Abstract
The aim of the present work was to compare the efficacies and levels of selectivity of different culture-dependent and -independent methods for analyzing bifidobacteria in human stool samples. The three different culture media used here significantly differed from each other, particularly with regard to the recovery of Bifidobacterium adolescentis. Bifidobacterium medium failed to recover B. adolescentis; Beerens medium recovered some B. adolescentis organisms (17% of total bifidobacteria), whereas tomato-Eugon medium recovered mainly B. adolescentis organisms (58% of total bifidobacteria). A culture-independent method that combines GC fractionation of bacterial community DNA and 16S rRNA sequencing indicated that B. adolescentis organisms accounted for 85% of all bifidobacteria. Methodological biases, such as those described in this paper, should be taken into account in interpreting earlier studies and designing future experiments.
The abundance of bifidobacteria is a frequently used biomarker in evaluating the efficacy of health-supporting pre- and probiotics. Numerous experiments and clinical studies have been published over the past 20 years on this topic (for reviews, see references 6, 7, 11, and 22). In spite of the important status of bifidobacteria, a standard enumeration procedure is still lacking. Selective plating has been widely used for enumeration of bifidobacteria, but the number of culture medium variations used in different studies is high (4, 8, 19, 21, 26). This fact is of concern since several studies have shown that medium composition affects the total number of bifidobacteria recovered (8, 15, 19). Also, some species of the genus Bifidobacterium are difficult to culture, whereas some grow readily on culture media when standard laboratory procedures are followed (25). Consequently, the use of different media may lead to inconsistent results and makes the direct comparison of results among different studies difficult. In this paper we compare the selectivities, recoveries of total bifidobacteria, and species distributions on three different randomly selected media which are among the ones that have been used in the literature for enumeration of bifidobacteria. Furthermore, we apply a culture-independent approach which combines GC fractionation of bacterial community DNA (1, 2) and 16S rRNA sequencing (3, 9) (GC-16S). The Ethics Committee of the Helsinki University Central Hospital, Department of Surgery, approved the study protocol.
Viability of bacteria in fecal samples.
Fresh fecal samples from 10 healthy Finnish adult volunteers (5 men and 5 women, 27 to 53 years of age) were collected and immediately transferred to an anaerobic glove box (Don Whitley, West Yorkshire, United Kingdom) (containing 80% N2, 20% CO2, and H2 for catalytic O2 removal) for further processing. To extract the bacteria, 0.5-g fecal samples were suspended in anaerobic phosphate buffer, mixed, and centrifuged at 200 × g; the supernatant with bacteria was recovered as described previously in detail for ileal samples from broiler chickens (1). The bacteria were pelleted by centrifugation and resuspended in 5 ml of anaerobic 0.9% NaCl solution. To maintain bacterial viability, the samples were kept strictly anaerobic throughout the protocol by performing all the steps in the glove box. Total bacterial numbers were determined by flow cytometry as described previously (3). The total fecal bacterial numbers in the samples analyzed ranged from 1.19 × 1011 to 3.03 ×1011 per g of feces (Table 1).
TABLE 1.
Count of live and dead bacteria in human fecesa
| Sample | Total no. of bacteria/g of feces (1011) | % Dead |
|---|---|---|
| 1 | 1.19 | 27 |
| 2 | 2.38 | 34 |
| 3 | 2.10 | 24 |
| 4 | 1.78 | 29 |
| 5 | 3.03 | 30 |
| 6 | 2.62 | 17 |
| 7 | 2.34 | 25 |
| 8 | 3.00 | 28 |
| 9 | 1.80 | 33 |
| 10 | 2.59 | 28 |
Bacteria were counted by flow cytometry by using Syto 24 stain for total bacteria and propidium iodide for dead bacteria. The average total number of bacteria per gram of feces (1011) was 2.28; the standard deviation was 0.58. The average percentage of dead bacteria was 28%; the standard deviation was 5%.
For the enumeration of dead bacteria, bacterial suspensions were appropriately diluted in oxygen-free physiological salt solution, and 1-ml samples were mixed with 5 μl of 0.6 mM propidium iodide (Molecular Probes Europe BV, Leiden, The Netherlands) and then incubated for 15 min in the dark. Propidium iodide is a high-affinity nucleic acid stain which labels only cells with compromised membranes that allow entry of the dye into the cell and thus identifies dead cells with no membrane potential. After being stained, the samples were removed from the anaerobic chamber and immediately analyzed with a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, N.J.). While it is possible that the viability of some bacteria decreased during the sample preparation, the results of the assay would change only if the bacteria lost their membranes. The individual samples analyzed differed substantially in their proportions of dead bacteria, which ranged from 17 to 34% (Table 1). The bacteria identified by staining as dead are permanently beyond any culture-based method. The demise of bacteria as they pass towards the distal colon may be due to several reasons. Readily metabolized carbohydrates for bacterial growth are depleted, and dry matter content increases as digesta moves from the proximal towards the distal colon (10). It is likely that some bacterial types tolerate these challenging conditions in the distal colon better than others and, therefore, would be effectively enriched when they are measured as a proportion of live bacteria. Clearly, potential biases should be carefully considered in designing strategies for clinical analyses, especially when assays that rely on bacterial viability are employed.
Recovery of fecal bifidobacteria on different selective media.
Fresh fecal samples from six healthy Finnish adult volunteers (three men and three women, 32 to 53 years of age) were thoroughly mixed in the anaerobic glove box to form a pooled sample which was used in all subsequent analyses. For selective plating, the sample was serially diluted in anaerobic brain heart infusion broth, and identical subsamples from the same dilution series were plated in duplicate on Bifidobacterium medium (BFM) (19), Beerens (BS) medium (4), and tomato-Eugon (TE) medium (13). It should be noted that the three media used in this study were arbitrarily chosen examples of the numerous “bifidoselective” media described in the literature (4, 8, 19, 21, 26, 27). Plates were incubated under anaerobic conditions at 37°C. Table 2 shows the colony counts after 7 days of incubation. The total bacterial density measured by flow cytometry, 1.6 × 1011 per g of feces, was used to calculate the recovery percentages of bacteria on the three different media. Viable counts on BFM, BS medium, and TE medium represented 1.1, 1.8, and 4.0% of the total bacteria, respectively, with the absolute numbers ranging from 1.7 × 109 to 6.5 × 109 per g (Table 2). All three media contained the same amount of propionic acid (0.5%, vol/vol), a factor that is believed to select for bifidobacteria. The highest recovery was found with TE medium, which also had the highest pH, 6.0.
TABLE 2.
Fecal bifidobacterial counts by selective plating
| Bifidoselective medium | Bacterial density (no./g [fresh wt]) | % of total bacteriaa |
|---|---|---|
| BFM | 1.7 × 109 | 1.1 |
| BS medium | 2.9 × 109 | 1.8 |
| TE medium | 6.5 × 109 | 4.0 |
Total counts were analyzed by flow cytometry. The count of total bacteria was 1.6 × 1011 (100% for the purpose of determining percentages of total bacteria).
Clinical studies so far have monitored total numbers of bifidobacteria, not individual species of the genus. If different bifidobacterial species have different health effects, this widely used approach is not meaningful. The importance and effects of different species can be assessed only when we have developed and employed improved identification methods for a period of time and accumulated sufficient information regarding individual species in clinical trials and epidemiological studies. In the present study we identified the organisms assumed to be bifidobacteria growing on the three selective media by randomly picking 15 colonies from each medium, subculturing them in the same medium, and purifying the total genomic DNA with a Dynabeads DNA Direct kit (Dynal A.S, Oslo, Norway) according to the instructions of the manufacturer. Partial 16S rRNA was sequenced (MWG Biotech, High Point, N.C.) with the highly conserved 16S and 18S rRNA-specific primers 536f and 907r (9). By sequencing 15 isolates, we could detect the bacteria that comprise ≥20% of all growth on the medium at a 95% confidence level. The resulting sequences were compared to the corresponding sequences of bifidobacterial type strains deposited in the Ribosomal Database Project II (http://rdp.cme.msu.edu/html/analyses_preview.html) and in GenBank (http://www.ncbi.nlm.nih.gov/BLAST/). Two different bifidobacterial phylotypes were found among the isolates (Table 3). One of them represented B. adolescentis, and the other one represented a Bifidobacterium longum-Bifidobacterium infantis-Bifodobacterium breve homology group. Isolates with 100% sequence homology to B. longum, B. infantis, and B. breve (the sequence for all these species is identical in this region) were further characterized by PCR using species-specific primers and the protocol published previously (16). All these isolates were identified as B. longum. BFM appeared to be strictly selective for bifidobacteria, whereas BS medium also recovered representatives of the genus Ruminococcus (20% of the colonies analyzed) and TE medium recovered representatives of the genus Eubacterium (8% of the colonies analyzed) in addition to bifidobacteria. BS medium, the least selective of the tested media, has been reported also by others to capture bacteria other than bifidobacteria and to underestimate total numbers of bifidobacteria (8). The most bifidoselective of the tested media, BFM (Table 3), did not recover any B. adolescentis organisms at all, whereas the other two media did. Although B. adolescentis organisms were detected on BS medium and not on BFM, the relative abundances of this bifidobacterial species on BFM and BS medium were not significantly different from each other (P > 0.05). However, TE medium captured significantly more B. adolescentis organisms than were captured by the other two media used (P < 0.05). Indeed, it is possible that B. adolescentis is more sensitive than B. longum to low pH and, therefore, was recovered well only on the high-pH TE medium.
TABLE 3.
Distribution of bifidobacterial species by different enumeration methods
| Species | % of total bifidobacteriaa
|
|||
|---|---|---|---|---|
| BFM | BS medium | TE medium | GC-16S | |
| B. longum | 100 | 83 | 42 | 15 |
| B. adolescentis | 0 | 17 | 58 | 85 |
At the 95% confidence level, the abundance of B. adolescentis on BFM and BS medium did not differ significantly, while the abundance on TE medium was higher than that on BFM or BS medium (P < 0.05). The GC-16S method allowed for a higher abundance of B. adolescentis than any of the culture-based methods (P < 0.05).
Bifidobacterial quantification by GC-16S.
In addition to using selective plating, we analyzed bifidobacteria from the same sample by a method totally independent of culturing, percent G+C fractionation combined with 16S rRNA sequencing. In percent G+C fractionation, different taxa sort to different positions during equilibrium density gradient centrifugation, and, thus, to different fractions, based on the percent G+C content of their chromosomal DNA. Any fraction with chromosomes with the selected percentages of G+C (in this study, 55 to 65%, which are relevant for bifidobacteria) can be further analyzed by the 16S rRNA sequencing approach to detect, at high resolution, the species comprising the selected percentages of G+C. The protocol used was previously described. In brief, bacteria were recovered by differential centrifugation (1) and lysed by a combination of physical (bead-beating), chemical (sodium dodecyl sulfate), and enzymatic (lysozyme and proteinase K) steps, a process which has been shown to lyse more than 99% of the bacteria present (1). The DNA isolated was fractionated according to GC percentage by bisbenzimidazole-CsCl equilibrium density gradient centrifugation (1). The fraction with 55 to 65% G+C, which contains DNA from the species of the genus Bifidobacterium (25), was subjected to further phylogenetic analysis. Partial 16S rRNA gene fragments were amplified with primers 536f and 907r and subsequently cloned and sequenced as described previously (3). All bifidobacterial 16S sequences detected represented the same B. longum and B. adolescentis phylotypes and were 100% identical to the sequences obtained from isolates on the selective plates.
The abundance of organisms with the specific bifidobacterial sequence Bifx (number of cells per gram [fresh weight]) in the fecal sample was calculated with the formula
 |
where TotFlowCount is the flow cytometric count of total bacteria per gram of feces (fresh weight), 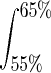 is the integral of the 55 to 65% fraction in the percent G+C profile,
is the integral of the 55 to 65% fraction in the percent G+C profile,  is the integral of the total percent G+C profile (20 to 80% G+C), BifxSeq55-65% is the number of copies of a specific bifidobacterial Bifx sequence in the 55 to 65% fraction, and TotSeq55-65% is the total number of sequences analyzed in the 55 to 65% fraction. The culture-independent GC-16S approach indicated an 85% relative abundance of bifidobacteria that were B. adolescentis, which was significantly higher than was obtained by any of the culture media tested (Table 3).
is the integral of the total percent G+C profile (20 to 80% G+C), BifxSeq55-65% is the number of copies of a specific bifidobacterial Bifx sequence in the 55 to 65% fraction, and TotSeq55-65% is the total number of sequences analyzed in the 55 to 65% fraction. The culture-independent GC-16S approach indicated an 85% relative abundance of bifidobacteria that were B. adolescentis, which was significantly higher than was obtained by any of the culture media tested (Table 3).
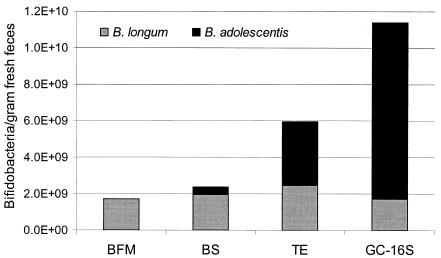
Absolute numbers of the two different bifidobacteria were calculated for different selective media by using the data presented in Tables 2 and 3. The estimated number of bacteria belonging to B. longum was close to 2 × 109 per g of feces and practically independent of the analytical approach used (Fig. 1). Conversely, the numbers of bacteria identified as B. adolescentis were highly dependent on the analytical method used, ranging from below the detection limit (<20% of total colonies) to 1010 per g of feces (Fig. 1). TE medium recovered bifidobacteria with the greatest efficiency of all media tested, recovering 3.7% of total bacteria or 6 × 109 per g of feces, which was more than twice the number obtained with the second-best BS medium but only half of the number of bifidobacteria estimated by the culture-independent GC-16S method.
FIG. 1.
Effect of method on observed number of major bifidobacteria in human feces. BFM, BS medium, and TE medium are different bifidoselective media. GC-16S values are based on direct extraction of DNA from feces, percent G+C fractionation, and 16S rRNA sequencing as described in detail previously (1, 3).
Species specificity of the bifidobacterial recovery.
When we categorized the types of bifidobacteria present, it became obvious that the differential recovery of total bifidobacteria was due to the fact that all of the culture media, to various degrees, failed to effectively recover B. adolescentis. The recovery rate for this bacterium on BFM was below detection; 100% of the isolates were identified as B. longum, whereas 85% of the total bifidobacteria were identified as B. adolescentis by the culture-independent approach. TE medium recovered B. adolescentis relatively well, indicating that 58% of the total bifidobacteria belonged to this species. We acknowledge that counting absolute numbers of bacteria by the GC-16S method may have biases arising from PCR and cloning. Nevertheless, enumeration of the B. longum cells was highly consistent; all the methods, including GC-16S, gave practically identical numbers, ∼2 × 109 cells per g of feces. We showed above that one-third of the total fecal bacteria were dead. It is possible that the fraction of injured bacteria is even higher and that the recovery of this fraction is particularly affected by the selection of the culture medium. The fact that the numbers of B. longum cells were the same by the culture-dependent and -independent methods suggests that B. longum represents a group of bacteria that tolerate passage through the colon well and can, therefore, be readily recovered also by culture-based methods. It is possible that this is not the case for B. adolescentis, and therefore its abundance has likely been underestimated where culture-based methods have been used, at least in the case of the media tested here. In some other reports B. adolescentis has been shown to grow on BFM. However, the percent recovery of B. adolescentis organisms on BFM was almost two logs lower than with some other bifidoselective media (19), and the species has been shown to be more troublesome to grow than other bifidobacteria (19, 27). Further, relative rates of recovery of fecal bacteria may also depend on the sampling procedure, freshness of samples, rate of transit of digesta in the colon, etc. With no standardized protocol for processing samples and synthesizing media, there might be subtle differences in experimental results among laboratories, even when the same medium is employed.
Molecular methods for bifidobacterial diagnostics.
PCR-based detection and quantification methods using species-specific 16S rRNA gene-based primers are gradually becoming common in the identification of bifidobacteria. Several papers have described molecular methods for the identification of bifidobacterial isolates from selective plates (14, 17, 18). However, those approaches do not eliminate concerns about bias caused by inefficient recovery when culture-based methods are used. To mitigate these concerns, analytical methods totally independent of culturability have also been developed. These are based on PCR, denaturing gradient gel electrophoresis, or fluorescent in situ hybridization analysis of bacteria or bacterial DNA directly extracted from fecal samples (5, 12, 16, 20, 23, 28).
Molecular techniques do not depend on having knowledge of the community composition and specific growth requirements of individual taxa and thus appear to have some advantages over culture-based methods for detecting the total diversity of intestinal bacterial populations. Further, bacterial populations of interest do not need to be viable or culturable as long as their nucleic acids are sufficiently intact to facilitate molecular detection strategies. It should be realized, however, that molecular approaches also have limitations. For example, many molecular techniques rely on the effective recovery of reasonably intact DNA from the constituent populations of the community. Yet, there is little knowledge available regarding rates of degradation of chromosomal DNA once a bacterium has died (i.e., the membrane potential has been lost). Thus, gastrointestinal microbial community DNA for analysis should be isolated from fresh samples as soon as possible. Methods of quantification based on the detected abundance of 16S rRNA genes should also consider different copy numbers of rRNA operons and genes in different bacterial chromosomes, which may affect the apparent relative abundance of bacteria in the sample. B. adolescentis has been reported to carry five copies of the 16S rRNA gene (23). The sequence of the total genome of B. longum was recently published and appeared to carry four copies of the 16S rRNA gene (24) (GenBank accession no. NC_004307). With the above-mentioned 16S rRNA copy numbers, the relative abundance of B. adolescentis organisms to B. longum organisms may be slightly overestimated (∼20%) when a 16S rRNA gene-based quantification method, such as GC-16S in this study, is applied.
The present study was motivated by inconsistent reports in the literature over the past 20 years of the effects of prebiotics on bifidobacteria. At this point, we cannot take any stand for or against the importance of bifidobacteria as health indicators; the evidence published to date based on a variety of media and/or techniques is insufficient. However, conclusive evidence can be obtained only if the methods used for bifidobacterial analysis are consistent and comparable and provide accurate information on the densities of different bifidobacterial species in clinical studies and epidemiological surveys.
Acknowledgments
We gratefully acknowledge Jaana Oksanen, Linda M. Schimmelpfennig, Harri Mäkivuokko, Osmo Siikanen, Jaana Larsson-Leskelä, and Brita Mäki for excellent technical assistance.
The work was financially supported by the Finnish National Technology Agency, Tekes.
REFERENCES
- 1.Apajalahti, J. H. A., L. K. Särkilahti, B. R. Mäki, J. P. Heikkinen, P. H. Nurminen, and W. E. Holben. 1998. Effective recovery of bacterial DNA and percent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl. Environ. Microbiol. 64:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apajalahti, J. H. A., A. Kettunen, M. R. Bedford, and W. E. Holben. 2001. Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens. Appl. Environ. Microbiol. 67:5656-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apajalahti, J. H. A., H. Kettunen, A. Kettunen, W. E. Holben, P. H. Nurminen, N. Rautonen, and M. Mutanen. 2002. Culture-independent microbial community analysis reveals that inulin in the diet primarily affects previously unknown bacteria in the mouse cecum. Appl. Environ. Microbiol. 68:4986-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beerens, H. 1991. Detection of bifidobacteria by using propionic acid as a selective agent. Appl. Environ. Microbiol. 57:2418-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brigidi, P., B. Vitali, E. Swennen, L. Altomare, M. Rossi, and D. Matteuzzi. 2000. Specific detection of bifidobacterium strains in a pharmaceutical probiotic product and in human feces by polymerase chain reaction. Syst. Appl. Microbiol. 23:391-399. [DOI] [PubMed] [Google Scholar]
- 6.Gibson, G. R. 1998. Dietary modulation of the human gut microflora using prebiotics. Br. J. Nutr. 80:S209-S212. [PubMed] [Google Scholar]
- 7.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 8.Hartemink, R., and F. M. Rombouts. 1999. Comparison of media for the detection of bifidobacteria, lactobacilli and total anaerobes from faecal samples. J. Microbiol. Methods 36:181-192. [DOI] [PubMed] [Google Scholar]
- 9.Holben, W. E., P. Williams, M. Saarinen, L. K. Särkilahti, and J. H. A. Apajalahti. 2002. Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon. Microb. Ecol. 44:175-185. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, I. T. 1996. Substrates for fermentation in the large bowel. Biochem. Soc. Trans. 24:824-828. [DOI] [PubMed] [Google Scholar]
- 11.Kirjavainen, P. V., and G. R. Gibson. 1999. Healthy gut microflora and allergy: factors influencing development of the microbiota. Ann. Med. 31:288-292. [DOI] [PubMed] [Google Scholar]
- 12.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langhout, D. J., J. B. Schutte, P. van Leeuwen, J. Wiebenga, and S. Tamminga. 1999. Effect of dietary high- and low-methylated citrus pectin on the activity of the ileal microflora and morphology of the small intestinal wall of broiler chicks. Br. Poult. Sci. 40:340-347. [DOI] [PubMed] [Google Scholar]
- 14.Lynch, P. A., B. J. Gilpin, L. W. Sinton, and M. G. Savill. 2002. The detection of Bifidobacterium adolescentis by colony hybridization as an indicator of human faecal pollution. J. Appl. Microbiol. 92:526-533. [DOI] [PubMed] [Google Scholar]
- 15.Martineau, B. 1999. Comparison of four media for the selection of bifidobacteria in dog fecal samples. Anaerobe 5:123-127. [Google Scholar]
- 16.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuki, T., K. Watanabe, R. Tanaka, and H. Oyaizu. 1998. Rapid identification of human intestinal bifidobacteria by 16S rRNA-targeted species- and group-specific primers. FEMS Microbiol. Lett. 167:113-121. [DOI] [PubMed] [Google Scholar]
- 18.McCartney, A. L., W. Wenzhi, and G. W. Tannock. 1996. Molecular analysis of the composition of the bifidobacterial and lactobacillus microflora of humans. Appl. Environ. Microbiol. 62:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nebra, Y., and A. R. Blanch. 1999. A new selective medium for Bifidobacterium spp. Appl. Environ. Microbiol. 65:5173-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Requena, T., J. Burton, T. Matsuki, K. Munro, M. A. Simon, R. Tanaka, K. Watanabe, and G. W. Tannock. 2002. Identification, detection, and enumeration of human Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 68:2420-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Resnick, I. G., and M. A. Levin. 1981. Quantitative procedure for enumeration of bifidobacteria. Appl. Environ. Microbiol. 42:427-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salminen, S., C. Bouley, M. C. Boutron-Ruault, J. H. Cummings, A. Franck, G. R. Gibson, E. Isolauri, M. C. Moreau, M. Roberfroid, and I. Rowland. 1998. Functional food science and gastrointestinal physiology and function. Br. J. Nutr. 80:S147-S171. [DOI] [PubMed] [Google Scholar]
- 23.Satokari, R. M., E. E. Vaughan, A. D. Akkermans, M. Saarela, and W. M. De Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sgorbati, B., B. Biavati, and D. Palenzona. 1995. The genus Bifidobacterium, p. 279-306. In B. J. B. Wood and W. H. Holzapfel (ed.), The genera of lactic acid bacteria. Chapman & Hall, Glasgow, Scotland.
- 26.Silvi, S., C. J. Rumney, and I. R. Rowland. 1996. An assessment of three selective media for bifidobacteria in faeces. J. Appl. Bacteriol. 81:561-564. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka, R., and M. Mutai. 1980. Improved medium for selective isolation and enumeration of Bifidobacterium. Appl. Environ. Microbiol. 40:866-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, R.-F., W.-W. Cao, and C. E. Cerniglia. 1996. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 62:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]