Abstract
Nuclear localization sequence (NLS)-dependent nuclear protein import is not conventionally held to require interaction with microtubules (MTs) or components of the MT motor, dynein. Here we report for the first time the role of sequences conferring association with dynein light chains (DLCs) in NLS-dependent nuclear accumulation of the rabies virus P-protein. We find that P-protein nuclear accumulation is significantly enhanced by its dynein light chain association sequence (DLC-AS), dependent on MT integrity and association with DLCs, and that P-protein-DLC complexes can associate with MT cytoskeletal structures. We also find that P-protein DLC-AS, as well as analogous sequences from other proteins, acts as an independent module that can confer enhancement of nuclear accumulation to proteins carrying the P-protein NLS, as well as several heterologous NLSs. Photobleaching experiments in live cells demonstrate that the MT-dependent enhancement of NLS-mediated nuclear accumulation by the P-protein DLC-AS involves an increased rate of nuclear import. This is the first report of DLC-AS enhancement of NLS function, identifying a novel mechanism regulating nuclear transport with relevance to viral and cellular protein biology. Importantly, this data indicates that DLC-ASs represent versatile modules to enhance nuclear delivery with potential therapeutic application.
INTRODUCTION
Nuclear protein transport is central to normal and aberrant cellular development and physiology, as well as the infectious cycles of intracellular pathogens (Poon and Jans, 2005; Hearps and Jans, 2006). The ability to exploit the cellular factors involved in nuclear targeting is of great therapeutic value as it permits the design of vehicles for the efficient delivery of drugs/therapeutic genes to the nuclear compartment (Chan and Jans, 2002; Mastrobattista et al., 2006a,b). All trafficking across the nuclear membrane occurs through the nuclear membrane embedded nuclear pore complex (NPC). The movement of molecules >45 kDa generally requires active transport, where proteins conventionally interact via a nuclear localization sequence (NLS) with import proteins (importins) which mediate docking to and transit through the NPC (Poon and Jans, 2005). Current understanding of this process focuses largely on these events at the NPC because investigation of nuclear import has conventionally used permeabilized cell systems with associated disruption of cytoplasmic structures. Thus, it is only through recent studies with live cells that it has become clear that nuclear import can involve additional levels of regulation in the cytoplasm involving the microtubule (MT) cytoskeleton (Giannakakou et al., 2000; Lam et al., 2002; Roth et al., 2007), a network composed of +/− polarized polymers of tubulin with the negative end at the MT organizing center (MTOC) located near the nucleus of nonpolarized cells. MTs are suggested to act as tracks for cargo delivery by molecular motors including the multiprotein complex motor, dynein, which mediates net movement of cargoes toward the MTOC (retrograde movement), providing directional transport from the cell periphery to the nucleus (Dohner et al., 2005).
Large bodies such as vesicles and viruses are known to rely on the cytoskeleton for trafficking in the cytoplasm, so that viruses including herpes simplex virus and human immunodeficiency virus are reliant on dynein function and MTs during infection (Dohner et al., 2005). Intriguingly, it has recently become clear that small, freely diffusible proteins including parathyroid hormone–related protein (PTHrP), p53 and retinoblastoma (Rb) protein are reliant on MTs and dynein for efficient nuclear entry (Giannakakou et al., 2000; Lam et al., 2002; Roth et al., 2007). Although the sequences responsible for p53, PTHrP, and Rb interaction with MTs/dynein remain largely undefined, short sequences from several other proteins have been identified, using the yeast two-hybrid system and proteomics approaches, which confer interaction with the dynein light chains (DLC), LC8 and Tctex-1. These sequences variously include the consensus motifs KSTQT (single letter amino acid code) and GIQVD for LC8, as well as less well-defined motifs for Tctex-1 interaction, including SKCSR (Raux et al., 2000; Alonso et al., 2001; Poisson et al., 2001; Rodriguez-Crespo et al., 2001; Mueller et al., 2002; Martinez-Moreno et al., 2003; Sugai et al., 2003; Lo et al., 2005). DLC-ASs have been suggested to mediate cargo association with dynein motor complexes to effect nuclear targeting, so that they could potentially be exploited for delivery of therapeutics to cell nuclei (Cohen et al., 2005; Mastrobattista et al., 2006a,b); however, this has never been directly demonstrated. Rather, it has been reported that DLC association can sequester proteins in the cytoplasm, inhibiting nuclear entry of protein (Ninomiya et al., 2005) or localization at the nuclear membrane (Bouillet and Strasser, 2002). Given the requirement for viral trafficking to the nucleus/perinuclear MTOC and the presence of LC8 binding DLC-ASs in numerous viral proteins, it has been conjectured that LC8 DLC-AS–containing proteins could direct viral particles to this site, although direct evidence for this is lacking (Alonso et al., 2001; Rodriguez-Crespo et al., 2001; Dohner et al., 2005; Greber and Way, 2006).
We set out to examine nuclear localization of the rabies virus phosphoprotein (P-protein [RPP]) in live cells. RPP's nucleocytoplasmic distribution is determined by an NLS and a nuclear export signal (NES). Although a DLC-AS region is present proximal to the NLS-containing domain (residues 139-174 and 174-297, respectively; see Figure 1; Raux et al., 2000; Poisson et al., 2001; Pasdeloup et al., 2005), its role in RPP subcellular localization is unclear. We find here that the DLC-AS, though having no intrinsic NLS activity, can synergize with the NLS to enhance nuclear accumulation both in situ in RPP and when expressed in fusion proteins with heterologous NLSs. This enhancement involves an increased rate of nuclear import and is dependent on both MTs and the ability to interact with DLC, with the effect also seen for several heterologous DLC-ASs. This is the first direct demonstration that DLC-ASs can facilitate NLS-dependent nuclear targeting, with important ramifications for the biology of many DLC-AS–containing viral/cellular factors, as well as identifying DLC-ASs as novel modules that may be exploited for DNA/drug delivery.
Figure 1.
Schematic diagram of (i) RPP and (ii–v) recombinant proteins used in this study. RPP139-297(*) and RPP139-174(**) were produced as wild-type protein and with D143 and Q147 mutated to alanine. GFP fusion proteins were also produced containing each individual DLC-AS and NLS module (iii–v).
MATERIALS AND METHODS
Constructs
NLS and DLC-AS modules were generated by PCR to produce fragments flanked by BamHI and BglII sites at the 5′ and 3′ ends, respectively, which in turn were flanked by Gateway recombination sites. Templates for parathyroid hormone receptor (PTHR), RPP, and p53-binding protein-1 (p53BP1) were supplied by J. Sukegawa (Tohoku University Graduate School of Medicine, Sendai, Japan), D. Blondel (Unite Mixté de Virologie Moléculaire et Structurale, Gif sur Yvette Cedex, France), and M. Zhang (Hong Kong University of Science and Technology, Hong Kong), respectively, and PCR amplified to produce PTHR478-511, RPP139-174, RPP139-297, RPP174-297, RPP139-151, and p53BP11117-1177 (positions of residues in protein amino acid sequence shown in lower case). PCR was used to mutate the D143 and Q147 residues of RPP139-174 and RPP139-297 to alanine (D143/Q147-A). NLS sequences from SV40 large T-antigen (T-ag111-135 [T-agNLS], T-ag111-135 containing the mutation K128T, and T-ag114-135), human cytomegalovirus UL44 protein (UL44422-433), and retinoblastoma protein (Rb860–876) were produced by PCR from previously described vectors (Efthymiadis et al., 1997; Alvisi et al., 2005; Wagstaff and Jans, 2006). Multimodular (DLC-AS-T-agNLS) cassettes were produced using BamHI/BglII cloning as previously described (Rosenkranz et al., 2003) and inserted into mammalian expression plasmids pEPI-GFP or pEGFPC1 fused in frame C-terminal to the coding sequence of green fluorescent protein (GFP; Figure 1). pExpress-dsRed-DLC-1 (dsRed-LC8; Finke et al., 2004) was supplied by K. Conzelmann (Ludwig Maximilians University, Munich, Germany).
Transfection and Drug Treatment
For transfection, COS-7, Vero, or HeLa cells were grown to 80% confluency on coverslips in DMEM with 10% FCS in 5% CO2 at 37°C. Transient transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, and cells were imaged 18–24 h after transfection. For cytoskeletal disruption, cells were incubated with 5 μg/ml nocodazole (NCZ; Sigma, St. Louis, MO) for 4 h or 0.5 μg/ml cytochalasin-D (CytD; Sigma) for 3 h as previously described (Roth et al., 2007). The effectiveness of these concentrations of drugs to disrupt cytoskeleton was evaluated by treating cells with or without drugs before washing in warm phosphate-buffered saline (PBS) and fixation with 4% paraformaldehyde/PBS at room temperature. Cells were then permeabilized with 0.1% Triton X-100 before staining with anti-β tubulin antibody (1/500 dilution, Cytoskeleton, Denver, CO) and Alexa 568–coupled secondary antibody (1/1000 dilution, Invitrogen, Carlsbad, CA) for MTs, or Alexa 594–labeled phalloidin (1/850 dilution, Invitrogen) for actin, before analysis by confocal laser scanning microscopy (CLSM) (below). For stabilization of the MT cytoskeleton, taxol was added to a final concentration of 1 μg/ml for 4 h as described previously (Roth et al., 2007).
CLSM and Image Analysis
Imaging of live cells was performed using a Bio-Rad MRC-600 CLSM (Richmond, CA) with a 40× water immersion objective and heated stage (Lam et al., 2002; Alvisi et al., 2005; Poon et al., 2005) or, for high-magnification images, an Olympus Fluoview 1000 with a 100× oil immersion objective and heated stage (Melville, NY; Roth et al., 2007). Fixed cells were viewed on a Bio-Rad MRC-600 CLSM with a 60× oil immersion objective (Harley et al., 2003). Analysis of digitized confocal files used Image J 1.62 public domain software (NIH) as previously described (Hubner et al., 1997; Xiao et al., 1997). Briefly, analysis was performed on confocal images of individual transfected cells. Image J software was used to select an area of the nucleoplasm equivalent to ∼30% of the nuclear area, and the mean fluorescence intensity for that region was calculated (mean nuclear fluorescence). An area of the cytoplasm of identical size was selected and the mean cytoplasmic fluorescence was obtained. Background fluorescence was acquired by performing the same operations on a neighboring nontransfected cell. The nuclear accumulation of protein was calculated by subtracting the background fluorescence from the nuclear and cytoplasmic fluorescence to obtain corrected nuclear (Fn) and cytoplasmic (Fc) fluorescence intensity values for the calculation of Fn/c for each individual cell. The Fn/c was calculated as the mean ± SEM for a transfected cell population (>60 cells). GraphPad Instat software (San Diego, CA) was used for statistical analysis. The p value comparing two populations of transfected cells was calculated using the Student's t test. Where SDs were significantly different, the analysis used the alternative Mann-Whitney test.
Fluorescence Recovery After Photobleaching
Fluorescence recovery after photobleaching (FRAP) was performed essentially as previously described (Roth et al., 2007). Briefly, COS-7 cells were transfected as above to express GFP-RPP139-174-T-agNLS or GFP-T-agNLS and visualized using an Olympus Fluoview 1000 microscope (100× oil immersion lens). To bleach the nucleus, an area covering ∼5–10% of the nucleus was selected by zooming 90-fold and was then bleached using 80% of the laser power with excitation of 488 nm (8 scans, 12.5 μs/pixel). After photobleaching of the nucleus, the cells were immediately scanned; images were collected using 3% total laser power with excitation at 488 nm (2.0× zoom, scanned 8 μs/pixel). The recovery of fluorescence was monitored by acquiring subsequent images at 20 s intervals. For some experiments, cells were treated with 5 μg/ml NCZ for 4 h before experimentation. Image analysis was performed as described above. To determine the rate of nuclear import, the results were expressed in Fn/c per second (Fn/c s−1) for the period 20–120 s.
Cell Lysis and Western Blotting
Cells were transfected as above before harvesting with trypsin, washing with PBS, and resuspension in lysis buffer (PBS containing 0.5% CHAPS and protease inhibitor cocktail (Complete, EDTA-free, Protease Inhibitor Cocktail Tablets; Roche, Indianapolis, IN). Cells were lysed over 30 min at 4°C on a rotary mixer before sedimentation at 13,000 × g at 4°C to remove insoluble material. Lysates were then subjected to SDS-PAGE (10% gel) and transferred to nitrocellulose that was probed with anti-GFP (clones 7.1 and 13.1; Roche) and goat anti-mouse IgG-HRP (A308P, Chemicon, Temecula, CA) before development using the Western Lightning Chemiluminescence Reagent (Perkin Elmer-Cetus, Wellesley, MA).
RESULTS
The RPP DLC-AS Mediates Association with LC8 In Vivo and Enhances Nuclear Accumulation by the RPP NLS Dependent on MTs
During rabies infection, “leaky scanning” using internal AUG start sites results in the production of several RPP truncated products, some of which show nuclear accumulation owing to deletion of an N-terminal NES and the presence of a NLS in the C-terminal domain (174–297; Pasdeloup et al., 2005). RPP has also been shown to interact with the DLC LC8 via a DLC-AS containing an archetypal LC8 association motif (KSTQT) in the region 139–174 using the yeast two-hybrid system (Raux et al., 2000; Poisson et al., 2001). A role in retrograde trafficking of RPP associated with dynein on MT has been suggested (Raux et al., 2000) but evidence for this is lacking, and no direct effect of the DLC-AS on transport into the nucleus has been described.
We generated the coding sequences for RPP residues 139-174 (DLC-AS–containing region), 139-297 (DLC-AS– and NLS-containing regions) and 174-297 (NLS-containing region) by PCR and cloned them downstream of GFP in the mammalian expression vector pEGFP-C1 (Figure 1). The constructs were then transfected into COS-7 cells and analyzed by CLSM. GFP-RPP139-174 showed a distribution not distinguishable from GFP alone, whereas GFP-RPP174-297 and GFP-RPP139-297 accumulated in the nucleus, but showed a significant amount of cytoplasmic localization (Figure 2A, single transfection). dsRed-LC8, by comparison, showed nuclear exclusion, with punctuate accumulation in perinuclear regions of some cells, presumably due to localization to the MTOC (Figure 2A, dual transfection). Coexpression of dsRed- LC8 with GFP-RPP139-174 and GFP-RPP139-297 resulted in a relocalization of the RPP protein to be excluded from the nucleus, with significant colocalization with dsRed-LC8 in the cytoplasm (Figure 2A, dual transfection, middle panel). However, the localization of GFP or GFP-RPP174-297 was unchanged by coexpression with dsRed-LC8 (Figure 2A, dual transfection, top and bottom panels). Thus, it appears that in live cells, RPP174-297 does not associate with dsRed-LC8 in contrast to RPP139-174 and RPP139-297, confirming that RPP139–174 confers association with LC8. Mutation of RPP residues D143 and Q147 to alanine within the DLC-AS has been reported to inhibit RPP interaction with LC8 (Poisson et al., 2001). Coexpression of GFP-RPP139-174 and GFP-RPP139-297 harboring these mutations [GFP-RPP139-174(D143/Q147-A) and GFP-RPP139-297(D143/Q147-A)] with dsRed-LC8 revealed no colocalization, confirming lack of interaction with LC8 in transfected cells (Figure 2A, dual transfection, bottom panel).
Figure 2.
GFP-fused RPP interacts via its DLC-AS region with LC8 and this region enhances NLS-dependent nuclear import via an MT-dependent mechanism. (A) Cells were transfected to express the indicated GFP fusion protein alone (single transfection) or to coexpress the indicated GFP fusion protein with dsRed-LC8 (dual transfection), and were imaged 18–24 h later by CLSM. GFP-RPP174-297 and GFP-RPP139-297 localize to the nucleus in transfected cells, in contrast to GFP-RPP139-174, which is diffusely distributed similar to GFP alone (single transfection), whereas dsRed-LC8 is predominantly cytoplasmic. GFP-RPP139-174 and GFP-RPP139-297 colocalize with dsRed-LC8 in the cytoplasm in cotransfected cells (dual transfection, middle panel), but GFP, GFP-RPP174-297, GFP-RPP139-174(D143/Q147-A) and GFP-RPP139-297(D143/Q147-A) are not affected (top and bottom panels). (B) Images such as those shown (A, single transfection) were analyzed to determine the nuclear to cytoplasmic fluorescence intensity (Fn/c), showing that RPP139-174 does not affect nuclear localization compared with GFP alone but can increase nuclear localization of RPP174-297. This effect is inhibitable by NCZ (added 4 h before imaging) but not CytD (added 3 h before imaging). Results are from single assays representative of three or more assays and are shown as mean ± SEM for >60 different cells. (C) CLSM images of cells treated with NCZ (4 h) or CytD (3 h) before appropriate staining show disruption of MT and actin cytoskeleton, respectively. (D) Image analysis of wild-type and D143/Q147-A mutated GFP-RPP139-297 show that the mutation inhibits nuclear accumulation. Results are for combined data from five assays and are shown as mean ± SEM for >350 different cells.
The extent of nuclear localization of GFP-RPP139-174, GFP-RPP174-297, and GFP-RPP139-297 was assessed by quantitative image analysis of digitized CLSM images as previously described (Hubner et al., 1997; Xiao et al., 1997). GFP-RPP139-174, which lacks the NLS region (174-297) showed no significant nuclear accumulation compared with GFP alone (Figure 2B), indicating that the DLC-AS region does not have NLS activity. Nuclear localization of GFP-RPP174-297 was increased compared with GFP alone confirming that the putative NLS of RPP (residues 174-297) can mediate nuclear accumulation (Figure 2B; Pasdeloup et al., 2005). Nuclear accumulation of GFP-RPP139-297 was significantly higher than GFP-RPP174-297 (Figure 2B). Thus, it appeared that the RPP DLC-AS (RPP139-174), though having no NLS activity per se, could facilitate nuclear import by the RPP NLS (RPP174-297). It has been suggested that DLC-ASs could assist movement toward the nucleus by mediating trafficking through associated dynein motors on MTs. We therefore tested the dependency of GFP-RPP139-297 nuclear accumulation on MT integrity by treating transfected cells with NCZ, which prevents MT assembly and, as a control, CytD, which affects the actin cytoskeleton. Accumulation by the GFP-RPP174-297 was not significantly affected by NCZ or CytD, indicating a lack of dependence of nuclear accumulation on cytoskeletal integrity (Figure 2B). However, nuclear accumulation of GFP-RPP139-297 was significantly decreased (p = 0.0011) by NCZ treatment to levels comparable to those of GFP-RPP174-297. In contrast, no decrease was seen with CytD (Figure 2B), indicating that the effect of the DLC-AS region was dependent on MT integrity but independent of the actin cytoskeleton. That the treatments with NCZ and CytD significantly affect COS-7 cell MT and actin cytoskeletal organization, respectively, was shown by immunostaining for tubulin and by using the filamentous actin-binding agent phalloidin (Figure 2C). We also assessed nuclear accumulation of GFP-RPP139-297(D143/Q147-A), finding that it was significantly reduced compared with wild type, supporting the idea of dependence of enhanced accumulation on LC8 association (Figure 2D). This data indicates that enhancement of RPP nuclear accumulation by RPP139-174 is dependent on DLC association and MT integrity.
RPP-LC8 Complexes Can Associate with MT in Live Cells
The above data indicated a functional link between RPP139-297, LC8 and MT that results in enhanced nuclear delivery of RPP139-297 compared with RPP174-297. However, physical interaction of RPP with MT via the DLC-AS has not previously been demonstrated. To investigate whether RPP-LC8 complexes can associate with MT, we used high-resolution CLSM to examine cells transfected to express GFP-RPP139-297 and GFP-RPP174-297 and cells cotransfected with dsRed-LC8. Transfected cells were treated with or without the MT stabilizing drug, taxol before imaging. No filamentous structures could be seen in cells transfected with GFP-RPP139-297 and GFP-RPP174-297 alone at 40× (Figure 2A, single transfection) or 100× (Figure 3, single transfection) magnification, with the proteins remaining diffusely distributed in cells treated with taxol (Figure 3, single transfection). However, in cells coexpressing dsRed-LC8, colocalization of dsRed-LC8 with GFP-RPP139-297 (but not GFP-RPP174-297) was seen at perinuclear regions (Figures 2A and 3, dual transfection) and, at 100× magnification, colocalization of a proportion of the GFP-RPP139-297 and dsRed-LC8 protein was observed on filamentous structures that emanated from the perinuclear region (Figure 3, dual transfection, top panel). It would appear that coexpression of dsRed-LC8 was acting to stabilize and enable visualization by CLSM of a transient interaction of GFP-RPP139-297 with endogenous LC8/MT.
Figure 3.
High magnification CLSM indicates that DLC-AS–containing RPP can form a complex with LC8 that interacts with the MT cytoskeleton in live cells. Cells were transfected to express the indicated proteins and analyzed by CLSM (100×) without (top panel) or with (bottom panel) pretreatment with the MT-stabilizing agent, taxol. GFP-RPP174-297 showed no evidence of association with MT cytoskeleton when expressed alone (left panel) or coexpressed with dsRed-LC8 (right panel), and no effect of taxol treatment was apparent. At this magnification, GFP-RPP139-297 did not show association with cytoskeletal structures when expressed alone. When GFP-RPP139-297 was coexpressed with dsRed-LC8, a proportion of the transfected proteins colocalized at a perinuclear region and on filamentous structures that converged at the perinuclear region (see yellow colorations in merged image). Treatment with taxol did not affect the appearance of cells expressing GFP-RPP139-297 alone, but in cells coexpressing GFP-RPP139-297 and dsRed-LC8, the filamentous structures on which the transfected proteins were colocalized became more defined.
The organization of the observed filaments is consistent with that of the MT network, with the MTOC located at a perinuclear region where the plus ends of MTs are anchored and the minus ends project outward (Dohner et al., 2005). The diffuse cytoplasmic appearance of protein in GFP-RPP174-297/dsRedLC8–cotransfected cells was unaffected by taxol, but in GFP-RPP139-297/dsRedLC8–cotransfected cells, the filaments on which the proteins colocalized formed more elongated, defined structures (Figure 3, dual transfection, bottom panel), strongly implying that the observed filamentous structures are indeed MTs.
Heterologous Fusion Proteins Containing DLC-AS and NLS Associate with DLC In Vivo
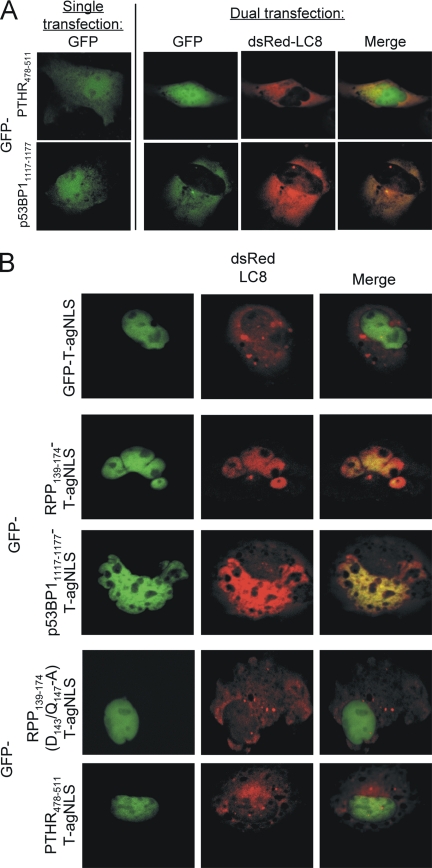
The data described above indicated that the RPP DLC-AS does not have intrinsic NLS activity, but can act in synergy with an NLS to enhance nuclear accumulation. To test the ability of DLC-ASs from alternative sources (PTHR residues 478-511 and p53BP1 residues 1117-1177; Sugai et al., 2003; Lo et al., 2005) to facilitate nuclear import, we cloned the relevant sequences downstream of GFP and expressed the proteins in COS-7 cells. As can be seen in Figure 4A (single transfection), the GFP-DLC-AS showed subcellular distributions not distinguishable from that of GFP alone (compare with Figure 2A); image analysis confirmed that the nuclear accumulation by both GFP-DLC-ASs was not increased compared with GFP alone (not shown). We confirmed that p53BP11117-1177 could associate with LC8 (Lo et al., 2005) by coexpression with dsRed-LC8, which resulted in colocalization in the cytoplasm (Figure 4A, dual transfection). PTHR478-511 did not colocalize with dsRed-LC8, as it is a DLC-AS for the light-chain Tctex-1 (Sugai et al., 2003; Figure 4A, dual transfection).
Figure 4.
Heterologous DLC-ASs are able to confer interaction with DLC to fusion proteins. Cells were transfected and analyzed by CLSM as described in the legend to Figure 2. (A) GFP-p53BP11117-1177 and GFP-PTHR478-511 show subcellular distribution that is not different from GFP alone (single transfection, compare with Figure 2A). In cotransfected cells, GFP-p53BP11117-1177, but not GFP-PTHR478-511, colocalizes with dsRed-LC8 in the cytoplasm (dual transfection). (B) GFP-DLC-AS fusion proteins containing the T-agNLS are localized to the nucleus of transfected cells. dsRed-LC8 protein colocalizes in the nucleus with GFP-RPP139-174-T-agNLS and GFP-p53BP11117-1177-T-agNLS (middle panel), but not GFP-T-agNLS, GFP-PTHR478-511-T-agNLS, or GFP-RPP139-174(D143/Q147-A)-T-agNLS (top and bottom panels).
To test whether DLC-ASs can generally affect nuclear localization mediated by an NLS, we produced DLC-ASs fused to a heterologous NLS. The coding sequences for the RPP, PTHR, and p53BP1 DLC-ASs were cloned in frame upstream of SV40 large T-antigen (T-ag) residues 111-135, which constitute the modular T-agNLS and which has previously been used out of context to facilitate the delivery of molecules of interest to the nucleus (Rosenkranz et al., 2003). When fused to the T-agNLS, all of the DLC-AS–containing fusion proteins were predominantly nuclear (Figure 4B). Coexpression of dsRed-LC8 with GFP-DLC-AS-T-agNLS resulted in clear colocalization of LC8 with GFP-RPP139-174-T-agNLS and GFP-p53BP11117-1177-T-agNLS (Figure 4B, middle panel), but not with the negative controls GFP-T-agNLS, GFP-RPP139-174(D143/Q147-A)-T-agNLS, or GFP-PTHR478-511-T-agNLS (Figure 4B, top and bottom panels), all of which are unable to bind to or show impaired binding to LC8. In contrast to GFP-RPP139-297, which colocalized with dsRed-LC8 in the cytoplasm (Figure 2A, middle panel), GFP-RPP139-174-T-agNLS and GFP-p53BP11117-1177-T-agNLS colocalized with dsRed-LC8 in the nucleus (Figure 4B, middle panel). Thus, GFP-RPP139-297 was able to associate with dsRed-LC8 via its DLC-AS region (RPP139-174), but was unable to localize the protein in the nucleus because the RPP-NLS is relatively weak; in contrast, the strong T-agNLS, when fused to the RPP or p53BP1 DLC-ASs, could “piggy-back” dsRed-LC8 into the nucleus.
DLC-AS Enhance Nuclear Localization Conferred by the T-agNLS
We next examined the capacity of RPP DLC-AS to influence nuclear localization conferred by the T-agNLS. Analysis of digitized CLSM images revealed a significant increase in the extent of nuclear accumulation of GFP-RPP139-174-T-agNLS compared with GFP-T-agNLS in transfected COS-7 (Figure 5, A and B), Vero and HeLa cells (not shown).
Figure 5.
RPP DLC-AS enhances nuclear localization mediated by T-agNLS dependent on MT integrity and LC8 association. Cells were transfected with the indicated GFP fusion protein and CLSM images were used to calculate the Fn/c ratio as described in the legend to Figure 2. (A) CLSM images of representative cells are shown, with the brightness of all images increased using Image J software. Cytoplasmic fluorescence can be discerned in GFP-T-agNLS with and without NCZ treatment, but little or no cytoplasmic fluorescence can be seen in cells expressing GFP-RPP139-174-T-agNLS unless the cells are treated with NCZ. (B) GFP-RPP139-174-T-agNLS accumulates in the nucleus of transfected cells to a greater extent than GFP-T-agNLS. This increase is inhibitable by NCZ treatment or mutation of D143/Q147-A. No effect by NCZ is observed on nuclear accumulation of GFP-T-agNLS or GFP-RPP139-174(D143/Q147-A)-T-agNLS. (C) CytD treatment does not affect nuclear accumulation of GFP-T-agNLS or GFP-RPP139-174-T-agNLS. (D) Western blot analysis of transfected cell lysates reveals that all GFP-fused proteins show similar expression levels/stability and NCZ treatment does not affect the RPP139-174-T-agNLS protein species. (E) RPP139-151 enhances nuclear localization conferred by T-agNLS to the same levels as RPP139-174 (GFP-RPP139-151-T-agNLS and GFP-RPP139-174-T-agNLS, respectively), and both are equally affected by NCZ treatment. Results are from single assays representative of three or more assays and are shown as mean ± SEM for >60 different cells.
We tested the effect of NCZ, CytD and D143/Q147-A mutation on T-agNLS–mediated nuclear accumulation. As reported previously (Roth et al., 2007), and in common with the RPP-NLS, accumulation conferred by the T-agNLS alone was not significantly affected by NCZ or CytD treatment (Figure 5, B and C). The enhanced nuclear accumulation of GFP-T-agNLS conferred by RPP139–174 was significantly inhibited by NCZ treatment or D143/Q147-A mutation, but not by CytD treatment (Figure 5, B and C). Nuclear accumulation of GFP-RPP139-174(D143/Q147-A)-T-agNLS was not significantly affected by NCZ treatment (Figure 5B). Thus, DLC-AS–dependent enhancement of nuclear localization conferred by the T-agNLS requires both MT integrity and interaction with DLC.
To exclude the possibility that the different levels of observed nuclear fluorescence might stem from degradation of the fusion proteins in the transfected cells, we prepared extracts from cells transfected to express GFP, GFP-T-agNLS, GFP-RPP139-174-T-agNLS, GFP-RPP139-174 (D143/Q147-A)-T-agNLS, as well as cells expressing GFP-RPP139-174-T-agNLS, which had been treated with NCZ. As can be seen in Figure 5D, the proteins were similarly expressed and produced comparable patterns in SDS-PAGE/Western blot analysis.
The key residues for LC8 association by RPP are found in the region 139-151 (Poisson et al., 2001), but functional sufficiency of this 13 residue fragment has not been demonstrated. We examined the effect of this fragment on nuclear localization mediated by the T-agNLS, finding that localization was not significantly different from that imparted by the larger RPP fragment, 139–174, and showed the same dependency on NCZ (Figure 5E). Thus, this 13-residue sequence is sufficient to mediate MT-dependent enhancement of NLS-dependent nuclear accumulation by the DLC-AS.
DLC-AS Increases the Rate of Nuclear Accumulation of the T-agNLS with Dependence on MT
The effects of NCZ described above suggested that MT disruption inhibits the ability of RPP139-174 to enhance nuclear import conferred by T-agNLS. Because the NCZ is added 4 h before CLSM (performed 18–24 h after transfection), it is likely that the effect of NCZ is to decrease the rate of nuclear import of newly synthesized protein that has not already accumulated in the nucleus.
To directly address whether RPP139-174 can enhance the rate of nuclear import conferred by the T-agNLS in live cells, we performed FRAP (Roth et al., 2007). The fluorescent protein in the nuclei of single COS-7 cells expressing either GFP-T-agNLS or GFP-RPP139-174-T-agNLS was photobleached using a 488-nm laser as described in Materials and Methods, and the accumulation of unbleached cytoplasmic fluorescent protein in the prebleached nucleus was followed by capturing CLSM images at 0 s after bleaching and then at 20 s intervals for periods up to 800 s. To examine the effects of MT on nuclear import kinetics, some samples were pretreated with NCZ for 4 h before FRAP.
To quantify the rate of nuclear accumulation, Fn/c values were calculated for cell images captured at each time point. We found that the accumulation of proteins in the nucleus was linear over the first 200 s after photobleaching (Figure 6A). As can be seen in Figure 6A, the Fn/c increased more rapidly for GFP-RPP139-174-T-agNLS than GFP-RPP139-174-T-agNLS after NCZ treatment or GFP-T-agNLS with or without NCZ treatment.
Figure 6.
RPP DLC-AS increases the rate of nuclear accumulation by T-agNLS dependent on MT integrity. Cells were transfected with the indicated GFP fusion protein for analysis by FRAP as described in Materials and Methods. Cells were treated with or without NCZ (added 4 h before analysis) and at 18–24 h after transfection, the nuclei of single cells expressing the protein were photobleached. CLSM images were then captured at 20 s time periods between 0 and 200 s. The Fn/c ratio at each time period was calculated to assess nuclear accumulation of unbleached cytoplasmic fluorescent protein (fluorescence recovery) over time. (A) shows Fn/c plotted linearly against time from single cells expressing the indicated proteins (the R2 for GFP-RPP139-174-T-agNLS, GFP-RPP139-174-T-agNLS+NCZ, GFP-T-agNLS and GFP-T-agNLS+NCZ were 0.98, 0.99, 0.86, and 0.99, respectively). (B) Data such as those shown in A were used to calculate the average rate of nuclear import (Fn/c s−1). The data are shown as mean ± SEM for GFP-RPP139-174-T-agNLS (data from eight different cells), GFP-RPP139-174-T-agNLS+NCZ (data from eight different cells), GFP-T-agNLS (data from 12 different cells), and GFP-T-agNLS+NCZ (data from nine different cells). The rate of nuclear import of GFP-RPP139-174-T-agNLS is significantly diminished by NCZ treatment to levels that are not different from that of GFP-T-agNLS treated with or without NCZ. Treatment of GFP-T-agNLS with NCZ did not affect the rate of nuclear accumulation.
The average rate of import (Fn/c s−1) for GFP-T-agNLS and GFP-RPP139-174-T-agNLS with or without NCZ treatment was calculated as described in Materials and Methods. This showed that nuclear import of GFP-RPP139-174-T-agNLS occurs at a significantly faster rate than that of GFP-T-agNLS (Figure 6B). Treatment with NCZ significantly decreased the rate of accumulation of GFP-RPP139-174-T-agNLS to levels not different from GFP-T-agNLS. Thus the DLC-AS–dependent increase in the rate of nuclear import of T-agNLS is dependent on the integrity of MT. The rate of nuclear import of GFP-T-agNLS was unaffected by NCZ, indicating that MT do not influence the rate of T-agNLS mediated nuclear import, this being consistent with previous observations (Roth et al., 2007).
Multiple DLC-ASs Can Affect NLS-dependent Nuclear Accumulation as Independent Modules
We next tested the ability of other DLC-ASs to facilitate T-agNLS–mediated nuclear import. Nuclear accumulation by the T-agNLS was found to be enhanced significantly by both PTHR478–511 and p53BP11117-1177 (Figure 7A). To test the modular nature of DLC-AS in enhancing NLS function, we compared the effect on nuclear accumulation of the RPP DLC-AS when it was present at either the 5′ (GFP-RPP139-174-T-agNLS) or 3′ (GFP-T-agNLS-RPP139-174) with respect to the T-agNLS. Both proteins showed similar enhancement of nuclear accumulation conferred by T-agNLS (Figure 7B). To further demonstrate that RPP139-174 can act as an independent module to enhance NLS function, we expressed GFP-fused proteins containing RPP139-174 and the NLSs from human cytomegalovirus UL44 (422-433, containing the minimal NLS sequence lacking enhancing phosphorylation sites; Alvisi et al., 2005) and Rb (amino acids 860-876, a bipartite NLS). Nuclear accumulation conferred by each NLS was significantly enhanced by the RPP DLC-AS (Figure 7C).
Figure 7.
Various different DLC-ASs can act as independent modules to enhance NLS-mediated nuclear association, and DLC-ASs can enhance the accumulation conferred by heterologous NLSs. Cells were transfected and analyzed as described in the legend to Figure 2. (A) Nuclear accumulation of GFP-T-agNLS in transfected cells is significantly enhanced by the DLC-ASs p53BP11117-1177 (GFP-p53BP11117-1177-T-agNLS) and PTHR478-511 (GFP-PTHR478-511-T-agNLS). (B) The RPP139-174 DLC-AS increases nuclear accumulation conferred by T-agNLS in both GFP-RPP139-174-T-agNLS and GFP-T-agNLS-RPP139-174. (C) RPP139-174 significantly enhances nuclear accumulation conferred by the NLSs from hCMV UL44 (left panel) and Rb (right panel). Results are from a single assay representative of three or more separate assays and are shown as mean ± SEM for >60 different cells.
Finally, we observed that RPP139–174 could enhance nuclear accumulation conferred by the T-agNLS harboring a K128T mutation, which is reduced in importin α/β recognition and thereby, nuclear import (Wagstaff and Jans, 2006), confirming that the functionality of weak NLSs can be enhanced by DLC-ASs. Similar observations were made using T-agNLS–truncated for residues 111–113, which lacks the serines phosphorylated by protein kinase CK2 that enhance α/β-importin recognition and nuclear targeting activity (Hubner et al., 1997; Xiao et al., 1997; not shown).
DISCUSSION
We show here for the first time that DLC-ASs containing consensus DLC association motifs can enhance NLS-dependent nuclear accumulation, dependent on MT integrity and DLC association, using a variety of NLSs and several distinct DLC-ASs. The enhanced nuclear accumulation is associated with an increase in the rate of nuclear import. We found that complexes of LC8 and RPP containing a DLC-AS can associate with the MT cytoskeleton of living cells, indicating that the MT-dependent effects on nuclear import involve physical interaction of DLC-AS-NLS protein, LC8, and MT.
A key feature of our study is that MT-dependent enhancement of nuclear accumulation appears to be modular in that short DLC-AS regions (13–60 residues) can be fused to heterologous proteins to confer this effect. In the absence of an NLS, DLC-AS–containing proteins did not show significant nuclear accumulation although they could associate with DLC. Thus, DLC-ASs contain no intrinsic NLS activity so that, although they are likely to associate with endogenous DLC, they do not mediate active import into the nucleus. DLC-ASs, however, clearly are able to enhance nuclear localization mediated by quite diverse NLSs: the RPP-NLS involves amino acids separated by 52 residues in the primary sequence but proximally located on the tertiary structure of the folded protein (Mavrakis et al., 2004; Pasdeloup et al., 2005); its interaction with importins is unknown. The T-ag and UL44 NLSs, conversely, are short, linear sequences (PKKKRKV and PNTKKQK, respectively), whereas the Rb NLS is bipartite in nature (KR-11 amino acid spacer-KKLR); all three of these NLSs mediate nuclear import through the action of the importin α/β heterodimer (Efthymiadis et al., 1997; Hubner et al., 1997; Xiao et al., 1997; Alvisi et al., 2005). These NLSs differ in their capacity to confer nuclear accumulation to a protein, with the RPP NLS being a much weaker NLS than the others, and the effect of the DLC-ASs on NLS-mediated import depends on the strength of the NLS used: the effect of RPP DLC-AS on RPP NLS-mediated import is quite subtle, whereas that of the same DLC-AS on T-agNLS–mediated import is considerably greater. This is consistent with a facilitating or synergistic activity of DLC-ASs with NLSs. We also found that the RPP DLC-AS is functional in terms of enhancing nuclear import whether 5′ or 3′ relative to the NLS, implying that DLC-ASs are wholly autonomous modules, able to enhance NLS-dependent nuclear accumulation.
Our data are consistent with the possibility that DLC-AS interaction with DLC could facilitate association with the dynein motor on MT, permitting retrograde transport to the nucleus, resulting ultimately in increased transport through the NPC and into the nuclear compartment. However, it remains possible that alternative mechanisms involving DLC and MT are involved in mediating enhanced nuclear import. There are reports of interaction of α-importins with dynein in neuronal cells and with MTs in plant cells (Smith and Raikhel, 1998; Hanz et al., 2003) so it is conceivable that the DLC-AS could bring NLS-containing proteins to the MTs, where their interaction with MT-associated importins is enhanced. However, general functional interaction of α-importins with dynein/MT would imply a universal dependence of α/β-importin cargo on MT for nuclear import. Our data indicate that this is not the case, as nuclear accumulation of numerous cargoes is unaffected by MT disruption, and, furthermore, we have been unable to detect importin β-MT association in COS-7 cells (see here and Roth et al., 2007).
The novel discovery that DLC-ASs act as modules to enhance nuclear accumulation indicates that they may be useful for therapeutic applications. This is of particular relevance to the development of “artificial viruses” (AVs), in which there is considerable research interest owing to the identification of significant pathogenesis in viral-mediated gene delivery/gene therapy trials (Glover et al., 2005; Mastrobattista et al., 2006b). Nonviral approaches should provide a safe alternative to viral gene delivery, but are believed to be inefficient, largely owing to the poor nuclear delivery of DNA (Chan and Jans, 2002; Glover et al., 2005). AVs are intended to overcome these problems by incorporating modules that mimic efficient nuclear targeting by viruses (e.g., cell-specific binding/uptake, endosomal escape, and nuclear import; Cohen et al., 2005; Glover et al., 2005; Alvisi et al., 2006; Mastrobattista et al., 2006a,b), while avoiding the pathogenicity associated with viral approaches. We have successfully utilized modular approaches to achieve nuclear drug delivery relevant to chemotherapy (Rosenkranz et al., 2003). It has been suggested that DLC-ASs could optimize nonviral delivery of therapeutics by mimicking the MT-dependent trafficking of viruses toward the nucleus (Cohen et al., 2005; Mastrobattista et al., 2006a,b), but this has been speculative given the prior lack of evidence that DLC association can enhance NLS activity. The data here indicates that DLC-AS may be incorporated into pre-existing or novel AVs along with other heterologous targeting modules to enhance nuclear delivery. Association with MTs may endow additional desirable properties for nonviral delivery of DNA, including protection from cytosolic degradation.
The impetus for this study came from our initial observations that nuclear import by the RPP NLS was enhanced by its DLC-AS. The demonstration of DLC-AS involvement in RPP nuclear import identifies a novel mechanism which, along with the previously identified RPP NLS and NES, regulates nucleocytoplasmic trafficking of RPP. Regulated nuclear accumulation of RPP may be important in evasion of antiviral mechanisms and the capacity of RPP to associate with LC8 has been shown to be important to infectious viral spread (Rasalingam et al., 2005; Chelbi-Alix et al., 2006). The DLC-AS motif of P-protein is highly conserved in the lyssavirus genus along with the capacity to bind LC8, with the NES and proposed NLS sequences also conserved (Jacob et al., 2000; Mavrakis et al., 2004; Pasdeloup et al., 2005), meaning that this trafficking mechanism is likely to be common among lyssavirus P-proteins as well as other viral/cellular DLC-AS/NLS–containing proteins.
In conclusion, the data here not only demonstrate an in situ function for DLC-ASs in nuclear targeting of RPP, but also the potential utility of DLC-ASs as modules to enhance NLS-dependent nuclear accumulation for nuclear targeted therapies. The application of DLC-ASs in modular AVs represents the basis of further research in this laboratory.
ACKNOWLEDGMENTS
The authors thank Dr. G. Alvisi for critical reading of the manuscript. This research was supported by the National Health and Medical Research Council, Australia (fellowship 143790/333013/384109 and project grant 384107). We also acknowledge D. Blondel, J. Sukegawa, M. Zhang, and K. Conzelmann for kindly supplying plasmid DNA.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0030) on June 13, 2007.
REFERENCES
- Alonso C., Miskin J., Hernaez B., Fernandez-Zapatero P., Soto L., Canto C., Rodriguez-Crespo I., Dixon L., Escribano J. M. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 2001;75:9819–9827. doi: 10.1128/JVI.75.20.9819-9827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvisi G., Jans D. A., Guo J., Pinna L. A., Ripalti A. A protein kinase CK2 site flanking the nuclear targeting signal enhances nuclear transport of human cytomegalovirus ppUL44. Traffic. 2005;6:1002–1113. doi: 10.1111/j.1600-0854.2005.00331.x. [DOI] [PubMed] [Google Scholar]
- Alvisi G., Poon I. K., Jans D. A. Tumor-specific nuclear targeting: promises for anti-cancer therapy? Drug Resist. Update. 2006;9:40–50. doi: 10.1016/j.drup.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Bouillet P., Strasser A. BH3-only proteins—evolutionarily conserved proapoptotic Bcl-2 family members essential for initiating programmed cell death. J. Cell Sci. 2002;115:1567–1574. doi: 10.1242/jcs.115.8.1567. [DOI] [PubMed] [Google Scholar]
- Chan C. K., Jans D. A. Using nuclear targeting signals to enhance non-viral gene transfer. Immunol. Cell Biol. 2002;80:119–130. doi: 10.1046/j.1440-1711.2002.01061.x. [DOI] [PubMed] [Google Scholar]
- Chelbi-Alix M. K., Vidy A., El Bougrini J., Blondel D. Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, Stat1, and PML nuclear bodies. J. Interferon Cytokine Res. 2006;26:271–280. doi: 10.1089/jir.2006.26.271. [DOI] [PubMed] [Google Scholar]
- Cohen R. N., Rashkin M. J., Wen X., Szoka F. C., Jr Molecular motors as drug delivery vehicles. Drug Discov. Today Technol. 2005;2:111–118. doi: 10.1016/j.ddtec.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Dohner K., Nagel C. H., Sodeik B. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol. 2005;13:320–327. doi: 10.1016/j.tim.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Efthymiadis A., Shao H., Hubner S., Jans D. A. Kinetic characterization of the human retinoblastoma protein bipartite nuclear localization sequence (NLS) in vivo and in vitro. A comparison with the SV40 large T-antigen NLS. J. Biol. Chem. 1997;272:22134–22139. doi: 10.1074/jbc.272.35.22134. [DOI] [PubMed] [Google Scholar]
- Finke S., Brzozka K., Conzelmann K. K. Tracking fluorescence-labeled rabies virus: enhanced green fluorescent protein-tagged phosphoprotein P supports virus gene expression and formation of infectious particles. J. Virol. 2004;78:12333–12343. doi: 10.1128/JVI.78.22.12333-12343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakakou P., Sackett D. L., Ward Y., Webster K. R., Blagosklonny M. V., Fojo T. p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat. Cell Biol. 2000;2:709–717. doi: 10.1038/35036335. [DOI] [PubMed] [Google Scholar]
- Glover D. J., Lipps H. J., Jans D. A. Towards safe, non-viral therapeutic gene expression in humans. Nat. Rev. Genet. 2005;6:299–310. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- Greber U. F., Way M. A superhighway to virus infection. Cell. 2006;124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Hanz S., Perlson E., Willis D., Zheng J. Q., Massarwa R., Huerta J. J., Koltzenburg M., Kohler M., van-Minnen J., Twiss J. L., Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Harley V. R., Layfield S., Mitchell C. L., Forwood J. K., John A. P., Briggs L. J., McDowall S. G., Jans D. A. Defective importin beta recognition and nuclear import of the sex-determining factor SRY are associated with XY sex-reversing mutations. Proc. Natl. Acad. Sci. USA. 2003;100:7045–7050. doi: 10.1073/pnas.1137864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearps A. C., Jans D. A. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta dependent mechanism. Biochem. J. 2006;398:475–484. doi: 10.1042/BJ20060466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner S., Xiao C. Y., Jans D. A. The protein kinase CK2 site (Ser(111/112)) enhances recognition of the Simian virus 40 large T-antigen nuclear localization sequence by importin. J. Biol. Chem. 1997;272:17191–17195. doi: 10.1074/jbc.272.27.17191. [DOI] [PubMed] [Google Scholar]
- Jacob Y., Badrane H., Ceccaldi P., Tordo N. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J. Virol. 2000;74:10217–10222. doi: 10.1128/jvi.74.21.10217-10222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. H., Thomas R. J., Loveland K. L., Schilders S., Gu M., Martin T. J., Gillespie M. T., Jans D. A. Nuclear transport of parathyroid hormone (PTH)-related protein is dependent on microtubules. Mol. Endocrinol. 2002;16:390–401. doi: 10.1210/mend.16.2.0775. [DOI] [PubMed] [Google Scholar]
- Lo K.W.H., Kan H. M., Chan L. N., Xu W. G., Wang K. P., Wu Z. G., Sheng M., Zhang M. J. The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation. J. Biol. Chem. 2005;280:8172–8179. doi: 10.1074/jbc.M411408200. [DOI] [PubMed] [Google Scholar]
- Martinez-Moreno M., Navarro-Leida I., Roncal F., Albar J. P., Alonso C., Gavilanes F., Rodriguez-Crespo I. Recognition of novel viral sequences that associate with the dynein light chain LC8 identified through a pepscan technique. FEBS Lett. 2003;544:262–267. doi: 10.1016/s0014-5793(03)00516-7. [DOI] [PubMed] [Google Scholar]
- Mastrobattista E., Bravo S. A., van der Aa M., Crommelin D.J.A. Nonviral gene delivery systems: from simple transfection agents to artificial viruses. Drug Discov. Today Technol. 2006a;2:103–109. doi: 10.1016/j.ddtec.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Mastrobattista E., van der Aa M., Hennink W. E., Crommelin D.J.A. Artificial viruses: a nanotechnological approach to gene delivery. Nat. Rev. Drug Discov. 2006b;5:115–121. doi: 10.1038/nrd1960. [DOI] [PubMed] [Google Scholar]
- Mavrakis M., McCarthy A. A., Roche S., Blondel D., Ruigrok R. W. Structure and function of the C-terminal domain of the polymerase cofactor of rabies virus. J. Mol. Biol. 2004;343:819–831. doi: 10.1016/j.jmb.2004.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S., Cao X., Welker R., Wimmer E. Interaction of the poliovirus receptor CD155 with the dynein light chain Tctex-1 and its implication for poliovirus pathogenesis. J. Biol. Chem. 2002;227:7897–7904. doi: 10.1074/jbc.M111937200. [DOI] [PubMed] [Google Scholar]
- Ninomiya K., Ishimoto T., Taguchi T. Subcellular localization of PMES-2 proteins regulated by their two cytoskeleton-associated domains. Cell. Mol. Neurobiol. 2005;25:899–911. doi: 10.1007/s10571-005-4955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasdeloup D., Poisson N., Raux H., Gaudin Y., Ruigrok R. W., Blondel D. Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology. 2005;334:284–293. doi: 10.1016/j.virol.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Poisson N., Real E., Gaudin Y., Vaney M. C., King S., Jacob Y., Tordo N., Blondel D. Molecular basis for the interaction between rabies virus phosphoprotein P and the dynein light chain LC8, dissociation of dynein-binding properties and transcriptional functionality of P. J. Gen. Virol. 2001;82:2691–2696. doi: 10.1099/0022-1317-82-11-2691. [DOI] [PubMed] [Google Scholar]
- Poon I., Oro C., Dias M., Zhang J., Jans D. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res. 2005;65:7059–7064. doi: 10.1158/0008-5472.CAN-05-1370. [DOI] [PubMed] [Google Scholar]
- Poon I. K., Jans D. A. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Rasalingam P., Rossiter J. P., Mebatsion T., Jackson A. C. Comparative pathogenesis of the SAD-L16 strain of rabies virus and a mutant modifying the dynein light chain binding site of the rabies virus phosphoprotein in young mice. Virus Res. 2005;111:55–60. doi: 10.1016/j.virusres.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Raux H., Flamand A., Blondel D. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 2000;74:10212–10216. doi: 10.1128/jvi.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Crespo I., Yelamos B., Roncal F., Albar J. P., de Montellano P.R.O., Gavilanes F. Identification of novel cellular proteins that bind to the LC8 dynein light chain using a pepscan technique. FEBS Lett. 2001;503:135–141. doi: 10.1016/s0014-5793(01)02718-1. [DOI] [PubMed] [Google Scholar]
- Rosenkranz A. A., et al. Recombinant modular transporters for cell-specific nuclear delivery of locally acting drugs enhance photosensitizer activity. FASEB J. 2003;17(9):1121–1123. doi: 10.1096/fj.02-0888fje. [DOI] [PubMed] [Google Scholar]
- Roth D. M., Moseley G. W., Glover D., Pouton C. W., Jans D. A. A microtubule-facilitated nuclear import pathway for cancer related proteins. Traffic (OnlineAccepted Articles) 2007 doi: 10.1111/j.1600-0854.2007.00564.x. doi: 10.1111/j.1600-0854.2006.00564.x. [DOI] [PubMed] [Google Scholar]
- Smith H. M., Raikhel N. V. Nuclear localization signal receptor importin alpha associates with the cytoskeleton. Plant Cell. 1998;10:1791–1799. doi: 10.1105/tpc.10.11.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai M., Saito M., Sukegawa I., Katsushima Y., Kinouchi Y., Nakahata N., Shimosegawa T., Yanagisawa T., Sukegawa J. PTH/PTH-related protein receptor interacts directly with Tctex-1 through its COOH terminus. Biochem. Biophys. Res. Commun. 2003;311:24–31. doi: 10.1016/j.bbrc.2003.09.157. [DOI] [PubMed] [Google Scholar]
- Wagstaff K. M., Jans D. A. Intramolecular masking of nuclear localization signals: analysis of importin binding using a novel AlphaScreen-based method. Anal. Biochem. 2006;348:49–56. doi: 10.1016/j.ab.2005.10.029. [DOI] [PubMed] [Google Scholar]
- Xiao C. Y., Hubner S., Jans D. A. SV40 large tumor antigen nuclear import is regulated by the double-stranded DNA-dependent protein kinase site (serine 120) flanking the nuclear localization sequence. J. Biol. Chem. 1997;272:22191–22198. doi: 10.1074/jbc.272.35.22191. [DOI] [PubMed] [Google Scholar]