Abstract
The human genome has three unique genes coding for kinesin-13 proteins called Kif2a, Kif2b, and MCAK (Kif2c). Kif2a and MCAK have documented roles in mitosis, but the function of Kif2b has not been defined. Here, we show that Kif2b is expressed at very low levels in cultured cells and that GFP-Kif2b localizes predominately to centrosomes and midbodies, but also to spindle microtubules and transiently to kinetochores. Kif2b-deficient cells assemble monopolar or disorganized spindles. Chromosomes in Kif2b-deficient cells show typical kinetochore-microtubule attachments, but the velocity of movement is reduced ∼80% compared with control cells. Some Kif2b-deficient cells attempt anaphase, but the cleavage furrow regresses and cytokinesis fails. Like Kif2a-deficient cells, bipolar spindle assembly can be restored to Kif2b-deficient cells by simultaneous deficiency of MCAK or Nuf2 or treatment with low doses of nocodazole. However, Kif2b-deficient cells are unique in that they assemble bipolar spindles when the pole focusing activities of NuMA and HSET are perturbed. These data demonstrate that Kif2b function is required for spindle assembly and chromosome movement and that the microtubule depolymerase activities of Kif2a, Kif2b, and MCAK fulfill distinct functions during mitosis in human cells.
INTRODUCTION
Chromosome segregation during mitosis and meiosis is mediated by a microtubule-based structure called the spindle (Compton, 2000; Wittman et al., 2001). The assembly of microtubules into bipolar spindles and many aspects of chromosome behavior occur through the concerted actions of motor and nonmotor microtubule-associated proteins. Nonmotor proteins such as NuMA, TPX2, TOGp, and TACC promote the formation of bipolar spindles with focused poles (Raff, 2002; Fant et al., 2004; Gard et al., 2004; Gruss and Vernos, 2004). In addition, at least 12 of the ∼41 members of the kinesin family of motor proteins identified in mammals have important functions in mitosis (Sharp et al., 2000; Zhu et al., 2005). For example, Kid and CENP-E influence chromosome behavior by generating polar ejection force on chromosome arms (Antonio et al., 2000; Funabiki and Murray, 2000; Levesque and Compton, 2001) and driving chromosome congression (Kapoor et al., 2006), respectively. Eg5 is essential for the establishment of bipolar spindles (Sawin et al., 1992), and Kif4a, Kif4b, MKLP1, and MKLP2 play crucial roles in the assembly and function of the central spindle during anaphase (Mishima et al., 2002; Kurasawa et al., 2004). Finally, minus end–directed microtubule motors, HSET and cytoplasmic dynein, cross-link and focus microtubule minus ends at spindle poles (Heald et al., 1996; Gaglio et al., 1997; Walczak et al., 1997; Mountain et al., 1999).
The dynamic properties of microtubules are also crucial for spindle assembly and chromosome movement (Kline-Smith and Walczak, 2004). Microtubules grow and shrink by addition and loss of tubulin subunits at their ends, and compounds that perturb microtubule dynamics are valuable chemotherapeutic agents because they impair the timely progression of mitosis by disrupting spindle assembly and chromosome movement (Jordan and Wilson, 2004). Chromosome motion is tightly coupled to microtubule dynamics as chromosomes follow the growing and shrinking ends of microtubules through their attachment site at kinetochores. In anaphase A, sister chromatids separate and move poleward as kinetochore-bound microtubules shorten by loss of tubulin subunits from both kinetochore-bound plus- and pole-associated minus ends (Sharp and Rogers, 2004).
Important regulators of microtubule dynamics during mitosis are the kinesin-13 proteins (Wordeman, 2005). This branch of the kinesin superfamily of proteins is defined by the localization of the conserved kinesin motor domain in the middle of the polypeptide (Lawrence et al., 2004). Kinesin-13 proteins are nonmotile and induce microtubule depolymerization by disassembling tubulin subunits from the polymer end (Desai et al., 1999). The human genome has three distinct genes encoding members of the kinesin-13 family that are called Kif2a (chromosome 5q12), Kif2b (chromosome 17q22), and MCAK/Kif2c (chromosome 1p34). MCAK is the best-characterized member of the family and localizes to spindle poles, spindle midzone, and kinetochores (Wordeman and Mitchison, 1995) in addition to associating with the tips of growing microtubules (Moore et al., 2005). Perturbation of MCAK function in cultured cells using dominant negative mutant expression (Maney et al., 1998; Kline-Smith et al., 2004) or RNA interference (RNAi; Ganem et al., 2005) has minimal effects on bipolar spindle assembly or chromosome movement, but leads to increases in the frequency of lagging chromatids at anaphase. This suggests that MCAK utilizes microtubule depolymerase activity to destabilize inappropriate (merotelic) microtubule attachments at kinetochores, a function that has been shown to be controlled by Aurora B kinase (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004; Knowlton et al., 2006). On the other hand, Kif2a is essential for both bipolar spindle assembly and chromosome movement (Ganem and Compton, 2004). It localizes to spindle poles in human cells, and cells lacking Kif2a form monopolar spindles instead of bipolar spindles in mitosis. Kif2a disassembles microtubules at their minus ends at spindle poles in association with poleward microtubule flux, and this activity makes a small contribution to poleward chromosome movement in anaphase (Ganem et al., 2005). In contrast to MCAK and Kif2a, the function of the remaining member of the kinesin-13 family, Kif2b, has not been explored. Here we test the mitotic function of Kif2b and demonstrate that it is essential for spindle assembly, chromosome movement, and cytokinesis. We also use functional criteria to show that each kinesin-13 family member fulfills a different function during mitosis in human cells.
MATERIALS AND METHODS
Cell Culture
Human U2OS and RPE-1 cells were maintained in Dulbecco's modified medium and DMEM:F12 medium, respectively, each containing 10% fetal bovine serum, 50 IU/ml penicillin, and 50 μg/ml streptomycin and grown at 37°C in a 5% CO2 atmosphere.
Antibodies
The region of the cDNA encoding the N-terminal 143 amino acids of human Kif2b (clone BM560261) was amplified using forward (5′-TCTCGAGCTCCATCACTCACCATGGC-3′) and reverse (5′-TGAATTCTTCATCAGACAAG GTTTGCTG-3′) primers and inserted into the XhoI/EcoRI sites of pRSET-C plasmid (Invitrogen, Carlsbad, CA). The His-tagged recombinant protein was expressed in BL21-Gold Escherichia coli and purified by affinity chromatography using a nickel-agarose column. Further purification of Kif2b was done by electroeluting the recombinant protein from an SDS-PAGE gel. Purified recombinant protein was dialyzed against phosphate-buffered saline (PBS) and used to immunize two rabbits (Covance Research Products, Richmond, CA). Other antibodies used in this study included the tubulin-specific mAb DM1α (Sigma-Aldrich, St. Louis, MO), Hec1-specific mAb (Novus Biologicals, Littleton, CO), GFP-specific rabbit serum (provided by William Wickner, Dartmouth Medical School), actin-specific mAb (provided by Harry Higgs, Dartmouth Medical School), CLASP1-specific rabbit antibody (Fedor Severin, Moscow State University), Kif2a-specific rabbit antibody (Ganem and Compton, 2004), MCAK-specific rabbit antibody (Mack and Compton, 2001), and CENP-E-specific mAb (Tim Yen, Fox Chase Cancer Center).
RNAi
Kif2a, MCAK, and Nuf2 levels were depleted using published sequences (Ganem et al., 2005). The sequence of the sense strands of the Kif2b siRNA duplexes were 5′-GGACCUGGAUAUCAUCACC-3′ and 5′-GGCAAGAAGAUUGACCUGG-3′. The sense strand of the CLASP1 small interfering RNA (siRNA) duplex was 5′-CGACACAUAUCAGUAUUAG-3′. All siRNA duplexes were synthesized with 3′ dTdT overhangs, HPLC purified, and were annealed (Ambion, Austin, TX). Approximately 30,000 U2OS cells were plated on coverslips in 6-well dishes or 20,000 cells on coverslips in 12-well dishes the day before transfection and grown without antibiotics. Double-stranded RNAs at a final concentration of 100–200 nM were transfected into cells using Oligofectamine reagent (Invitrogen) as described previously (Ganem and Compton, 2004; Manning and Compton, 2007). Cells were analyzed 48–72 h after transfection by either indirect immunofluorescence or immunoblot analysis.
Immunoblotting
For immunoblots, cultured cells were solubilized directly in 1× SDS-PAGE sample buffer. Total cell protein was then separated by size using SDS-PAGE and transferred to polyvinylidene difluoride membrane (Millipore, Bedford, MA). Primary antibodies were incubated for 1 h at room temperature in 1% milk Tris-buffered saline (TBS). Primary antibody was then detected using horseradish peroxide (HRP)-conjugated secondary antibodies (Bio-Rad, Richmond, CA) diluted in TBS for an additional 1 h. at room temperature. The signal was then detected using chemiluminescence. Total protein extracts of human brain, heart, skeletal muscle, liver, lung, kidney, spleen, placenta, ovary, and testis were obtained from Clontech (Palo Alto, CA).
Differential Interference Contrast and Indirect Immunofluorescence Microscopy
For live cell imaging, glass coverslips seeded with human U2OS cells were mounted on a stainless steel modified Rose chamber containing growth medium and sealed with VALAP (Vaseline, lanolin, and paraffin wax in a 1:1:1 mass ratio). The growth chamber was placed on a heated stage that was prewarmed to 37°C. Differential interference contrast (DIC) images were captured at 1-min intervals with a Hamamatsu Orca II CCD (charge-coupled device) camera (Bridgewater, NJ) mounted on a Zeiss Axioplan 2 microscope (Thornwood, NY) using a 63×, 1.4 NA objective. Chromosome traces and velocities were determined by measuring the movement of the central region of single chromosomes over time by DIC microscopy as previously described (Ganem et al., 2005). For indirect immunofluorescence, U2OS cells were extracted in either microtubule-stabilizing buffer (4 M glycerol, 100 mM PIPES, pH 6.9, 1 mM EGTA, 5 mM MgCl2, and 0.5% Triton X-100), or calcium-containing buffer (100 mM PIPES, pH 6.8, 1 mM MgCl2, 0.1% Triton X-100, 1 mM CaCl2) followed by fixation in 1% glutaraldehyde (microtubule and Hec1 staining), 3.5% paraformaldehyde (Kif2a and MCAK antibodies), or cold methanol (Kif2b, CENP-E, and CLASP1 antibodies). Subsequent antibody incubations and washes were done in TBS-BSA (10 mM Tris, pH 7.5, 150 mM NaCl, and 1% bovine serum albumin). Primary antibodies were detected using species-specific fluorescein– or Texas red–conjugated secondary antibodies (Vector Laboratories, Burlingame, CA). DNA was detected with 0.2 μg/ml DAPI (Sigma-Aldrich). Coverslips were mounted with ProLong Antifade mounting medium (Molecular Probes, Eugene, OR). Fluorescent images of fixed cells were captured with a Hamamatsu Orca ER cooled CCD camera mounted on a Nikon TE-2000E microscope (Melville, NY) with a 60×, 1.4 NA objective or a Hamamatsu Orca II CCD camera mounted on an Axioplan 2 Zeiss microscope with a 63×, 1.4 NA objective. A series of 0.25-μm optical sections were collected in the z-axis for each channel (DAPI, fluorescein, and/or Texas red). Iterative Restoration was performed on images using Phylum software (Improvision, Lexington, MA) for images acquired on the Nikon, or Openlab software (Improvision) for images acquired on the Zeiss microscope. Selected planes from the z-series were then overlaid to generate the final image. Areas occupied by chromosomes on monopolar spindles were measured in DAPI-stained cells from multiple, individual z-planes using the lasso tool in the Openlab software to circumscribe all chromosomes.
Quantification of MT Depolymerase Activity
Human Kif2b was amplified by PCR from IMAGE consortium clone 5170969 and ligated into the EcoRI and SmaI sites of pEGFP-C1. Clone (pmx230) integrity was verified by DNA sequencing. The microtubule quantification was performed as described in Ovechkina et al. (2002). Briefly, EGFP, EGFP-MCAK, and EGFP-Kif2b fusion constructs were transfected into cultured cells, fixed, and stained for tubulin. Cells expressing a level of EGFP-MCAK in which all microtubule polymer was absent by visual inspection were selected for quantitative comparison to EGFP-Kif2b. All transfected cells were matched for fluorescent green fluorescent protein (GFP) levels. Cells transfected with a control enhanced GFP (EGFP) construct showed normal levels of microtubule (MT) polymer and were assigned an MT polymer value of 1.0. Microtubule polymer content was determined in mitotic cells after transfection with siRNA after extraction, fixation, and staining for tubulin as described above. A series of 0.25-μm optical sections were collected in the z-axis for DAPI and fluorescein channels with a Hamamatsu Orca ER cooled CCD camera mounted on a Nikon TE-2000E microscope with a 60×, 1.4 NA objective. The lasso tool in Phylum software (Improvision) was used to circumscribe microtubules and total voxel intensities were obtained. Total voxel intensities were averaged in 50 cells for each condition.
Microinjection
Interphase U2OS cells growing on photoetched alphanumeric glass coverslips (Bellco Glass, Vineland, NJ) were microinjected using an automated injector (Femtojet, Eppendorf, Fremont, CA). IgG was purified from whole serum for microinjection by affinity chromatography using protein A–conjugated agarose (Roche Molecular Biochemicals, Indianapolis, IN). PD-10 Sephadex G-25 columns (Amersham Pharmacia Biochemicals, Piscataway, NJ) were used for buffer exchange to microinjection buffer (100 mM KCl, 10 mM KPO4, pH 7.0), followed by concentration using Centricon spin columns (Millipore). The following antibody concentrations refer to their concentrations in the microinjection needle: 10 mg/ml NuMA/15 mg/ml HSET (mixed) and 19 mg/ml nonimmune IgG. Interphase cells were injected 48 h after transfection and processed for imaging 12 h later.
Phylogenetic Tree
The phylogenetic tree was prepared using amino acid sequences for MCAK (human, NM_006845.2; mouse, BC006841.1), Kif2a (human, NM_004520.1; mouse, BC006803.1) and Kif2b (human, NM_032559.3; mouse, BC100484.1) for both Homo sapiens and Mus musculus, and Klp10A (NM_132459.2), KLP59C (NM_137915.3), and KLP59D (NM_137918.2) for Drosophila melanogaster. The sequence of budding yeast Kip3 (NC_001139.7) was used as an outgroup to root the tree. Standard Clustal X parameters were used for alignment (Jeanmougin et al., 1998). The tree branches are not to scale.
RESULTS
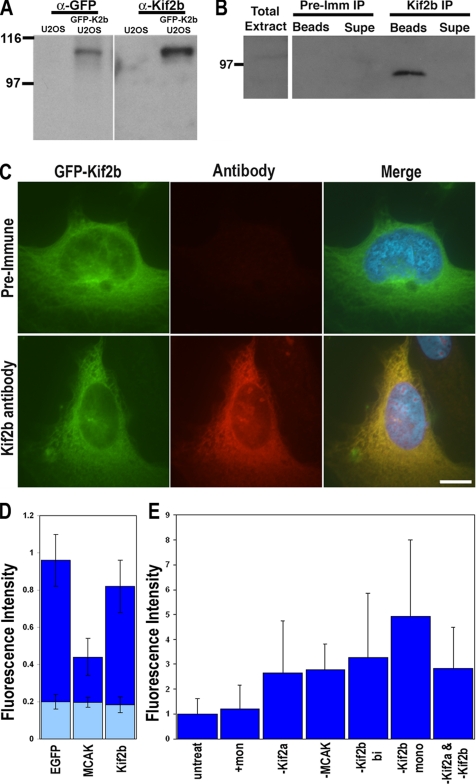
To characterize human Kif2b, we generated Kif2b-specific antibodies by immunizing rabbits with the unique N-terminal 143 amino acids. Immunoblots of extracts prepared from human U2OS cells over expressing GFP-Kif2b show a reactive protein of ∼105 kDa with the Kif2b antibodies only in cells expressing GFP-Kif2b (Figure 1A). GFP-specific antibodies also specifically detect this protein, demonstrating that it is the GFP-Kif2b fusion protein. The Kif2b-specific antibodies also show colocalization with GFP in cells expressing GFP-Kif2b (Figure 1C). Thus, our antibodies recognize Kif2b, but immunoblots of total cell extracts from cultured human cells (HeLa or U2OS) do not show detectable signal without overexpression (Figure 1, A and B). Immunoblots with defined quantities of purified recombinant protein demonstrate that these antibodies could detect Kif2b if it were present at >1000 copies per cell (data not shown), indicating that Kif2b may not be abundant. To test if endogenous Kif2b is below detection levels of immunoblotting total cell extracts of these cells, we used our antibodies to immunoprecipitate extracts prepared from 107 mitotic HeLa cells and blotted the precipitates with the Kif2b-specific antibody. With this approach the immune pellets represent a 100-fold increase in cell content relative to the supernatant fraction or total cell extracts. Although precipitation with a preimmune antibody shows no detectable signal, precipitation with the Kif2b-specific antibody shows a reactive band of ∼80 kDa (Figure 1B), consistent with the predicted size of Kif2b. Immunoblots of total cell protein from different human tissues also show a reactive protein of ∼80 kDa (Supplementary Figure 1), although some tissues show a slightly different size protein (∼75 kDa). The signal intensity is undetectable in some tissues, weak in many other tissues, and strong in lung. These data demonstrate that human Kif2b expression is variable in different tissues and very low in the human cultured cell lines HeLa and U2OS consistent with RT-PCR data previously shown by Zhu et al. (2005).
Figure 1.
Kif2b expression in cultured cells. (A) Total cell extract from untransfected U2OS cells (U2OS) or U2OS cells transfected with the plasmid encoding GFP-Kif2b (GFP-K2b U2OS) were separated by size by SDS-PAGE and blotted with GFP-specific antibodies (α-GFP) and the Kif2b polyclonal serum (α-Kif2b). (B) Extracts prepared from ∼107 mitotic HeLa cells were precipitated with either preimmune serum (Pre-Imm IP) or Kif2b polyclonal serum (Kif2b IP) and separated into soluble (Supe) and antibody-bound bead (Beads) fractions. The entire pellet fraction, and only ∼1% of the supernatant and total cell extract fractions were separated by size by SDS-PAGE and blotted with Kif2b polyclonal serum. Positions of β-galactosidase (116) and phosphorylase B (97) are shown in kiloDaltons. (C) U2OS cells were transfected with the plasmid encoding GFP-Kif2b, fixed, and stained with DAPI and either preimmune serum or Kif2b polyclonal serum. Scale bar, 5 μm. (D) Fluorescence intensity for EGFP, MCAK, and Kif2B are reported as a ratio of tubulin fluorescence of transfected cells to that of untransfected cells within the same field of view (dark blue bars). The average GFP fluorescence in the transfected cells was measured for each transfected cell and arbitrarily normalized as an indication of the level of expressed transfected protein (light blue bars). Error bars, SDs for each dataset. The microtubule polymer level in the Kif2b transfected cells was significantly lower than that of EGFP control cells (p < 0.01), indicating that Kif2b is a microtubule depolymerizing kinesin. (E) Fluorescent intensities of tubulin polymer content in mitotic U2OS cells that were untreated, treated with 100 μM monastrol (+mon), or transfected with MCAK-, Kif2a-, and/or Kif2b-specific siRNA (Kif2b). Error bars, SDs.
All kinesin-13 family members studied to date catalyze the disassembly of microtubules. To determine if Kif2b can disassemble microtubules, we quantified tubulin levels in CHO cells expressing GFP-Kif2b using the assay developed by Ovechkina et al. (2002). Chinese hamster ovary (CHO) cells transfected with GFP-Kif2b show significantly reduced tubulin fluorescence after 16 h relative to cells transfected with GFP alone (Figure 1D; t test, p < 0.05). The magnitude of reduction in tubulin fluorescence levels in cells expressing GFP-Kif2b in this assay is similar to that measured previously for neuronally expressed GFP-Kif2a-α (Moore et al., 2005). Next, we extracted nonpolymerized tubulin from cells before fixation and quantified microtubule polymer levels in mitotic cells after transfection with Kif2b-specific siRNA (details on siRNA are described below). Kif2b-deficient mitotic cells have, on average, three- to fourfold greater microtubule polymer than untreated control cells with bipolar spindles or mitotic cells with monopolar spindles induced by inhibition of Eg5 activity (Figure 1E). This is a significant increase in microtubule polymer content (t test, p < 0.05) and is observed in Kif2b-deficient cells with either monopolar or bipolar spindles. These results are consistent with Kif2b having microtubule depolymerization activity like other kinesin-13 family members MCAK and Kif2a.
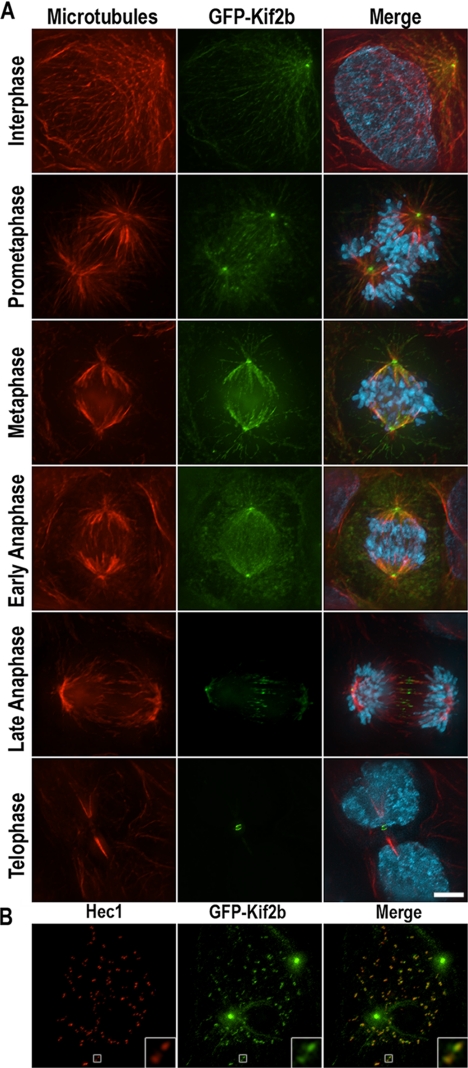
When expressed in U2OS cells, GFP-Kif2b localizes predominantly to centrosomes in interphase and throughout mitosis (Figure 2A). It also associates with spindle microtubules and demonstrates pronounced localization to the spindle midzone and midbody in late anaphase and telophase. Finally, it localizes to punctate structures near chromosomes in prometaphase. Costaining with Hec1, a component of the Ndc80 kinetochore complex that localizes to outer kinetochores, demonstrates that these punctate structures are kinetochores and that GFP-Kif2b colocalizes with Hec1 at outer kinetochores (Figure 2B). Kinetochore localization of GFP-Kif2b tends to be strongest immediately after nuclear envelope breakdown, but is detected in some cells as late as metaphase. In addition, kinetochore localization of GFP-Kif2b is abolished in Nuf2-deficient cells (Supplementary Figure 5B). These data demonstrate that Kif2b can localize to several key spindle structures during mitosis, although low expression levels prevent us from definitively localizing endogenous Kif2b. Because of the potential for inappropriate localization of overexpressed GFP-Kif2b, these cellular localizations should be considered provisional.
Figure 2.
Localization of GFP-Kif2b. (A) Human U2OS cells expressing GFP-Kif2b during interphase and at various stages of mitosis as indicated. Cells were stained for microtubules in red, DNA in blue, and GFP-Kif2b in green. (B) A prometaphase cell just after nuclear envelope breakdown is stained for DNA in blue, the kinetochore protein Hec1 in red, and GFP-Kif2b in green. The inset in the lower right corner is a 4× magnification of a kinetochore pair (white box). Scale bar, 5 μm.
Kif2b Is Essential for Spindle Assembly and Chromosome Movement
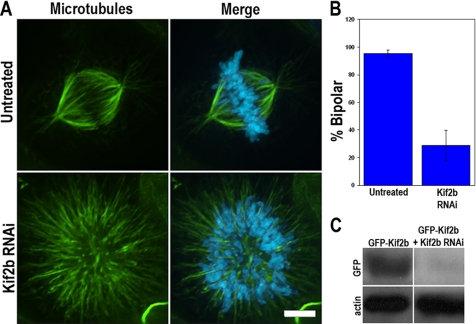
To determine the role of Kif2b in mitosis, we used siRNA to knockdown Kif2b protein levels in human U2OS cells. We used U2OS cells for these experiments because they are amenable to siRNA transfection and we have previously tested the role of other members of the kinesin-13 family in these cells (Ganem et al., 2005). Approximately 48 h after transfection with nonspecific siRNA the mitotic index of cell cultures is ∼3%. In contrast, cells transfected with Kif2b-specific siRNA sequences have a mitotic index of ∼40% at this same time point indicating that Kif2b deficiency delays progression through mitosis. We also observe a threefold increase in binucleate cells and an increase in dead cells after transfection with Kif2b-specific siRNA compared with mock-transfected cells. Most (>90%) mitotic cells in untreated cultures or cells transfected with nonspecific siRNA have bipolar spindles. However, <30% of mitotic cells transfected with Kif2b-specific siRNA have bipolar spindles (Figure 3, A and B). Kif2b-deficient mitotic cells lacking bipolar spindles generally have monopolar spindles (∼80%), but some have disorganized spindles (∼20%), and the same outcome is observed with two different Kif2b-specific siRNA sequences, eliminating the possibility of off-target effects. Because Kif2b abundance in U2OS cells is below detection limits for immunoblotting, we verified knockdown using cells expressing GFP-Kif2b (Figure 3C). Immunoblots for GFP-Kif2b show knockdown efficiencies ranging from 60 to over 80% (Figure 3C) 48 h after transfection with Kif2b-specific siRNA. These blots probably underestimate the efficiency of knockdown of endogenous Kif2b because GFP-Kif2b is over expressed relative to the endogenous protein. These data indicate that Kif2b is essential for bipolar spindle assembly in human cells.
Figure 3.
Kif2b is essential for spindle bipolarity. (A) Untreated U2OS or U2OS cells transfected with Kif2b-specific siRNA were fixed and stained for microtubules in green and DNA in blue. Scale bar, 5 μm. (B) Quantification of the percentage of mitotic cells with bipolar spindles after transfection with a control siRNA (untreated) or Kif2b-specific siRNA. Error bars, SD. (C) Total cell extracts prepared from GFP-Kif2b expressing U2OS cells that were either untreated or transfected with Kif2b-specific siRNA were separated by size by SDS-PAGE and blotted with antibodies specific to GFP and actin as indicated.
We performed several experiments to verify the specificity of Kif2b deficiency. First, immunoblots of untreated and Kif2b-deficient total cell extracts with antibodies specific for Kif2a and MCAK show no appreciable change in their abundance in cells transfected with Kif2b-specific siRNA (Supplementary Figure 2A). Similarly, the pole localization of Kif2a and the centromere and spindle localization of MCAK are not disrupted in Kif2b-deficient cells (Supplementary Figure 2B). The appropriate localization of Hec1 and CENP-E to kinetochores in Kif2b-deficient cells indicates no significant disruption of kinetochore structure in Kif2b-deficient cells (see Figure 5 and Supplementary Figure 2B). We also show that the localization of GFP-Kif2b to spindles is not perturbed in cells lacking either Kif2a or MCAK (Supplementary Figure 2C). Next, we transfected human hTERT-immortalized RPE-1 cells with siRNA specific to Kif2a, Kif2b, or MCAK and examined spindle structure in mitotic cells (Supplementary Figure 3). Similar to U2OS cells, Kif2b is insufficiently abundant to detect in immunoblots of whole cell extracts of RPE-1 cells, but Kif2a and MCAK are detectable and show efficient reduction upon transfection with specific siRNAs (Supplementary Figure 3B). Also similar to U2OS cells, a significant percentage of Kif2a- and Kif2b-deficient RPE-1 cells display monopolar spindles in mitosis, and MCAK-deficient RPE-1 cells have predominantly bipolar spindles but with exaggerated astral microtubule lengths. These data demonstrate that the three kinesin-13 proteins can be knocked down in human U2OS cells independently of each other and that deficiency of each kinesin-13 protein yields a similar outcome in both human RPE-1 and U2OS cells.
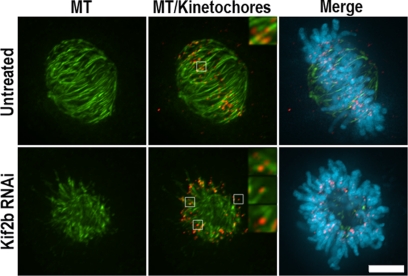
Figure 5.
Kif2b-deficient cells form stable kinetochore microtubule attachments. U2OS cells were either untreated or transfected with Kif2b-specific siRNA as indicated. Cells were extracted in buffer containing 1 mM calcium for 4 min before fixation to induce depolymerization of nonstable microtubules. Microtubules are shown in green, DNA in blue, and kinetochores (Hec1 staining) in red. The insets in the top right corners of each panel are 3× magnifications of kinetochore pairs (white box). Scale bar, 5 μm.
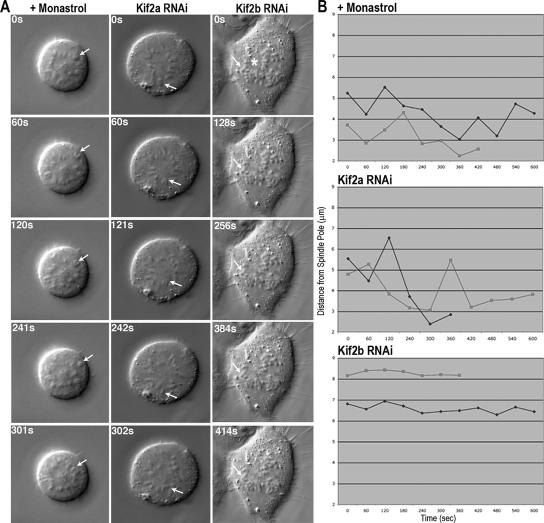
To test if Kif2b contributes to chromosome movement, we used time-lapse DIC microscopy to examine chromosome behavior in cells transfected with Kif2b-specific siRNA (Figure 4). Because most Kif2b-deficient cells form monopolar spindles, we compared chromosome behavior and velocity in Kif2b-deficient cells with cells with monopolar spindles induced by either Eg5 inhibition with 100 μM monastrol or Kif2a deficiency. Chromosomes on monopolar spindles induced by Eg5 inhibition oscillate toward and away from the pole with an average velocity of 1.98 ± 0.70 μm/min (n = 104; Figure 4, A and B; Supplementary Movie 1). Chromosomes on monopolar spindles in Kif2a-deficient cells also oscillate toward and away from the pole with an average velocity of 1.75 ± 0.70 μm/min (n = 119; Figure 4, A and B; Supplementary Movie 2). Chromosomes on monopolar spindles in Kif2b-deficient cells show little directed movement and do not oscillate (Supplementary Movie 3). Some Kif2b-deficient cells with disorganized spindles remain relatively flat in mitosis, making it easier to track chromosome movement (Figure 4A; Supplementary Movie 4). In these cells we observe initial rapid poleward movement of a few chromosomes consistent with dynein-driven movement associated with kinetochore interactions with microtubule sidewalls. However, most chromosomes show little directed movement and do not oscillate (Figure 4, A and B). The average velocity of these chromosomes is 0.46 ± 0.20 μm/min (n = 59). This is a significant reduction in velocity compared with chromosome movement on monopolar spindles induced by inhibition of Eg5 (t test, p < 0.05) and represents only 20% the rate of chromosome movement in prometaphase of untreated U2OS cells with bipolar spindles (2.28 ± 0.09 μm/min). Thus, prometaphase chromosome movement is suppressed in Kif2b-deficient cells.
Figure 4.
Chromosome velocities are suppressed in Kif2b-deficient cells. (A) U20S cells were treated with monastrol or transfected with Kif2a- or Kif2b-specific siRNA as indicated and imaged by time-lapse DIC microscopy during mitosis. Times are shown in seconds. White arrows identify individual chromosomes over time, and the asterisk in the Kif2b RNAi cell denotes the position of a centrosome as judged by GFP-tubulin distribution (not shown). (B) The position of two individual chromosomes relative to the spindle pole were tracked over time in mitotic cells treated with monastrol or transfected with Kif2a- or Kif2b-specific siRNA as indicated.
One explanation for altered chromosome behavior in Kif2b-deficient cells is impaired attachment of microtubules to kinetochores. To test this, we examined Kif2b-deficient cells for calcium-stable microtubules (Figure 5). Untreated cells display numerous calcium-stable microtubule bundles that terminate at kinetochores as judged by the localization of the kinetochore protein Hec1. Kif2b-deficient cells also possess numerous calcium stable microtubules that terminate at kinetochores. In addition, the distance between sister centromeres stained by CREST sera in Kif2b-deficient cells are, on average, 1.6 ± 0.4 μm (n = 60). This distance is shorter than sister centromere distance on chromosomes in untreated cells with bipolar spindles (2.5 ± 0.4 μm, n = 124), but comparable to sister centromere distances on monopolar spindles induced by inhibition of Eg5 (1.4 ± 0.3 μm, n = 158) and Kif2a-deficient cells (1.3 ± 0.4 μm, n = 158). All of these distances exceed sister centromere distances in mitotic cells that lack microtubules because of nocodazole treatment (1.0 ± 0.3 μm, n = 99). These data demonstrate that the lack of chromosome oscillation and reduction in chromosome velocity is not caused by failure in microtubule attachment to kinetochores.
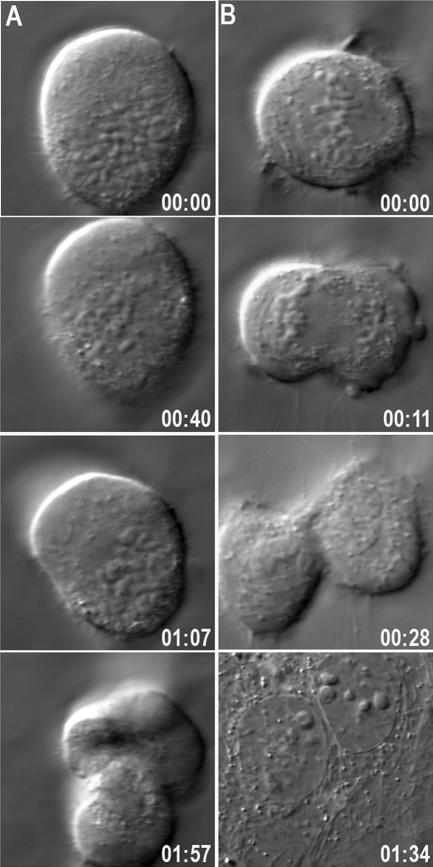
Although there is an increase in cell death in Kif2b-deficient cells, DIC time-lapse microscopy reveals that some of these cells exit mitosis and attempt anaphase and cytokinesis (Figure 6). Some Kif2b-deficient cells with monopolar spindles attempt anaphase, as judged by the appearance of cleavage furrows in the membrane. Multiple furrows are frequently observed and the cell fails to complete cytokinesis properly (Figure 6A, Supplementary Movie 5). Cytokinesis also fails in rare Kif2b-deficient cells that build bipolar spindles. These cells may represent partial reductions in Kif2b protein levels because chromosomes oscillate and align on bipolar spindles. Nevertheless, these cells fail to complete cytokinesis resulting in a binucleate daughter cell (Figure 6B, Supplementary Movie 6). Thus, some Kif2b-deficient cells exit mitosis despite spindle abnormalities, and those that exit mitosis with bipolar spindles fail to complete cytokinesis properly.
Figure 6.
Cytokinesis is impaired in Kif2b-deficient cells. Time-lapse DIC microscopy of Kif2b-deficient cells with either monopolar (A) or bipolar (B) spindles failing to complete cytokinesis. Times are in hours:minutes.
Kif2a, Kif2b, and MCAK Have Functionally Distinct Roles in Mitosis
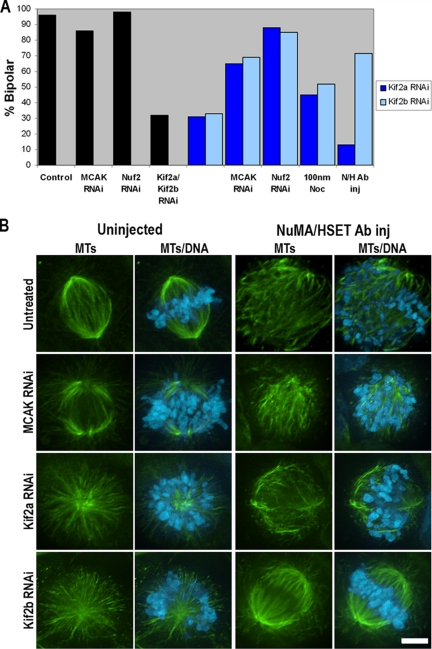
MCAK-deficient cells build bipolar spindles in mitosis (Figure 7, A and B; Ganem et al., 2005), demonstrating that MCAK activity is not needed for spindle assembly. However, both Kif2a- (Ganem and Compton, 2004) and Kif2b-deficient (Figures 3 and 7) mitotic cells assemble predominantly monopolar spindles demonstrating that the activity of each is required for spindle bipolarity. Thus, bipolar spindle formation provides a criterion that functionally discriminates MCAK activity from both Kif2a and Kif2b.
Figure 7.
Kif2a, Kif2b, and MCAK have distinct functions during mitosis. (A) Percentage of mitotic cells with bipolar spindles in cells transfected with MCAK-, Nuf2-, or Kif2a- and Kif2b-specific siRNA are represented by black bars. Percentage of mitotic cells with bipolar spindles in cells transfected with Kif2a- (dark blue bars) or Kif2b-specific (light blue bars) alone or in combination with other treatments including transfection with MCAK- or Nuf2-specific siRNA, treatment with 100 nm nocodazole (100 nm Noc), or injection with antibodies specific to NuMA and HSET (N/H Ab inj). (B) U2OS cells were fixed and stained for microtubules in green and DNA in blue. Cells were either untreated or transfected with MCAK-, Kif2a-, or Kif2b-specific siRNA. Subsequent to transfection, cells were either fixed without injection (uninjected) or injected with NuMA- and HSET-specific antibodies (NuMA/HSET Ab inj). Scale bar, 5 μm.
To determine if Kif2a and Kif2b fill functionally distinct roles during mitosis, we tested how Kif2b-deficient cells respond to experimental conditions shown to rescue bipolar spindle formation in Kif2a-deficient cells (Ganem and Compton, 2004). A majority of Kif2a-deficient cells build bipolar spindles if they enter mitosis in the presence of low doses of nocodazole or if they are simultaneously deficient in MCAK or Nuf2 activities (Figure 7A). The percentage of Kif2b-deficient mitotic cells that assemble bipolar spindles under each of these experimental conditions closely mimics that seen in Kif2a-deficient cells (Figure 7A). Moreover, simultaneous knockdown of both Kif2a and Kif2b does not further decrease the percentage of mitotic cells with bipolar spindles relative to knockdown of each protein alone (Figure 7A). Thus, spindle bipolarity is not a criterion that discriminates between the functions of Kif2a and Kif2b during mitosis and suggests that there may be some overlap in how Kif2a and Kif2b contribute to spindle bipolarity.
Next, we tested the capacity of Kif2b-deficient cells to generate force on spindle microtubules. First, we examined the position of chromosomes on monopolar spindles in Kif2b-deficient cells. Steady state chromosome positioning on monopolar spindles is determined by both poleward kinetochore force and antipoleward ejection force (Cassimeris et al., 1994; Levesque and Compton, 2001). Chromosomes in Kif2b-deficient cells occupy a significantly greater area on monopolar spindles than chromosomes on monopolar spindles generated by either Eg5 inhibition or Kif2a deficiency (Supplementary Figure 4; t test, p < 0.05). This indicates a decrease in poleward kinetochore force acting on chromosomes in Kif2b-deficient cells. Next, we examined how Kif2b- or Kif2a-deficient mitotic cells behave if activities that focus microtubule minus ends at spindle poles are inhibited. We recently showed that focused spindle poles form in the absence of minus end–directed motor activities associated with NuMA and HSET when the ability of kinetochores to exert sustained force on spindle microtubules is inhibited (Manning and Compton, 2007). Thus, if Kif2a or Kif2b contribute to force generation at kinetochores, then cells deficient in their activities should organize microtubules at spindle poles independently of NuMA and HSET activities. Kif2a disassembles microtubule ends at spindle poles in association with poleward microtubule flux (Ganem et al., 2005) and is not known to play a direct role in kinetochore activity. Consistently, very few (8%, n = 24 from this and previously published data; Manning and Compton, 2007) Kif2a-deficient mitotic cells injected with NuMA and HSET antibodies assemble spindles with focused poles (Figure 7, A and B). Likewise, few MCAK-deficient mitotic cells injected with NuMA and HSET antibodies (3%, n = 36 from this and previously published data; Manning and Compton, 2007) assemble spindles with focused poles (Figure 7B). In contrast, a majority (69%, n = 83) of Kif2b-deficient cells injected with NuMA and HSET antibodies assemble bipolar spindles with focused poles (Figure 7, A and B). These data show that Kif2b contributes to force that impacts spindle pole organization and that bipolar spindle assembly in Kif2b-deficient cells can be restored if cells enter mitosis in the absence of pole focusing activities associated with NuMA and HSET. Taken together, these data indicate that Kif2b contributes to poleward kinetochore force providing a criterion that functionally discriminates Kif2b activity from both MCAK and Kif2a.
DISCUSSION
Here we show that the kinesin-13 protein Kif2b is essential for mitosis in human cultured cells. Despite its low expression level, Kif2b participates in several key events during mitosis including bipolar spindle assembly, cytokinesis, and chromosome movement, and we discuss each below. Interestingly, failure of these mitotic processes in Kif2b-deficient cells bears a striking resemblance to cultured cells lacking CLASP2 (Pereira et al., 2006). CLASP-deficient murine cells are impaired in bipolar spindle assembly, show reduced rates of poleward chromosome movement, enter anaphase with unaligned chromosomes, and are impaired in completing cytokinesis. Although a smaller percentage of CLASP-deficient mitotic cells display mitotic defects compared with Kif2b-deficient cells, the similarities in the type of mitotic defects observed raises the possibility that Kif2b and CLASP cooperate in mitosis. In support of this idea, we find that GFP-Kif2b fails to localize to kinetochores in CLASP1-deficient cells (Supplementary Figure 5). It remains unknown whether these proteins physically associate or merely act through the same functional pathways.
Kif2b is required for spindle bipolarity and our data raise the possibility that the function of Kif2b may overlap that of Kif2a to promote bipolar spindle assembly. Deficiencies in either or both proteins yield similar percentages of mitotic cells with monopolar spindles, and spindle bipolarity is restored to similar extents in cells lacking either protein by several different secondary treatments (low dose nocodazole, Nuf2 RNAi, or MCAK RNAi). Despite these similarities, the functions of Kif2b and Kif2a are not identical because deficiency of each protein reduces chromosome velocity to different extents and inhibition of the pole focusing activities of NuMA and HSET restores spindle bipolarity to Kif2b-deficient cells but not Kif2a-deficient cells. Thus, the data imply two possibilities. First, Kif2a and Kif2b contribute to spindle bipolarity through a shared, overlapping function, but each protein also fills other, nonoverlapping functions during mitosis. Second, Kif2a and Kif2b share no overlapping functions and each promotes spindle bipolarity through independent pathways. In this scenario, it is coincidental that spindle bipolarity can be rescued in either Kif2a- or Kif2b-deficient cells by nocodazole treatment, Nuf2 deficiency, or MCAK deficiency. The former possibility seems more plausible at this time, but further functional assays are needed to discriminate between these two possibilities.
Kif2a, Kif2b, and MCAK all localize to spindle midzones in late anaphase and telophase (Figure 2; Wordeman and Mitchison, 1995; Ganem and Compton, 2004). However, using RNAi as a tool to reduce protein expression, we only observe defects in cytokinesis and anaphase onset with monopolar spindles in Kif2b-deficient cells (Ganem et al., 2005). Thus, to date, Kif2b is unique among kinesin-13 proteins to be required for cytokinesis. Kif2b may be needed directly to disassemble the microtubule bundle linking daughter cells or may disrupt cytokinesis indirectly by impairing the targeting of critical midzone components or causing lagging chromatids to be trapped in the cleavage furrow.
Finally, we show that Kif2b contributes to force generation on spindle microtubules. That force influences the molecular requirements for spindle pole organization and contributes to both poleward chromosome velocity and chromosome position on monopolar spindles. Kif2b might exert force on spindle microtubules independently of its microtubule depolymerase activity. Support for this view comes from mutant forms of MCAK that lack robust microtubule depolymerase activity in vitro yet support efficient spindle assembly in frog egg extracts (Ems-McClung et al., 2007). In this context, Kif2b may use high-affinity association with microtubule plus ends, similar to that demonstrated for MCAK (Hunter et al., 2003), to assemble structures (Moores et al., 2006; Tan et al., 2006) that contribute to force generation on spindle microtubules. Alternatively, Kif2b might exert force on spindle microtubules through its microtubule depolymerase activity. Microtubule disassembly can generate force and has been shown to drive chromosome movement under in vitro conditions (Coue et al., 1991; Grishchuk et al., 2005). In cultured vertebrate cells, poleward chromosome movement is dominated by disassembly of microtubule plus ends attached to kinetochores (Gorbsky et al., 1987). In this context, Kif2b could promote the disassembly of microtubule plus ends at kinetochores, resulting in force being applied to spindle microtubules. This view is consistent with the significant reduction in poleward chromosome velocity in Kif2b-deficient cells, the localization of GFP-Kif2b to kinetochores in early prometaphase, and a potential functional relationship between Kif2b and CLASP (Supplementary Figure 5). Both views are tempered by our inability to detect endogenous Kif2b, and it is puzzling why GFP-Kif2b only transiently associates with kinetochores. Perhaps GFP impairs the ability of Kif2b to remain associated with kinetochores when microtubules attach. Alternatively, Kif2b may use a “hit and run” mechanism where it only needs to associate with kinetochores briefly to exert force. Kinetochores have a propensity to maintain attachment to depolymerizing microtubule ends (Zhai et al., 1995), and there is a preponderance of depolymerizing microtubule ends at kinetochores observed by electron microscopy (VandenBeldt et al., 2006), raising the possibility that Kif2b may not need to dwell at kinetochores to influence kinetochore microtubules.
By using RNAi to reduce expression of each member of the kinesin-13 family in human U2OS cells, we show that all three kinesin-13 proteins are required for mitosis, but that each fills a different functional role. MCAK is the only kinesin-13 to show microtubule plus tip-tracking activity (Moore et al., 2005) and uses microtubule depolymerizing activity to correct improper microtubule attachments at kinetochores (Maney et al., 1998; Ganem and Compton, 2004; Kline-Smith et al., 2004). Deficiency of MCAK does not significantly impair bipolar spindle assembly or chromosome movement (Maney et al., 1998; Kline-Smith and Walczak, 2004; Ganem and Compton, 2004). Kif2a is required for bipolar spindle assembly and utilizes microtubule depolymerizing activity to disassemble microtubule minus ends at spindle poles to drive poleward microtubule flux during mitosis (Ganem and Compton, 2004; Ganem et al., 2005). Kif2a-dependent flux activity makes a minor contribution to poleward chromosome velocity (Ganem et al., 2005). Finally, Kif2b is required for bipolar spindle assembly, plays a role in cytokinesis, and makes a major contribution to poleward chromosome velocity during prometaphase.
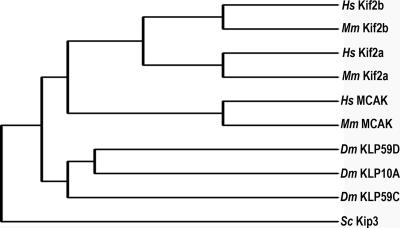
The lack of significant functional overlap among three different kinesin-13 proteins during mitosis in human cells demonstrates that each protein fills a distinct functional niche. Kinesin-13 proteins also fill nonoverlapping roles during mitosis in other metazoan species including fruit flies (Rogers et al., 2004) and frogs (Gaetz and Kapoor, 2004). For example, in Drosophila embryos, Klp10A is required for disassembly of microtubule minus ends associated with flux, Klp59C is necessary for poleward chromosome movement, and Klp59D appears dispensable (Rogers et al., 2004). In this context, it seems logical to assume that gene duplication to generate this gene family was driven by the need to fill nonoverlapping functions in mitosis. Interestingly, phylogenetic comparison of protein sequences among kinesin-13 family members reveals that the three kinesin-13 proteins in Drosophila are more related to one another than to the mammalian proteins (Figure 8). This indicates that gene duplication generating nonoverlapping functional members of this gene family occurred separately in mammals and flies after speciation. Consequently, assigning homologous functions to kinesin-13 genes in different organisms based on sequence comparison alone is not valid.
Figure 8.
Phylogenetic comparison of kinesin-13 genes. Amino acid sequences from three kinesin-13 proteins encoded by fruit fly (Dm), mouse (Mm), and human (Hs) are compared using Clustal X. The sequence of yeast Kip3 is used as an outgroup to root the tree. Line lengths are not to scale.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grants GM51542 to D.A.C. and GM69429 to L.W.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0110) on May 30, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Andrews P. D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J. R. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Antonio C., Ferby I., Wilhelm H., Jones M., Karsenti E., Vernos I. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 2000;102:425–435. doi: 10.1016/s0092-8674(00)00048-9. [DOI] [PubMed] [Google Scholar]
- Cassimeris L., Rieder C. L., Salmon E. D. Microtubule assembly and kinetochore directional instability in vertebrate monopolar spindles: implications for the mechanism of chromosome congression. J. Cell Sci. 1994;107:285–297. doi: 10.1242/jcs.107.1.285. [DOI] [PubMed] [Google Scholar]
- Compton D. A. Spindle assembly in animal cells. Annu. Rev. Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- Coue M., Lombillo V. A., McIntosh J. R. Microtubule depolymerization promotes particle and chromosome movement in vitro. J. Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T. J., Walczak C. E. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Ems-McClung S. C., Hertzer K. M., Zhang X., Miller M. W., Walczak C. E. The interplay of the N- and C-terminal domains of MCAK control microtubule depolymerization activity and spindle assembly. Mol. Biol. Cell. 2007;18:282–294. doi: 10.1091/mbc.E06-08-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fant X., Merdes A., Haren L. Cell and molecular biology of spindle poles and NuMA. Int. Rev. Cytol. 2004;238:1–57. doi: 10.1016/S0074-7696(04)38001-0. [DOI] [PubMed] [Google Scholar]
- Funabiki H., Murray A. W. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Gaetz J., Kapoor T. M. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T., Dionne M. A., Compton D. A. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J. Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Compton D. A. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Upton K., Compton D. A. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- Gard D. L., Becker B. E., Josh Romney S. MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int. Rev. Cytol. 2004;239:179–272. doi: 10.1016/S0074-7696(04)39004-2. [DOI] [PubMed] [Google Scholar]
- Gorbsky G. J., Sammak P. J., Borisy G. G. Chromosomes move poleward in anaphase along stationary microtubules that coordinately disassemble from their kinetochore ends. J. Cell Biol. 1987;104:9–18. doi: 10.1083/jcb.104.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E. L., Molodtsov M. I., Ataullakhanov F. I., McIntosh J. R. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- Gruss O. J., Vernos I. The mechanism of spindle assembly: functions of Ran and its target TPX2. J. Cell Biol. 2004;166:949–955. doi: 10.1083/jcb.200312112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hunter A. W., Caplow M., Coy D. L., Hancock W. O., Diez S., Wordeman L., Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F., Thompson J. D., Gouy M., Higgins D. G., Gibson T. J. Multiple sequence alignment with Clustal X. Trends Biochem. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Jordan M. A., Wilson L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- Kapoor T. M., Lampson M. A., Hergert P., Cameron L., Cimini D., Salmon E. D., McEwen B. F., Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S. L., Walczak C. E. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Kline-Smith S. L., Khodjakov A., Hergert P., Walczak C. E. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton A. L., Lan W., Stukenberg P. T. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- Kurasawa Y., Earnshaw W. C., Mochizuki Y., Dohmae N., Todokoro K. Essential roles of Kif4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S. L., Rosasco S. E., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Walczak C. E., Stukenberg P. T. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Lawrence C. J., et al. A standardized kinesin nomenclature. J. Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque A. A., Compton D. A. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 2001;154:1135–1146. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack G. J., Compton D. A. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc. Natl. Acad. Sci. USA. 2001;98:14434–14439. doi: 10.1073/pnas.261371298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T., Hunter A. W., Wagenbach M., Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A. L., Compton D. A. Mechanisms of spindle-pole organization are influenced by kinetochore activity in mammalian cells. Curr. Biol. 2007;17:260–265. doi: 10.1016/j.cub.2006.11.071. [DOI] [PubMed] [Google Scholar]
- Mishima M., Kaitna S., Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- Moore A. T., Rankin K. E., von Dassow G., Peris L., Wagenbach M., Ovechkina Y., Andrieux A., Job D., Wordeman L. MCAK associates with the tips of polymerizing microtubules. J. Cell Biol. 2005;169:391–397. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores C. A., Cooper J., Wagenbach M., Ovechkina Y., Wordeman L., Milligan R. A. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 2006;5:1812–1815. doi: 10.4161/cc.5.16.3134. [DOI] [PubMed] [Google Scholar]
- Mountain V., Simerly C., Howard L., Asako A., Schatten G., Compton D. A. The kineisin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 1999;147:351–365. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J., Mitchison T. J. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovechkina Y., Wagenbach M., Wordeman L. K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant. J. Cell Biol. 2002;159:557–562. doi: 10.1083/jcb.200205089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A. L., et al. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol. Biol. Cell. 2006;17:4526–4542. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff J. W. Centrosomes and cancer: lessons from a TACC. Trends Cell Biol. 2002;12:222–225. doi: 10.1016/s0962-8924(02)02268-7. [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Schwimmer T. A., Ems-McClung S. C., Walczak C. E., Vale R. D., Scholey J. M., Sharp D. J. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Sawin K. E., LeGuellec K., Philippe M., Mitchison T. J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Rogers G. C., Scholey J. M. Microtubule motors in mitosis. Nature. 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Rogers G. C. A KinI-dependent Pacman-flux mechanism for anaphase A. Cell Cycle. 2004;3:707–710. [PubMed] [Google Scholar]
- Tan D., Asenjo A. B., Mennella V., Sharp D. J., Sosa H. Kinesin-13's form rings around microtubules. J. Cell Biol. 2006;175:25–31. doi: 10.1083/jcb.200605194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBeldt K. J., Barnard R. M., Hergert P. J., Meng X., Maiato H., McEwen B. F. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr. Biol. 2006;16:1217–1223. doi: 10.1016/j.cub.2006.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C. E., Verma S., Mitchison T. J. XCTK2, a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J. Cell Biol. 1997;136:859–870. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittman T., Hyman A., Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat. Cell Biol. 2001;3:E28–E34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- Wordeman L. Microtubule-depolymerizing kinesins. Curr. Opin. Cell Biol. 2005;17:82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Wordeman L., Mitchison T. J. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Kronebusch P. J., Borisy G. G. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J. D., Bossy-Wetzel E., Fan J.-B., Abraham R. T., Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.