linking studies of IR and GM3, and live cell studies using total internal reflection fluorescence microscopy and fluorescence recovery after photobleaching techniques. We found that (i) IR form complexes with Cav1 and GM3 independently; (ii) in GM3-enriched membranes the mobility of IR is increased by dissociation of the IR–Cav1 interaction; and (iii) the lysine residue localized just above the transmembrane domain of the IR β-subunit is essential for the interaction of IR with GM3. Because insulin metabolic signal transduction in adipocytes is known to be critically dependent on caveolae, we propose a pathological feature of insulin resistance in adipocytes caused by dissociation of the IR–Cav1 complex by the interactions of IR with GM3 in microdomains.
Keywords: adipocyte, caveolae microdomain, lipid rafts, live cell imaging, type 2 diabetes
Interaction of gangliosides and insulin receptors (IR) was originally elaborated by Nojiri et al. (1), demonstrating the ganglioside-mediated inhibition of insulin-dependent IR activation. Later, we presented evidence that transformation to a insulin-resistant state induced in adipocytes by TNFα may depend on increased biosynthesis of the ganglioside GM3 after up-regulated gene expression of GM3 synthase, indicating that GM3 may function as an inhibitor of insulin signaling during chronic exposure to TNFα (2).
Caveolae, a subset of membrane microdomains, are particularly abundant in adipocytes (3). In these cells, insulin metabolic signal transduction is critically dependent on the caveolae, as demonstrated by the study results summarized in Table 1. Initial evidence suggesting a major role for caveolae and the resident protein caveolin-1 (Cav1) in insulin signaling came from experiments in which gold-labeled insulin was endocytosed by rat adipocytes via clathrin-independent, uncoated invaginations (4). ImmunoGold electron (5) and immunofluorescence microscopy further established that IR are highly concentrated in caveolae. Moreover, in the β-subunit of IR, Couet et al. (6) identified a motif capable of binding to the scaffold domain of Cav1. In fact, Cav1-null mice developed insulin resistance when placed on a high-fat diet (7, 8). Interestingly, insulin signaling, as measured by IR phosphorylation and its downstream targets, was selectively decreased in the adipocytes of these animals, whereas signaling in both muscle and liver cells was normal (7). This signaling defect was attributed to a 90% decrease in IR protein content in the adipocytes, with no changes in mRNA levels, indicating that Cav1 functions to stabilize the IR protein (7, 8). These studies clearly indicate the critical importance of the interaction between Cav1 and IR in executing successful insulin signaling in adipocytes (Table 1).
Table 1.
Localization of the IR in caveolae microdomains is essential for the metabolic signaling of insulin
| Function | Evidence |
|---|---|
| Direct binding of IR and Cav1 | IR has caveolin binding domain (6) |
| Coimmunoprecipitation of IR and Cav1 (18) | |
| Colocalization of IR and Cav1 | IR and Cav1 are recovered in light-density fractions in a sucrose density flotation assay (12, 39) |
| Fluorescence microscopy (5) | |
| Electron microscopy (40, 41) | |
| Insulin signaling via caveolae | Stimulation of Cav1 tyrosine phosphorylation by insulin (9, 42) |
| Cav1-deficient mice exhibit insulin resistance due to accelerated degradation of IR in adipose tissue (7, 43) | |
| Cholesterol depletion disrupts caveolae and metabolic signaling of insulin (44, 45) | |
| Accumulated GM3 eliminates IR from DRM and inhibits IR–IRS-1 signaling (12) |
Saltiel and colleagues (9) found that insulin stimulation of 3T3-L1 adipocytes is associated with the tyrosine phosphorylation of Cav1. However, in their assay conditions they found only trace levels of IR in the detergent-resistant membrane microdomains (DRM), leading them to speculate on the presence of intermediate molecule(s) bridging IR and Cav1 (10). Dissociation of IR from Cav1-containing DRM was also observed by Gustavsson et al. (5) after similar detergent treatment. However, comparisons of protein and lipid content in DRM prepared with various detergents have revealed considerable differences yet have identified Triton X-100 as being the most reliable detergent (11). Using this information we were able to establish that in normal adipocytes IR can localize to the DRM (12). In the presence of TNFα, IR is selectively eliminated from the DRM, while Cav1 remains (12). Dissociation of IR from the DRM by TNFα treatment was due to an excessive accumulation of GM3 in these microdomains, as illustrated by attenuation of the dissociation upon eliminating GM3 biosynthesis with the glucosylceramide synthase inhibitor D-PDMP (12).
GM3 and other glycosphingolipids are important components of glycosphingolipid-enriched microdomains (GEM). Technically, caveolar membrane and GEM are both Triton X-resistant, low-density membranes, collectively termed “raft,” but GEM may be independent of microdomains associated with caveolae, because they are present in cells not containing caveolae (13) and can be separated from caveolar membranes (14, 15). Recently, various indispensable approaches including the strategy used here have been developed to understand the microorganization of raft microdomains including caveolae and GEM (16, 17).
In the study presented here we have explored molecular mechanisms involved with interactions among IR, Cav1, and GM3 using coimmunoprecipitation, cross-linking, and live cell imaging. We demonstrate the dynamic segregation of raft-associated IR between caveolae and GEM and present a mechanism behind the dissociation of the IR–Cav1 complex by GM3 during the state of insulin resistance.
Results and Discussion
IR Can Form Distinct Complexes with Cav1 and GM3.
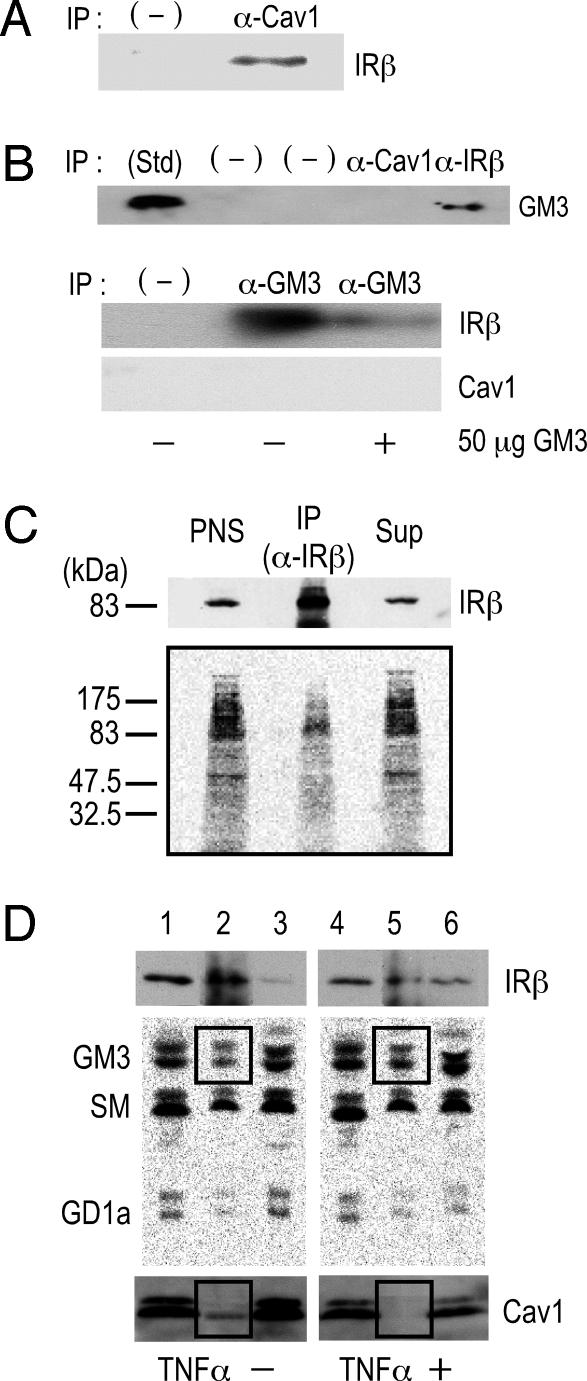
To examine interactions among IR, Cav1, and GM3 in 3T3-L1 adipocytes, we initially performed coimmunoprecipitation assays. Cav1 has a scaffolding domain to which IR and other functional transmembrane proteins bind through a caveolin binding domain in their cytoplasmic region (6, 18). As expected from another study (18), IR was coprecipitated with Cav1 (Fig. 1A). GM3 was coprecipitated with IR but not with Cav1 (Fig. 1B Upper). In addition, IR but not Cav1 was coprecipitated with GM3 (Fig. 1B Lower). Thus, IR can bind both Cav1 and GM3, but there is no interaction between GM3 and Cav1, suggesting that IR can form distinct complexes with each. The association between IR and GM3 was abolished by the presence of 50 μg of GM3, confirming the specific binding ability of the anti-GM3 antibody to GM3 in the immunoprecipitation medium (Fig. 1B Lower).
Fig. 1.
The IR forms distinct complexes with Cav1 and GM3 in 3T3-L1 adipocytes. (A) Interaction of Cav1 and IR. PNS of whole-cell lysates were immunoprecipitated with an anti-Cav1 antibody or anti-mouse IgG (−), and the precipitates were subjected to SDS/PAGE followed by immunoblotting with an anti-IRβ antibody. (B) GM3 associates with IR but not with Cav1. (Upper) PNS were immunoprecipitated with an anti-Cav1 antibody, an anti-IRβ antibody, or an negative control antibody (−). The precipitates were subjected to TLC followed by immunostaining with the anti-GM3 antibody M2590 as described in Materials and Methods. (Lower) Immunoprecipitation was performed with the anti-GM3 antibody DH2, in the presence or absence of 50 μg of GM3 or with anti-mouse IgG (−). The precipitates were then subjected to SDS/PAGE, followed by immunoblotting with an anti-IRβ or anti-Cav1 antibody. (C) GM3 directly binds to IR. A cross-linking assay of GM3 and IR in adipocytes was performed by using photoactivatable 3H-labeled GM3 described in Materials and Methods. After cross-linking, cells were then lysed and subjected to immunoprecipitation with an anti-IRβ antibody. PNS, anti-IRβ immunoprecipitates (IP), and the supernatant from the immunoprecipitation (Sup) were subjected to SDS/PAGE followed by immunoblotting with an anti-IRβ antibody and autoradiography. (D) Formation of the IR–GM3 complex is increased relative to that of the IR–Cav1 complex in adipocytes in a state of TNFα-induced insulin resistance. Glycosphingolipids in adipocytes were metabolically labeled with [3H]sphingosine, and the cells were left untreated or treated with 0.1 nM TNFα. A coimmunoprecipitation assay was performed on cell lysates by using an anti-IRβ antibody. Samples (equivalent in radioactivity) of PNS, anti-IRβ immunoprecipitate, and the corresponding supernatant, obtained from cells untreated (lanes 1–3) and treated with TNFα (lanes 4–6), were subjected to SDS/PAGE followed by immunoblotting with an anti-IRβ antibody (Top) or an anti-Cav1 antibody (Bottom). Radioactive lipids were extracted and separated by HPTLC and visualized by autoradiography. In all experiments, reproducible results were obtained, and representative data are presented.
We next examined GM3–protein interactions occurring within the plasma membrane of living cells by performing a cross-linking assay using a photoactivatable radioactive derivative of GM3 (19). Adipocytes were preincubated with [3H]GM3(N3), then irradiated to induce cross-linking of GM3. Target proteins were then separated by SDS/PAGE and visualized by autoradiography. A broad range of radioactivity reflecting GM3–protein complexes could be detected from 80 kDa to 200 kDa (Fig. 1C), suggesting a close association between GM3 and a variety of cell surface proteins, including IR. Moreover, a specific radioactive band corresponding to the 90-kDa IRβ subunit was immunoprecipitated with anti-IRβ antibodies, confirming the direct association of GM3 and IR.
In previous studies using 3T3-L1 adipocytes, we found a significant increase in cellular GM3 levels during a state of insulin resistance induced by TNFα (2, 12). Therefore, we compared the relative amounts of IR and of GM3 coimmunoprecipitated with IR in untreated cells and those treated with TNFα, using cells metabolically labeled with [3H]sphingosine. As shown in Fig. 1D, the association of GM3 with IR was clearly increased in the TNFα-treated cells. In fact, in the anti-IR immunoprecipitate obtained from control cells 15% of total lipid radioactivity is represented by the ganglioside GM3, whereas in the immunoprecipitated obtained from TNFα-treated cells the radioactivity associated with GM3 increased to 21%. Thus, the ratio between GM3 and IR in the immunoprecipitate of treated cells is 3.5-fold higher than in the immunoprecipitate of control cells. Notably, however, the association between Cav1 and IR was decreased in the treated cells.
Taken together, these results and our previous observation that GM3 regulates IR localization in DRM (12) strongly suggest the presence of distinct membrane subdomains comprising an IR–Cav1 complex in caveolae and an IR–GM3 complex in GEM. Moreover, the data indicate that the association of IR and GM3 is increased in the state of insulin resistance.
The Localization of IR in Caveolae Is Controlled by both Cav1 and GM3 in Living Cells.
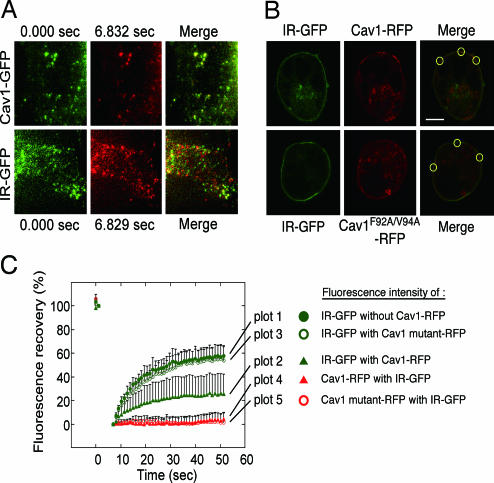
To monitor the actual fluidity and interaction of IR and Cav1 in living cells we constructed fluorescently labeled proteins and expressed them in HEK293 cells, which normally express Cav1 at low levels (7). In addition to GFP-tagged IR, GFP-tagged Cav1, and RFP-tagged Cav1, we constructed an RFP-tagged Cav1 mutant (Cav1F92A/V94A-RFP) carrying in its scaffolding domain F92A and V94A point mutations that render it unable to bind IR through its caveolin binding domain (18). We then used total internal reflection fluorescence microscopy (TIR-FM). This technique allows selective visualization of the plasma membrane and the cortical cytoplasm at the bottom surface of adherent cells (20, 21). Reportedly, the majority of Cav1-GFP molecules at the plasma membrane are static in TIR-FM (22). We confirmed that most of the Cav1-GFP molecules in our cultured cells were static during 7 sec of recording [Fig. 2A Upper and supporting information (SI) Movie 1], indicating that they were correctly sorted to caveolae. In contrast, ≈50% of the IR-GFP molecules exhibited rapid lateral movement during the same period, indicating that IR proteins are resident in both mobile and immobile fractions of the plasma membrane (Fig. 2A Lower and SI Movie 2).
Fig. 2.
Immobilization of IR by Cav1 in living cells. (A) TIR-FM analyses. Cav1-GFP and IR-GFP were expressed in HEK293 cells, and time-lapse images at the cell surface were taken (SI Movie 1 for Cav1-GFP and SI Movie 2 for IR-GFP). Selected frames in the same area between 0 sec (green) and 7 sec (pseudored) and their merged images are shown. (B) FRAP analyses. Cav1-RFP or Cav1F92A/F94A-RFP was coexpressed with IR-GFP in HEK293 cells. The area at the cell surface of aggregated Cav-1 or mutant protein was identified by confocal images (Upper), and the areas were bleached. (C) The fluorescence recovery of IR-GFP expressed alone (plot 1) or coexpressed with Cav1-RFP (plot 2) or the Cav1-RFP mutant (plot 3), and the fluorescence recovery of Cav1-RFP (plot 4) or the Cav1-RFP mutant (plot 5) coexpressed with IR-GFP were measured.
The immobile nature of some IR is likely due to an interaction with Cav1. To verify this we used a fluorescence recovery after photobleaching (FRAP) technique to measure the actual mobilities of IR and Cav1 at the plasma membrane in living cells. This method involves tagging the membrane protein of interest with a specific fluorescent group, bleaching the fluorescent group in a small area by laser, then measuring the time required for the adjacent unbleached membrane proteins to diffuse into the bleached area. In the TIR-FM experiments the Cav1 molecules in the caveolae were stable (Fig. 2A), so if the relative amounts of the IR binding to Cav1 in the bleached area increased compared with those of freely moving IR, the fluorescence recovery of IR-GFP in the area would decrease. In HEK293 cells coexpressing IR-GFP with Cav1-RFP or the Cav1-RFP mutant, IR was expressed at the cell surface uniformly, and Cav1 and its mutant were expressed as aggregates in the plasma membranes (Fig. 2B Upper). We performed FRAP analysis at the area of the aggregated Cav1, which we regarded to be caveolae, and found that, as expected, the fluorescence recovery of Cav1-RFP and the Cav1-RFP mutant was very low (<10%) (Fig. 2B Lower). In contrast, in HEK293 cells expressing only IR-GFP, the fluorescence recovery of IR 60 sec after bleaching was 60% (Fig. 2B Lower). However, the coexpression of both IR-GFP and Cav1-RFP resulted in a significant decrease in the IR recovery (25%). Importantly, there was no decrease in IR mobility in cells cotransfected with IR and the Cav1 mutant, confirming the specific interaction of the Cav1 scaffolding domain with IR in caveolae microdomains. This demonstrates the formation of an IR fraction immobilized by its binding to Cav1 in living cells.
A Lysine Residue at IR944 Is Essential for the Interaction of IR with GM3.
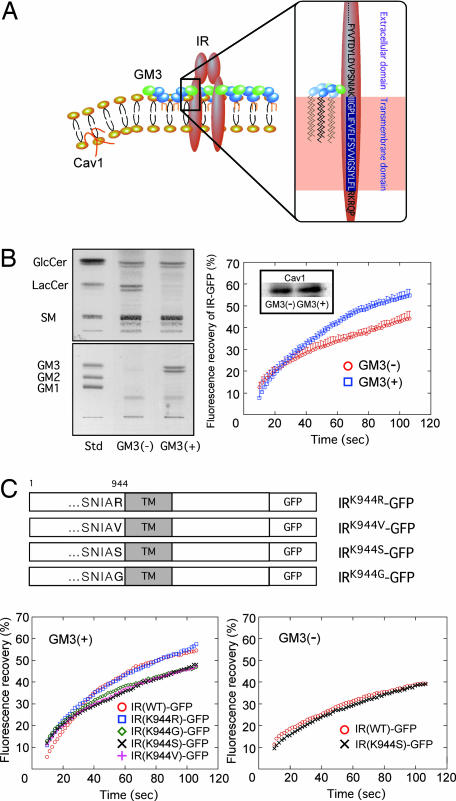
Lipids are asymmetrically distributed in the outer and inner leaflets of plasma membranes. In typical mammalian cells, most acidic phospholipids are located in the inner leaflet, and only acidic glycosphingolipids such as sulfatides and gangliosides are in the outer leaflet. The binding of proteins to lipid membranes is often mediated by electrostatic interactions between the proteins' basic domains and acidic lipids. Gangliosides, which bear sialic acid residues, exist ubiquitously in the outer leaflet of the vertebrate plasma membrane. GM3 is the most abundant ganglioside and the primary ganglioside found in adipocytes (23). Glycosphingolipids, including gangliosides, share a common minimum energy conformational structure in which the oligosaccharide chain is oriented at a defined angle to the axis of the ceramide (24). In addition, GM3 spontaneously forms clusters with its own saturated fatty acyl chains, regardless of any repulsion between the negatively charged units in the sugar chains (15). Thus, GM3 clusters with other cell surface gangliosides generate a negatively charged environment just above the plasma membrane. Conversely, IR has a sequence in its transmembrane domain, homologous among mammals, that allows presentation of the basic amino acid lysine (IR944) just above the transmembrane domain (SI Fig. 5). Therefore, during lateral diffusion an electrostatic interaction between the lysine residue at IR944 and the GM3 cluster could occur because of their proximity on the plasma membrane (Fig. 3A).
Fig. 3.
The lysine residue IR944 is essential for the interaction of IR with GM3. (A) Schematic representation of the proposed interaction of a lysine residue at IR944, which is located just above the transmembrane domain, and GM3 at the cell surface. (B) Enhanced mobility of IR in GM3-enriched membrane. (Left) Glycosphingolipid analysis of GM3-reconstituted cells [GM3 (+)] and mock cells [GM3 (−)]. Glycosphingolipids extracted from these cells, corresponding to 1 mg of cellular protein, were separated on HPTLC plates and stained with resorcinol–HCl reagent to visualize gangliosides, or with cupric acetate–phosphoric acid reagent for neutral glycosphingolipids as described in Materials and Methods. (Right) FRAP analyses. Shown is fluorescence recovery of IR-GFP in GM3 (−) and GM3 (+) cells expressing equal levels of Cav1 (Inset). (C) Specificity of the interaction between lysine at IR944 and GM3 by FRAP analyses. (Upper) Schematic structure of IR-GFP mutants in which the lysine at IR944 is replaced with basic and neutral amino acids. (Lower Left) Fluorescence recovery of IR-GFP mutants in GM3 (+) cells. (Lower Right) Fluorescence recovery of IR-GFP mutants in GM3 (−) cells.
We previously developed GM3-reconstituted cells by stably transfecting the GM3 synthase (SAT-I) gene into GM3-deficient cells (25) (Fig. 3B Left). Using the FRAP technique we examined the mobility of IR in the plasma membranes of GM3-reconstituted [GM3 (+)] cells and mock [GM3 (−)] cells expressing equal levels of Cav1 (Fig. 3B Right Inset). The mobility of IR-GFP expressed in the GM3 (+) cells was statistically (10%) higher than that in the GM3 (−) cells (Fig. 3B Right), providing further evidence that GM3 is able to enhance IR mobility by dissociating the Cav1 and IR complex in living cells.
The binding between IR and Cav1 has been studied in detail (18). To similarly understand interactions between IR and GM3 we constructed several mutants of IR in which the lysine at IR944 was replaced with the basic amino acid arginine or with the neutral amino acid valine, serine, or glutamine (Fig. 3C Upper). We first confirmed that all of the mutants were expressed on the plasma membrane similar to the wild-type protein (SI Fig. 6), then performed FRAP analysis in transfected GM3 (+) or GM3 (−) cells. The fluorescence recovery of IR(K944G), IR(K944S), and IR(K944V) 100 sec after bleaching was decreased by 10% compared with those of IR(WT) and IR(K944R) in GM3 (+) cells (Fig. 3C Lower Left). However, in GM3 (−) cells, no such difference in the mobility between IR(WT) and IR(K944S) was observed (Fig. 3C Lower Right). This demonstrates that the lysine in the wild type is essential for its binding to GM3 because of its basic charge.
Concluding Remarks
A growing body of evidence implicates glycosphingolipids including gangliosides in the pathogenesis of insulin resistance. We previously demonstrated that, in 3T3-L1 adipocytes in a state of TNF-induced insulin resistance, the inhibition of insulin metabolic signaling was associated with an accumulation of the ganglioside GM3, and, moreover, the pharmacological inhibition of GM3 biosynthesis by the glucosylceramide synthase inhibitor D-PDMP resulted in the nearly complete recovery of TNF-induced suppression of insulin signaling, suggesting a new target for therapy against insulin resistance and type 2 diabetes (2). The importance of GM3 (rather than other glycosphingolipids) in insulin signaling was further demonstrated by evidence that mice lacking GM3 synthase exhibit enhanced insulin signaling and are resistant to insulin resistance induced by a high-fat diet (26). Recently, an improved PDMP analog (27) and another type of glucosylceramide synthase inhibitor (28) were proven to have therapeutic value by oral administration in diabetic rodent models.
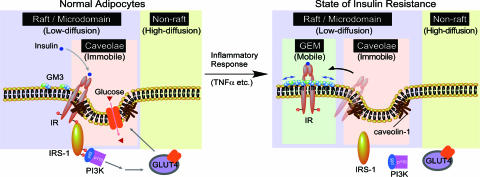
Accordingly, we have been investigating the mechanism by which GM3 inhibits insulin signaling. We previously found that elimination of IR from the caveolae microdomains was associated with an accumulation of GM3 (12). Here we have presented live cell studies of real-time lateral interactions among IR, Cav1, and GM3 at the plasma membrane, as well as relevant biochemical studies, which all together provide evidence of the dynamic segregation of IR from caveolae microdomains into GEM during the state of insulin resistance. Thus, our current and previous (2, 12) observations demonstrate that IR can form complexes with GM3 or Cav1 and that the ratio of such complexes in rafts/microdomains determines the level of insulin metabolic signaling in adipocytes (Fig. 4). In addition, our data substantiate a rationale for designing novel therapies against type 2 diabetes and related diseases based on inhibition of ganglioside biosynthesis.
Fig. 4.
Proposed mechanism behind the shift of IR from the caveolae to the GEMs in adipocytes during a state of insulin resistance. Shown is a schematic representation of raft/microdomains comprising caveolae and noncaveolae rafts such as GEM. Caveolae and GEM reportedly can be separated by an anti-Cav1 antibody (14). IR may be constitutively resident in caveolae via its binding to the scaffolding domain of Cav1 through the caveolin binding domain in its cytoplasmic region. Binding of IR and Cav1 is necessary for successful insulin metabolic signaling (Table 1). In adipocytes the localization of IR in the caveolae is interrupted by elevated levels of the endogenous ganglioside GM3 during a state of insulin resistance induced by TNFα (12). The present study has proven a mechanism, at least in part, in which the dissociation of the IR–Cav1 complex is caused by the interaction of a lysine residue at IR944, located just above the transmembrane domain, and the increased GM3 clustered at the cell surface.
Many receptor tyrosine kinases, including EGF receptor, PDGF receptor, and IR, have been shown to be localized in lipid rafts; all of these carry a caveolin binding motif in the cytoplasmic region (6). Although it has been reported that the localization of these receptors in caveolae is interrupted by elevated levels of endogenous gangliosides, the precise mechanism of this phenomenon has not been determined (reviewed in ref. 29). Interestingly, like IR, some of these other growth factor receptors present basic amino acids just above their transmembrane domains, providing spatial proximity to GEM (SI Fig. 7). We are currently pursuing studies into interactions between these receptor tyrosine kinases and the ganglioside GM3, including any pathophysiological relevance, based on our concept demonstrated here (Fig. 4).
Materials and Methods
Cell Line and Culture Conditions.
Murine 3T3-L1 preadipocytes were cultured, maintained, and differentiated as described previously (30). For chronic cytokine treatment studies, fully differentiated adipocytes were incubated for 96 h in maintenance medium in the absence or presence of 0.1 nM human TNFα (Genzyme-Techne, Minneapolis, MN) as described previously (31).
The J5 subclone of the murine 3LL Lewis lung carcinoma cell line has been described previously (32). J5 cells were maintained in RPMI medium 1640 containing 10% FBS. Cells transfected with the GM3 synthase (SAT-I) gene and mock-transfected cells (25) were cultured in the same medium containing also 300 μg/ml zeocin (Invitrogen, Carlsbad, CA). The HEK293 cell line was maintained in DMEM containing 10% FBS.
Immunoprecipitation and Immunoblotting.
3T3-L1 adipocytes were washed three times with cold PBS and scraped in cold immunoprecipitation buffer (50 mM Hepes, pH 7.0/150 mM NaCl/10% glycerol/1.0% Triton X-100/1.5 mM MgCl2/1 mM EGTA/10 mM sodium pyrophosphate/100 mM NaF/1 mM phenylmethylsulfonyl fluoride/0.15 unit/ml aprotinin/10 μg/ml leupeptin/10 μg/ml pepstatin A/1 mM sodium orthovanadate), then sonicated in ice for 5 min. The lysates were centrifuged (2,400 × g for 3 min), and their postnuclear supernatant (PNS) was obtained by removing the fat and nuclear debris, then transferred into new tubes and assayed for protein content.
Aliquots (500 μl containing 1 mg of protein) of PNS were precleared for nonspecific binding by incubating for 2 h at 4°C with 150 μl of protein G-coupled magnetic beads (Dynabeads Protein G; Invitrogen). Precleared samples were added to protein G-coupled magnetic beads (150 μl) preincubated with anti-IRβ (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Cav1 (BD Transduction Laboratories, Lexington, KY), or anti-GM3 (DH2; kindly provided by S.H.) antibodies (20 μl), and the mixture was incubated for 2 h at 4°C. The immune complexes were washed three times with immunoprecipitation buffer, the beads were recovered by centrifugation, and the bound proteins were eluted by boiling in Laemmli sample buffer for 2 min. Proteins were separated by SDS/PAGE and immunoblotted with the antibodies above. Bound lipids were eluted by chloroform/methanol (1:2, vol/vol) followed by TLC immunostaining. TLC immunoblotting was performed by the method of Taki et al. (33) using anti-GM3 IgM antibody (M2590; Nippon Biotest Laboratory, Tokyo, Japan).
Treatment of Adipocytes with a Photoactivable Radioactive Derivative of the GM3 Ganglioside.
3T3-L1 differentiated adipocytes (two 100-mm dishes per experiment) untreated or treated with 0.1 nM TNFα were incubated for 6 h with a mixture of 10 μM GM3 and 10 μM [11-3H(Neu5Ac)]GM3-N3{[3H]GM3(N3)} (34) in serum-free DMEM. After the incubation, the cells were washed five times with culture medium containing 10% FBS, then further incubated for 12 h in 10% FBS-containing culture medium without gangliosides. The cells were washed five times with cold PBS, and then 4 ml of cold PBS were added and the cells were illuminated for 45 min under UV light (λ = 360 nm). All procedures before exposure to the UV light were performed under a red safelight. Cells were collected and lysed by incubating in 2 ml of immunoprecipitation buffer and sonicating in ice for 5 min. The lysate was centrifuged (2,400 × g for 3 min), and the PNS was removed from the fat and transferred into new tubes.
[1-3H]Sphingosine Metabolic Labeling.
Fully differentiated adipocytes maintained as above were incubated in the absence or presence of 0.1 nM TNFα. On the second day of TNFα treatment cells were incubated in the presence of [1-3H]sphingosine in DMEM supplemented with 10% FBS for a 2-h pulse followed by a 48-h chase. Under these conditions, free radioactive sphingosine was barely detectable in the cells, and all cell sphingolipids were metabolically radiolabeled (35). At the end of chase period cells were washed twice with ice-cold PBS, then scraped in PBS and lysed in immunoprecipitation buffer.
Plasmid Construction.
To construct the cDNA of IR-GFP, the coding region of the human IR type A (kindly provided by Y. Kaburagi, Research Institute International Medical Center of Japan, Tokyo, Japan), with its stop codon replaced by GGA, was subcloned in frame with pEGFP-Nl (Clontech, Palo Alto, CA). For Cav1-GFP and Cav1-RFP, the cDNA coding region of the mouse Cav1, with its stop codon replaced by CTG, was subcloned in frame with pEGFP-Nl or replaced by GFP in tandem with an mRFP (36) fusion vector. For Cav1F92A/V94A-RFP, F92A and V94A point mutations were introduced into the scaffolding domain of Cav1-RFP using the mutagenic oligonucleotide 5′-TGC GAC AAA ATA C-3′ and the LA-PCR in vitro mutagenesis kit (TaKaRa, Shiga, Japan).
For IR-GFP mutants, K944R, K944V, K944G, and K944S point mutants of IR-GFP were generated by using the mutagenic oligonucleotides 5′-GTCCCGTCAAATATTGCACGGATTATC-3′, 5′-GTCCCGTCAAATATTGCAGTGATTATC-3′, 5′-GTCCCGTCAAATATTGCAGGAATTATC-3′, and 5′-GTCCCGTCAAATATTGCATCCATTATC-3′, respectively.
Lipid Analysis.
The lipids were extracted from the cell pellet, fractionated, and separated by HPTLC as described previously (37, 38).
FRAP Analysis.
For microscopy, cells were grown on LAB-TEK eight-chambered glass slides (Nalge Nunc International) and transfected by using Lipofectamine 2000 (Invitrogen). Cells were maintained in phenol-free medium containing 10 mM Hepes and 10% FBS. The confocal microscope used in the FRAP experiments was a Leica TCS SP2 equipped with a Tempcontrol 37°C stage, an argon–krypton laser, and HCX PL APO CS ×63/N.A. 1.20 water immersion objective. Bleaching of outlined regions of interest was performed at 37°C with an open pinhole, with the 488- or 594-nm laser line at full power. Recovery was observed at full laser power and 14% transmission. Leica confocal software was used to measure pixel intensity in the regions of interest. The recovery values are a percentage of prebleach values.
TIR-FM Analysis.
Cell surface dynamics of Cav1 and IR in regions of the cell closest to the coverslip were recorded for 7 sec. The images were acquired on an Olympus total internal reflection illumination system attachment for the IX-70 with an argon–krypton laser at 488 nm and a ×100/N.A. 1.45 oil immersion objective for TIR-FM, and by using Metamorph 4.0 software (Universal Imaging, Downingtown, PA).
Supplementary Material
Acknowledgments
We thank Dr. K. Saito and Y. Anada for technical assistance and Dr. R. Tsien (Department of Pharmacology, University of California at San Diego, La Jolla, CA) for providing the mRFP1 plasmid. This work was supported by research grants from Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Agency (JST) “Academic Frontier” Project for Private Universities; matching fund subsidy from MEXT (Ministry of Education, Culture, Sports, Science, and Technology) 2006–2011; and Mizutani Foundation for Glycoscience. T.S. is a Research Fellow of the Japan Society for the Promotion of Science.
Abbreviations
- FRAP
fluorescence recovery after photobleaching
- TIR-FM
total internal reflection fluorescence microscopy
- IR
insulin receptor
- GEM
glycosphingolipid-enriched microdomain
- DRM
detergent-resistant membrane microdomain
- PNS
postnuclear supernatant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703650104/DC1.
References
- 1.Nojiri H, Stroud M, Hakomori S. J Biol Chem. 1991;266:4531–4537. [PubMed] [Google Scholar]
- 2.Tagami S, Inokuchi J-i, Kabayama K, Yoshimura H, Kitamura F, Uemura S, Ogawa C, Ishii A, Saito M, Ohtsuka Y, et al. J Biol Chem. 2002;277:3085–3092. doi: 10.1074/jbc.M103705200. [DOI] [PubMed] [Google Scholar]
- 3.Razani B, Woodman SE, Lisanti MP. Pharmacol Rev. 2002;54:431–467. doi: 10.1124/pr.54.3.431. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg RI, Smith RM, Jarett L. J Cell Physiol. 1987;133:203–212. doi: 10.1002/jcp.1041330202. [DOI] [PubMed] [Google Scholar]
- 5.Gustavsson J, Parpal S, Karlsson M, Ramsing C, Thorn H, Borg M, Lindroth M, Peterson KH, Magnusson KE, Stralfors P. FASEB J. 1999;13:1961–1971. [PubMed] [Google Scholar]
- 6.Couet J, Li S, Okamoto T, Ikezu T, Lisanti MP. J Biol Chem. 1997;272:6525–6533. doi: 10.1074/jbc.272.10.6525. [DOI] [PubMed] [Google Scholar]
- 7.Cohen AW, Razani B, Wang XB, Combs TP, Williams TM, Scherer PE, Lisanti MP. Am J Physiol. 2003;285:C222–C235. doi: 10.1152/ajpcell.00006.2003. [DOI] [PubMed] [Google Scholar]
- 8.Cohen AW, Combs TP, Scherer PE, Lisanti MP. Am J Physiol. 2003;285:E1151–E1160. doi: 10.1152/ajpendo.00324.2003. [DOI] [PubMed] [Google Scholar]
- 9.Mastick CC, Brady MJ, Saltiel AR. J Cell Biol. 1995;129:1523–1531. doi: 10.1083/jcb.129.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mastick CC, Saltiel AR. J Biol Chem. 1997;272:20706–20714. doi: 10.1074/jbc.272.33.20706. [DOI] [PubMed] [Google Scholar]
- 11.Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. Proc Natl Acad Sci USA. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabayama K, Sato T, Kitamura F, Uemura S, Kang BW, Igarashi Y, Inokuchi J. Glycobiology. 2005;15:21–29. doi: 10.1093/glycob/cwh135. [DOI] [PubMed] [Google Scholar]
- 13.Fra AM, Williamson E, Simons K, Parton RG. J Biol Chem. 1994;269:30745–30748. [PubMed] [Google Scholar]
- 14.Iwabuchi K, Handa K, Hakomori S. Methods Enzymol. 2000;312:488–494. doi: 10.1016/s0076-6879(00)12933-7. [DOI] [PubMed] [Google Scholar]
- 15.Sonnino S, Mauri L, Chigorno V, Prinetti A. Glycobiology. 2007;17:1R–13R. doi: 10.1093/glycob/cwl052. [DOI] [PubMed] [Google Scholar]
- 16.Jacobson K, Mouritsen OG, Anderson RG. Nat Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 17.Parton RG, Hancock JF. Trends Cell Biol. 2004;14:141–147. doi: 10.1016/j.tcb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Nystrom FH, Chen H, Cong LN, Li Y, Quon MJ. Mol Endocrinol. 1999;13:2013–2024. doi: 10.1210/mend.13.12.0392. [DOI] [PubMed] [Google Scholar]
- 19.Ono M, Handa K, Sonnino S, Withers DA, Nagai H, Hakomori S. Biochemistry. 2001;40:6414–6421. doi: 10.1021/bi0101998. [DOI] [PubMed] [Google Scholar]
- 20.Lanni F, Waggoner AS, Taylor DL. J Cell Biol. 1985;100:1091–1102. doi: 10.1083/jcb.100.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steyer JA, Horstmann H, Almers W. Nature. 1997;388:474–478. doi: 10.1038/41329. [DOI] [PubMed] [Google Scholar]
- 22.Pelkmans L, Zerial M. Nature. 2005;436:128–133. doi: 10.1038/nature03866. [DOI] [PubMed] [Google Scholar]
- 23.Ohashi M. Lipids. 1979;14:52–57. doi: 10.1007/BF02533566. [DOI] [PubMed] [Google Scholar]
- 24.Hakomori SI. Proc Natl Acad Sci USA. 2002;99:225–232. doi: 10.1073/pnas.012540899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uemura S, Kabayama K, Noguchi M, Igarashi Y, Inokuchi J. Glycobiology. 2003;13:207–216. doi: 10.1093/glycob/cwg022. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita T, Hashiramoto A, Haluzik M, Mizukami H, Beck S, Norton A, Kono M, Tsuji S, Daniotti JL, Werth N, et al. Proc Natl Acad Sci USA. 2003;100:3445–3449. doi: 10.1073/pnas.0635898100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H, Przybylska M, Wu IH, Zhang J, Siegel C, Komarnitsky S, Yew NS, Cheng SH. Diabetes. 2007;56:1210–1218. doi: 10.2337/db06-0719. [DOI] [PubMed] [Google Scholar]
- 28.Aerts JM, Ottenhoff R, Powlson AS, Grefhorst A, van Eijk M, Dubbelhuis PF, Aten J, Kuipers F, Serlie MJ, Wennekes T, et al. Diabetes. 2007;56:1341–1349. doi: 10.2337/db06-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inokuchi J, Kabayama K. Comprehensive Glycoscience. 2007;3:733–744. [Google Scholar]
- 30.Rubin CS, Lai E, Rosen OM. J Biol Chem. 1977;252:3554–3557. [PubMed] [Google Scholar]
- 31.Guo D, Donner DB. J Biol Chem. 1996;271:615–618. doi: 10.1074/jbc.271.2.615. [DOI] [PubMed] [Google Scholar]
- 32.Inokuchi J, Jimbo M, Kumamoto Y, Shimeno H, Nagamatsu A. Clin Exp Metastasis. 1993;11:27–36. doi: 10.1007/BF00880063. [DOI] [PubMed] [Google Scholar]
- 33.Taki T, Handa S, Ishikawa D. Anal Biochem. 1994;221:312–316. doi: 10.1006/abio.1994.1418. [DOI] [PubMed] [Google Scholar]
- 34.Mauri L, Prioni S, Loberto N, Chigorno V, Prinetti A, Sonnino S. Glycoconjugate J. 2004;20:11–23. doi: 10.1023/B:GLYC.0000016738.37102.03. [DOI] [PubMed] [Google Scholar]
- 35.Prinetti A, Chigorno V, Prioni S, Loberto N, Marano N, Tettamanti G, Sonnino S. J Biol Chem. 2001;276:21136–21145. doi: 10.1074/jbc.M010666200. [DOI] [PubMed] [Google Scholar]
- 36.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 37.Macher BA, Klock JC. J Biol Chem. 1980;255:2092–2096. [PubMed] [Google Scholar]
- 38.Ledeen RW, Yu RK, Eng LF. J Neurochem. 1973;21:829–839. doi: 10.1111/j.1471-4159.1973.tb07527.x. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto M, Toya Y, Schwencke C, Lisanti MP, Myers MG, Jr, Ishikawa Y. J Biol Chem. 1998;273:26962–26968. doi: 10.1074/jbc.273.41.26962. [DOI] [PubMed] [Google Scholar]
- 40.Karlsson M, Thorn H, Danielsson A, Stenkula KG, Ost A, Gustavsson J, Nystrom FH, Stralfors P. Eur J Biochem. 2004;271:2471–2479. doi: 10.1111/j.1432-1033.2004.04177.x. [DOI] [PubMed] [Google Scholar]
- 41.Foti M, Porcheron G, Fournier M, Maeder C, Carpentier JL. Proc Natl Acad Sci USA. 2007;104:1242–1247. doi: 10.1073/pnas.0610523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimura A, Mora S, Shigematsu S, Pessin JE, Saltiel AR. J Biol Chem. 2002;277:30153–30158. doi: 10.1074/jbc.M203375200. [DOI] [PubMed] [Google Scholar]
- 43.Capozza F, Cohen AW, Cheung MW, Sotgia F, Schubert W, Battista M, Lee H, Frank PG, Lisanti MP. Am J Physiol. 2005;288:C677–C691. doi: 10.1152/ajpcell.00232.2004. [DOI] [PubMed] [Google Scholar]
- 44.Parpal S, Karlsson M, Thorn H, Stralfors P. J Biol Chem. 2001;276:9670–9678. doi: 10.1074/jbc.M007454200. [DOI] [PubMed] [Google Scholar]
- 45.Gustavsson J, Parpal S, Stralfors P. Mol Med. 1996;2:367–372. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.