Abstract
The septation initiation network (SIN) and mitotic exit network (MEN) signaling pathways regulate cytokinesis and mitotic exit in the yeasts Schizosaccharomyces pombe, and Saccharomyces cerevisiae, respectively. One function of these pathways is to keep the Cdc14-family phosphatase, called Clp1 in S. pombe, from being sequestered and inhibited in the nucleolus. In S. pombe, the SIN and Clp1 act as part of a cytokinesis checkpoint that allows cells to cope with cytokinesis defects. The SIN promotes checkpoint function by 1) keeping Clp1 out of the nucleolus, 2) maintaining the cytokinetic apparatus, and 3) halting the cell cycle until cytokinesis is completed. In a screen for suppressors of the SIN mutant cytokinesis checkpoint defect, we identified a novel nucleolar protein called Dnt1 and other nucleolar proteins, including Rrn5 and Nuc1, which are known to be required for rDNA transcription. Dnt1 shows sequence homology to Net1/Cfi1, which encodes the nucleolar inhibitor of Cdc14 in budding yeast. Like Net1/Cfi1, Dnt1 is required for rDNA silencing and minichromosome maintenance, and both Dnt1 and Net1/Cfi1 negatively regulate the homologous SIN and MEN pathways. Unlike Net1/Cfi1, which regulates the MEN through the Cdc14 phosphatase, Dnt1 can inhibit SIN signaling independently of Clp1, suggesting a novel connection between the nucleolus and the SIN pathway.
INTRODUCTION
As in higher eukaryotes, cytokinesis in the fission yeast Schizosaccharomyces pombe occurs through the use of an actomyosin contractile ring. A spindle pole body (SPB)-based regulatory network, called septation initiation network (SIN), is required to initiate cytokinesis in late anaphase (for review, see Guertin et al., 2002; Simanis, 2003). A similar signaling cascade exists in budding yeast called the mitotic exit network (MEN). The MEN is essential for mitotic exit, and it plays an important but nonessential role in cytokinesis (for review, see Simanis, 2003; Stegmeier and Amon, 2004). Both pathways function together with the Cdc14 phosphatase, which is called Clp1 in S. pombe. Cdc14 phosphatases reverse phosphorylation by cyclin-dependent kinase (Cdk) (Visintin et al., 1998; Kaiser et al., 2002; Wolfe and Gould, 2004; Wolfe et al., 2006). The activities of Cdc14 and Clp1 are controlled at least in part by regulated sequestration and release from the nucleolus (Shou et al., 1999; Visintin et al., 1999; Cueille et al., 2001; Trautmann et al., 2001). In budding yeast, Cdc14 is kept sequestered and inactive in the nucleolus throughout most of the cell cycle by binding to the Net1/Cfi1 protein in the nucleolus (Shou et al., 1999; Visintin et al., 1999). In early anaphase, Cdc14 is released from the nucleolus by the combined action of a set of proteins termed the FEAR network and Cdk1 phosphorylation of Net1 (Stegmeier et al., 2002; Azzam et al., 2004). The MEN then acts to maintain the release of Cdc14 from the nucleolus, which is sufficient to trigger mitotic exit. The essential function of the MEN is to keep Cdc14 out of the nucleolus so it can promote mitotic exit. Moreover, disruption of nucleolar localization of Cdc14 by mutation in the nucleolar inhibitor Net1/Cfi1 can rescue deletions of MEN pathway components (Shou et al., 1999, 2001).
Regulation of Clp1 in fission yeast has similarities as well as important differences from Cdc14 regulation in budding yeast (Cueille et al., 2001; Trautmann et al., 2001). As with budding yeast Cdc14, Clp1 localizes to the nucleolus throughout interphase. However unlike budding yeast, but similar to mammalian Cdc14B (Cho et al., 2005), Clp1 is released from the nucleolus upon mitotic entry. Then, in anaphase, much like the MEN in budding yeast, the SIN acts to keep Clp1 out of the nucleolus until cytokinesis is complete. The mechanism governing release of Clp1 from the nucleolus in early mitosis is not known, but it does not require homologues of the budding yeast FEAR pathway components (Chen et al., 2006). How Clp1 is inhibited in the nucleolus is not known. An S. pombe homologue of Net1/Cfi1 has not been identified.
In fission yeast, Clp1 and the SIN each promote the others activity as part of a positive feedback loop that stays active until completion of cytokinesis (Cueille et al., 2001; Trautmann et al., 2001). Clp1 keeps the SIN active, and the SIN keeps Clp1 out of the nucleolus. This positive feedback loop functions as part of a surveillance mechanism, termed the cytokinesis checkpoint, that halts further cell cycle progression until cytokinesis is complete (Liu et al., 2000; Mishra et al., 2004). Under normal growth conditions, the checkpoint is not essential for viability. However, when cytokinesis is slowed, for example by perturbation of the actomyosin ring, the checkpoint becomes essential for viability (Mishra et al., 2004). The checkpoint blocks further rounds of nuclear division, and it maintains the cytokinetic apparatus, so the cells can eventually divide and retain normal ploidy. Cells with weakened SIN signaling, or a deletion of Clp1, are defective for the checkpoint, and they are unable to deal with defects in cytokinesis and become multinucleate and die when cytokinesis is delayed.
Here, we describe a genetic screen for suppressors of the cytokinesis checkpoint defect in a weakened SIN mutant. As described below, our screen identified a protein similar to Net1/Cfi1 as well as several other nucleolar proteins. Unlike Net1/Cfi1, which regulates mitotic exit and MEN signaling through the Cdc14 phosphatase, Dnt1 affects the SIN independently of Clp1, suggesting a new and unexpected link between the nucleolus and the SIN signaling pathway.
MATERIALS AND METHODS
Yeast Media, Strains, and Genetic Manipulations
The fission yeast strains used in this study are listed in Table 1. Genetic crosses and general yeast techniques were performed as described previously (Moreno et al., 1991). S. pombe strains were grown in rich medium (yeast extract [YE]) or Edinburgh minimal medium (EMM) with appropriate supplements (Moreno et al., 1991). EMM with 5 μg/ml thiamine was used to repress expression from the nmt1 promoter. YE containing 100 mg of Geneticin (G-418; Sigma-Aldrich, St. Louis, MO) per liter was used for selecting Kanr cells. For latrunculin B treatment in cytokinesis checkpoint experiments, a final concentration of 2–5 μM was used. YE containing 5 μg/ml trichostatin A (Sigma-Aldrich) was used to test the sensitivity of mutant cells. For serial dilution drop tests for growth, three serial 10-fold dilutions were made, and 5 μl of each dilution was spotted on plates with the starting cell number of 104. Cells were pregrown in liquid YE or EMM at 25°C and then spotted onto YE or EMM plates at the indicated temperatures and incubated for 3–5 d before photography. In rDNA silencing assays, N/S refers to EMM with glutamate, containing adenine, leucine, and uracil at 75 mg/l each. 5-Fluroorotic acid (5-FOA; Toronto Research Chemicals, North York, Ontario, Canada) plates contain N/S supplemented with 2 g of 5-FOA per liter. Ura− is N/S medium without uracil. The minichromosome loss assay was performed as described previously (Allshire et al., 1995). The rate of minichromosome loss was determined by the frequency of half-sectored colonies. S. cerevisiae strain PJ69-4A was used as the host strain in two-hybrid analyses (James et al., 1996), and it was transformed using the LiAc/PEG procedure (Gietz et al., 1995). Leu+ and Trp+ transformants were selected and scored for positive interactions by spotting onto synthetic dextrose plates lacking histidine. Synchronous populations of S. pombe cells at early G2 were generated by centrifugal elutriation using a Beckmann JE 5.0 rotor (Beckman Coulter, Fullerton, CA).
Table 1.
S. pombe strains used in this study
| Strain | Genotype |
|---|---|
| YDM105 | leu1-32 ura4-D18 ade6-210 h-− |
| YDM106 | leu1-32 ura4-D18 ade6-210 h+ |
| YDM116 | sid4-A1 leu1-32 ura4-D18 ade6 h− |
| YDM2719 | dnt1Δ::ura4+sid4-A1 leu1-32 ura4-D18 ade6 h? |
| YDM275 | cdc11-123 leu1-32 ura4-D18 ade6-210 h− |
| YDM2718 | dnt1Δ::ura4+cdc11-123 ura4-D18 h? |
| YDM430 | spg1-106 leu1-32 ura4-D18 ade6-210 h+ |
| YDM2720 | dnt1Δ::ura4+spg1-106 leu1-32 ura4-D18 ade6-210 h? |
| YDM1239 | cdc7-24 h+ |
| YDM2721 | dnt1Δ::ura4+cdc7-24 leu1-32 ura4-D18 ade6-210 h? |
| YDM76 | sid1-239 leu1-32 ade6 h− |
| YDM2723 | dnt1Δ::ura4+sid1-239 leu1-32 ura4-D18 ade6 h? |
| YDM445 | sid1-125 leu1-32 ura4-D18 ade6-210 h+ |
| YDM2724 | dnt1Δ::ura4+sid1-125 leu1-32 ura4-D18 ade6-210 h? |
| YDM272 | cdc14-118 ura4-D18 ade6-M210 h− |
| YDM2710 | dnt1Δ::ura4+cdc14-118 clp1-GFP-kanR h? |
| YDM429 | sid2-250 leu1-32 ura4-D18 ade6 h+ |
| YDM2725 | dnt1Δ::ura4+sid2-250 leu1-32 ura4-D18 ade6 h? |
| YDM670 | mob1-1 leu1-32 ura4-D18 ade6 his3-D1 h− |
| YDM2726 | dnt1Δ::ura4+mob1-1 leu1-32 ura4-D18 ade6 h? |
| YDM1311 | myo2-E1 leu1-32 ura4-D18 ade6 h− |
| YDM2709 | dnt1Δ::ura4+myo2-E1 clp1-GFP-kanR h? |
| YDM1380 | myo2-E1 clp1-GFP-kanR h? |
| YDM1314 | myo2-E1 cdc14-118 clp1-GFP-kanR leu1-32 ura4-D18 ade6 h? |
| YDM2737 | rrn5-S6 clp1-GFP-kanR h+ |
| YDM2738 | rrn5-S6 myo2-E1 cdc14-118 clp1-GFP-kanR h+ |
| YDM2749 | dnt1-(4-3) leu1-32 ura4-D18 h− |
| YDM3445 | dnt1-(4-3) myo2-E1 cdc14-118 h− |
| YDM2379 | dnt1Δ::ura4+leu1-32 ura4-D18 ade6-210 h− |
| YDM2711 | dnt1Δ::ura4+myo2-E1 cdc14-118 clp1-GFP-kanR h? |
| YDM3484 | sdc4-12 leu1-32 ura4-D18 ade6 h+ |
| YDM3688 | sdc4-12 myo2-E1 cdc14-118 leu1-32 ura4-D18 ade6 h? |
| YDM3454 | nuc1-632 leu1-32 ura4-D18 h− |
| YDM3527 | nuc1-632 myo2-E1 cdc14-118 h? |
| YDM2333 | dnt1-GFP-kanR leu1-32 ura4-D18 ade6-210 h+ |
| YDM2430 | dnt1-13myc-kanR gar2-GFP-kanR leu1-32 ura4-D18 ade6-210 h+ |
| YDM3409 | rrn5-S6 dnt1-GFP-kanR h+ |
| YDM3483 | sdc4-12 dnt1-GFP-kanR leu1-32 ura4-D18 ade6 h+ |
| YDM3455 | nuc1-632 dnt1-GFP-kanR leu1-32 ura4-D18 h? |
| YDM3431 | nuc1-GFP-kanR h− |
| YDM3677 | rrn5-S6 nuc1-GFP-kanR h? |
| YDM3473 | dnt1Δ::ura4+nuc1-GFP-kanR ura4-D18 h? |
| YDM3659 | sdc4-12 nuc1-GFP-kanR h? |
| YDM824 | clp1-GFP-kanR leu1-32 ura4-D18 ade6-216 h− |
| YDM2737 | rrn5-S6 clp1-GFP-kanR h+ |
| YDM2629 | dnt1Δ::ura4+clp1-GFP-kanR leu1-32 ura4-D18 ade6-216 h− |
| YDM3601 | sdc4-12 clp1-GFP-kanR leu1-32 ura4-D18 ade6 h? |
| YDM3614 | nuc1-632 clp1-GFP-kanR h? |
| YDM2381 | Yip2.4pUCura4.7 ura4-DS/E leu1-32 ade6-216 h+ (ura4+inserted at rDNA repeat) |
| YDM2926 | dnt1Δ::kanR Yip2.4pUCura4.7 ura4-D18 leu1-32 ade6-210 h− |
| YDM821 | clp1Δ::ura4+leu1-32 ura4-D18 leu1-32 ade6-216 h+ |
| YDM2741 | clp1Δ::ura4+cdc14-118 ura4-D18 ade6 h? |
| YDM2742 | dnt1Δ::ura4+clp1Δ::ura4+cdc14-118 ura4-D18 ade6 h? |
| YDM648 | cdc7-GFP-ura4+leu1-32 ura4-D18 ade6 h− |
| YDM986 | clp1Δ::ura4+cdc7-GFP-ura4+leu1-32 ura4-D18 h? |
| YDM3001 | dnt1Δ::kanR cdc7-GFP-ura4+leu1-32 ura4-D18 ade6 h? |
| YDM2996 | dnt1Δ::kanR clp1Δ::ura4+cdc7-GFP-ura4+leu1-32 ura4-D18 ade6 h? |
Isolation of Suppressors of Cytokinesis Checkpoint and Cloning of rrn5+
The cdc14-118 myo2-E1 strain was grown at 25°C, and then it was plated at 30°C to select for random suppressors. In total, 68 colonies were picked and crossed to wild type to determine whether they represented single mutations, and whether they had phenotypes on their own. Most suppressors only weakly suppressed either the cdc14-118 or myo2-E1 single mutants and were not analyzed further. However, nine mutants could suppress cdc14-118 and myo2-E1 single and double mutants. These mutants fell into three complementation groups: group I (6 members: 14, 16, 2-12, 2-13, 3-8, and 4-3), group II (2 members: 6 and 3-3) and group III (1 member: sdc4-12). In the course of mapping group I mutations, we also crossed them to dnt1Δ, because, although the dnt1Δ strain was generated as part of a separate study, we suspected dnt1 might be identified in our screen, and we knew that dnt1Δ could rescue cdc14-118 myo2-E1. Group I mutations were all tightly linked to dnt1+, with no double mutants with dnt1Δ::ura4+ isolated out of >20 complete tetrads dissected. This suggested that group I mutations might be alleles of dnt+. Subsequently, we amplified the dnt1+ gene from the group I mutants by polymerase chain reaction (PCR), and we sequenced the PCR products, confirming that three of the group I mutants carried mutations in the open reading frame (ORF) of the dnt1+ gene: in suppressor 14, it had a 17-nucleotide insertion between nucleotides 230 and 231 in the coding region, which introduced a premature stop codon TAA; in sup16, one nucleotide is deleted at base 1503, causing a frameshift and a stop codon 20 amino acids before the C-terminal end of the protein; in suppressor 4–3, nucleotide 492 is deleted, causing a premature stop codon at nucleotide 500. For the other three group I mutants (2-12, 2-13, and 3-8) obtained from our screen, sequencing of the ORF of the dnt1+ gene did not reveal any mutations, suggesting that they may carry mutations outside the coding region that affect gene expression or RNA stability. Suppressor 6 was cloned by complementation of its temperature-sensitive phenotype by using an S. pombe genomic library (Clontech, Mountain View, CA). Several plasmids were identified, and one gene called rrn5+ was determined to be responsible for the rescuing activity. Therefore, we renamed suppressor 6 as rrn5-S6.
Epitope Tagging, Gene Deletion, and Cloning
Carboxy-terminal green fluorescent protein (GFP) and 13-myc epitope tagging of Dnt1 was done by PCR-based gene targeting (Bahler et al., 1998). To construct the dnt1 deletion strains, the entire dnt1 coding region was replaced with the ura4+ gene or kanR cassette by homologous recombination.
For complementation experiment of budding yeast NET1/CFI1, NET1/CFI1 gene was amplified by PCR from wild-type budding yeast genomic DNA. The full-length NET1/CFI1 open reading frame was first cloned into the entry vector of the Gateway System (Invitrogen, Carlsbad, CA), and then it was cloned into destination vectors based on pREP41X and pREP3X, which were designed for expression in S. pombe (Choi and McCollum, unpublished data).
Microscopy
GFP-fusion proteins were observed in cells after fixation with −20°C methanol or in live cells. DNA was visualized with 4′, 6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) at 2 μg/ml. Immunofluorescence microscopy of Gar2-GFP and Dnt1–13myc was done as described previously (Balasubramanian et al., 1997). Briefly, cells expressing Gar2-GFP and Dnt1–13myc fusions were fixed with methanol and digested with lysing enzymes (Sigma-Aldrich), followed by indirect immunofluorescence with polyclonal rabbit anti-GFP (Invitrogen) and monoclonal mouse anti-myc (a gift from Dr. Kathy Gould, Vanderbilt University). Secondary anti-mouse Texas Red and Alexa 594-immunoglobulin (Ig)G (Invitrogen) were used. Photomicrographs were obtained using a Nikon Eclipse E600 fluorescence microscope coupled to a cooled charge-coupled device camera (ORCA-ER; Hamamatsu, Bridgewater, NJ), and image processing and analysis were carried out using IPLab Spectrum software (Signal Analytics, Vienna, VA).
RESULTS
A Genetic Screen for Suppressors of the Cytokinesis Checkpoint Defect of SIN Mutants
To further understand the mechanisms controlling the SIN and the cytokinesis checkpoint in fission yeast, we carried out a genetic screen for suppressors of the cytokinesis checkpoint defects in SIN mutants. This screen took advantage of the fact that temperature-sensitive SIN mutants are viable at semipermissive temperatures but that they have reduced SIN function and a defective cytokinesis checkpoint (Mishra et al., 2004). Due to lack of the checkpoint, these mutations become lethal when the actomyosin ring is slightly perturbed. Thus, these cells fail cytokinesis and they die as multinucleate cells. This can be observed when examining the phenotype of single and double mutants between cdc14-118 (note that S. pombe cdc14 encodes a subunit of the Sid1p kinase and is not a homologue of budding yeast Cdc14) and the type II myosin mutant myo2-E1. The temperature-sensitive SIN mutant cdc14-118 is viable at 30°C, but it is largely defective for the cytokinesis checkpoint (Mishra et al., 2004). The temperature-sensitive type II myosin mutant myo2-E1 is also viable at 30°C, but it is slow in completing cytokinesis, and its viability at this temperature depends on the cytokinesis checkpoint (Mishra et al., 2004). However, the cdc14-118 myo2-E1 double mutant strain is dead at 30°C, because of the combined delay in cytokinesis and defective cytokinesis checkpoint. As shown previously, these cells either fail to make septa, or they make incomplete aberrant septa and become multinucleate (Mishra et al., 2004).
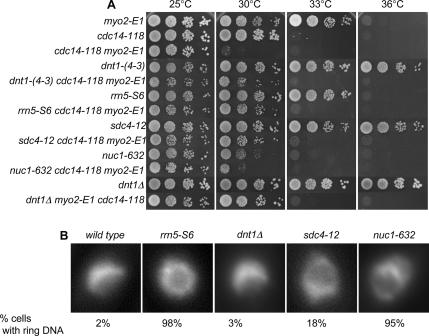
The cdc14-118 myo2-E1 strain was screened for spontaneous suppressing mutants at 30°C. From this screen, we identified many spontaneous suppressors, and we picked different-sized colonies, which were backcrossed to wild type. The weaker suppressors only poorly suppressed one of the single mutants, and the stronger suppressors suppressed both cdc14-118 and myo2-E1 (Figure 1A; data not shown). The best suppressors we identified fell into three complementation groups: group I (6 members: 14, 16, 2-12, 2-13, 3-8, and 4-3), group II (2 members: 6 and 3-3), and group III (1 member: 4-12 called sdc4 for suppressor of defective checkpoint). The first two groups are later referred to as dnt1 and rrn5, respectively (for reasons, see Materials and Methods). Outcrossing revealed that although the dnt1 mutant cells showed no obvious growth defect, the two alleles of rrn5 were temperature sensitive on their own, and the sdc4-12 strain grew slowly at all temperatures (Figure 1A).
Figure 1.
Suppressors of cdc14-118 myo2-E1 double mutant. (A) Selected suppressor single mutants and triple mutants with cdc14-118 myo2-E1 were grown in YE at 25°C, and then serial dilutions were spotted on YE plates. Plates were incubated at different temperatures as indicated for 3–5 d before photography. (B) Nuclear phenotype of the suppressors. Wild-type cells and suppressor mutants were first grown in YE at 25°C, and then they were shifted to 30°C for 4 h. Cells were fixed and stained with DAPI. Example nuclei from interphase cells are shown. Note the crescent-shaped nuclear DNA in wild type and dnt1Δ cells and ring-shaped nuclear DNA phenotype in rrn5-S6, sdc4-12, and nuc1-632 mutants. The percentage of cells with the ring-shaped nuclear DNA phenotype is indicated (n > 100).
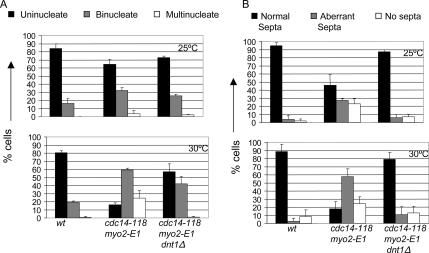
Examination of the cdc14-118 myo2-E1 cells carrying the different suppressor mutations showed that they all had similar morphology and that they had recovered the ability to form complete septa at 30°C (Figure 2; data not shown). To characterize the phenotype more closely, we examined cdc14-118 myo2-E1 and cdc14-118 myo2-E1 dnt1 cells after shift from 25 to 30°C for 4 h. In the cdc14-118 myo2-E1 cells, the number of mononucleate cells decrease and the cells become bi- and tetranucleate presumably due to cytokinesis defects caused by their inability to make proper septa (Figure 2). In contrast, the cdc14-118 myo2-E1 dnt1 cells seem to have at least partially restored cytokinesis checkpoint function, because these cells show a delay as binucleate cells but eventually can divide, because they maintain a mononucleate population and do not accumulate tetranucleate cells (Figure 2A). These cells also recover the ability to make proper septa (Figure 2B).
Figure 2.
dnt1Δ promotes proper completion of cytokinesis in the cdc14-118 myo2-E1 cells. Asynchronous cells of the indicated genotypes were grown at 25°C to log phase cell density, and portions were shifted to 30°C for 4 h. Cells were methanol fixed and stained simultaneously with DAPI and calcofluor to score for the number of nuclei per cell (A) and septum formation (B), respectively. Cells were scored for the presence of normal, aberrant, or lack of septa between nuclei. At least 100 cells were examined for each strain, and mean values (n = 3) were plotted.
Suppressors Encode Nucleolar Proteins
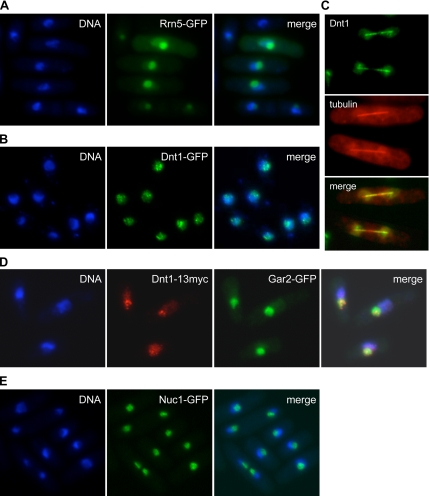
The group II suppressor gene was cloned by complementation of its temperature-sensitive phenotype and determined to be the rrn5+ gene (see Materials and Methods). The rrn5+ gene encodes an upstream activating factor (UAF) for RNA polymerase I involved in transcription of rRNA (Liu et al., 2002). Examination of the phenotype of the rrn5-S6 mutant at restrictive temperature revealed an unusual nuclear architecture. In wild-type cells, the nuclear DNA forms a crescent shape on one side of the nucleus, with the nucleolus occupying the other portion of the nucleus. However, at its restrictive temperature of 36°C the rrn5-S6 cells often had a ring-shaped nuclear DNA pattern (Figure 1B). The only other mutants reported to date with this phenotype are the topoisomerase I and II double mutants, and the temperature-sensitive mutant nuc1-632, which carries mutation in the largest subunit of RNA polymerase I (Hirano et al., 1986, 1989). Interestingly, the sdc4-12 mutant also displays the ring-shaped nuclear DNA phenotype (Figure 1B), which, together with genetic analysis described below, suggests that it might also affect RNA polymerase I transcription. Both point mutations and null mutations in the dnt1 did not cause the ring-shaped chromatin phenotype. To determine the localization of Rrn5, we expressed plasmid-borne and GFP-tagged Rrn5 in wild-type cells. Not surprisingly, similar to Nuc1, Rrn5 showed nucleolar localization as judged by colocalization with the non-DAPI staining region of the nucleus (Figure 3A).
Figure 3.
Nucleolar localization of suppressor proteins. The indicated strains were grown at 30°C. (A) Cells transformed with pREP42-rrn5+-GFP were first grown in EMM with thiamine (repressed), and then they were induced for 18 h in EMM without thiamine. Cells were fixed and DAPI stained. Note that the GFP signal varies due to variation in the plasmid copy number between cells. (B) Cells carrying integrated dnt1-GFP were fixed and DAPI stained. (C) Cells carrying integrated dnt1-GFP and mCherry-tubulin expressed from a plasmid were imaged live by fluorescence microscopy. A montage of two late anaphase cells is shown. (D) Cells expressing both integrated dnt1-13myc and gar2-GFP were fixed, and then they were subjected to immunofluorescence with antibodies against Myc and GFP. Dnt1–13myc foci were concentrated in the nucleolus as marked by Gar2-GFP. (E) Cells carrying integrated nuc1-GFP were fixed and DAPI stained.
To investigate whether general perturbation of the RNA polymerase (Pol) I transcription complex can suppress cdc14-118 myo2-E1 cells, we tested whether the nuc1-632 could also rescue this double mutant at 30°C. Although the nuc1-632 cells grew very poorly on their own at 30°C, the nuc1-632 mutation could weakly rescue cdc14-118 myo2-E1 cells at this temperature (Figure 1A).
The group I suppressors turned out to be in the dnt1 gene that had been identified in our laboratory as part of an unrelated proteomics screen by using mass spectrometry (see Materials and Methods; Jin and McCollum, unpublished observations). dnt1Δ deletion mutants were viable, and they grew at rates similar to wild-type cells. The dnt1Δ mutation also rescued the growth defect of cdc14-118 myo2-E1 cells at 30°C (Figure 1A). Interestingly, Dnt1 is also a nucleolar protein, because Dnt1-GFP is localized in the nucleolus as two or more punctate dots throughout the cell cycle (Figure 3, B and D). Dnt1-GFP signals can also be observed faintly in the rest of the nucleoplasm. In late anaphase, Dnt1 localizes to the ends of the mitotic spindle (Figure 3C).
Functional Interdependence of Dnt1 and Suppressors Involved in rDNA Transcription
Three of our suppressors, rrn5-S6, nuc1-632, and sdc4-12, showed a characteristic ring-shaped DNA phenotype, and they all grew slowly even at permissive temperature, probably due to reduced rDNA transcription. Although dnt1Δ mutants did not have a reduced growth rate or defects in nucleolar positioning, genetic analysis suggested that they may also have a role in RNA Pol I transcription. Double mutant analysis revealed that all combinations of double mutants between nuc1-632, rrn5-S6, sdc4-12, and dnt1Δ resulted in either synthetic lethality or very sick and slow-growing cells (Table 2). These data suggest that Dnt1 and Sdc4 might function in rDNA transcription like Rrn5 and Nuc1.
Table 2.
Negative genetic interactions between suppressorsa
| rrn5-S6 | sdc4-12 | nuc1-632 | |
|---|---|---|---|
| dnt1Δ | Sick | Sick | Lethal |
| rrn5-S6 | Lethal | Sick | |
| sdc4-12 | Sick |
a All the crosses were done at 25°C.
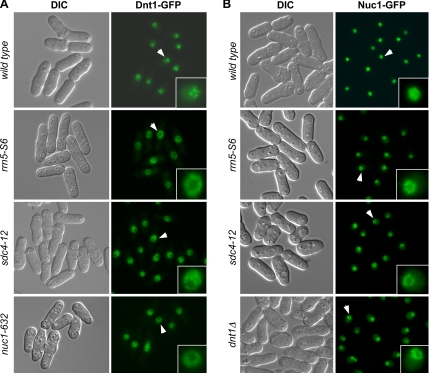
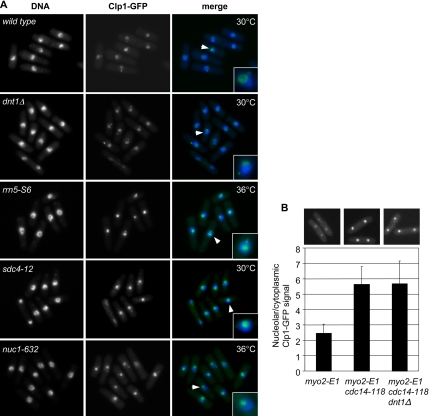
Further evidence also suggested an interaction between RNA Pol I and Dnt1. First, localization of Dnt1 to the nucleolus was disrupted in rrn5-S6, sdc4-12, and nuc1-632 cells (Figure 4A). At 30°C, the temperature where all these mutations can suppress the growth defects of cdc14-118 myo2-E1, Dnt1 is not enriched in the nucleolus in these mutants; instead, it was found in the nucleoplasm surrounding the nucleolus. This effect was not due to a global disruption of the nucleolus, because the nucleolar protein Gar2 still localized normally to the displaced nucleolus in these mutants at 30°C (Supplemental Figure S1). This disrupted localization of Dnt1 was observed in rrn5-S6 and nuc1-632 mutants even at 25°C, the permissive temperature for these mutants (data not shown). We also found that Dnt1 is required for maintaining the exclusive nucleolar localization of Nuc1, because examination of Nuc1-GFP localization in dnt1Δ cells showed signals not only in nucleolus but also in nucleoplasm (Figure 4B). This localization pattern is distinct from wild-type cells, in which Nuc1-GFP is almost exclusively found in nucleolus (Figure 4B). We noticed that Nuc1-GFP dnt1Δ cells grew very poorly, although strains carrying either individual allele grew well. This might be because GFP-tagged Nuc1 is not completely functional; thus, nuc1-GFP dnt1Δ cells demonstrate a negative genetic interaction (data not shown). Consistent with the genetic interactions we observed between all suppressors, we also found that nucleolar Nuc1-GFP localization is slightly disrupted in rrn5-S6 and sdc4-12 mutants. Like in dnt1Δ cells, strong Nuc1-GFP signals were found at the periphery of the nucleolus and weaker signals in the nucleoplasm (Figure 4B).
Figure 4.
Nucleolar localization of Dnt1 and Nuc1 is disrupted in suppressor mutants. Wild-type and mutant cells with the indicated genotypes were first grown at 25°C, and then they were shifted to 30°C for 4 h before being visualized by fluorescence microscopy. Arrows indicate the nuclei enlarged in insets.
S. pombe Dnt1: A Homologue of Budding Yeast Net1/Cfi1?
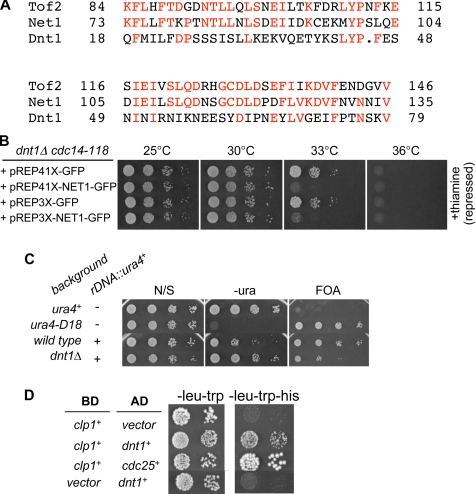
So why might these mutations in nucleolar proteins suppress cdc14-118 myo2-E1? A similar screen in S. cerevisiae identified Net1/Cfi1, the nucleolar inhibitor of the Cdc14 phosphatase (Shou et al., 1999). Our basic database searches had not revealed any obvious homologues of Net1/Cfi1 in S. pombe. However, in the course of this study, more sophisticated database searching (with advice from Dr. Aaron Neiman, SUNY, Stony Brook, NY) identified a candidate S. pombe protein called SPBC25D12.02c, which corresponds to Dnt1. Among Net1/Cfi1 homologues in related budding yeast, only the N-terminal 180 amino acids are conserved. This region is also conserved in a second nucleolar protein in S. cerevisiae called Tof2 (Huang et al., 2006) (Figure 5A). Psi-Blast searches with this region identified SPBC25D12.02c as the best homologue in S. pombe of the S. cerevisiae NET1/CFI1 and TOF2 genes (Figure 5A).
Figure 5.
Relationship between S. pombe Dnt1 and S. cerevisiae Net1/Cfi1. (A) Sequence alignment between Dnt1, Net1, and Tof2 at the N termini of the three proteins. (B) Rescue of cdc14-118 by dnt1Δ can be reversed by expression of budding yeast NET1. dnt1Δ cdc14-118 cells transformed with either empty vectors (pREP41X-GFP or pREP3X-GFP) or the NET1-expressing plasmid (pREP41X-NET1-GFP or pREP3X-NET1-GFP) were first grown at 25°C in EMM medium plus thiamine, and then they were washed, diluted, and spotted on EMM plates with or without (data not shown) thiamine. Plates were incubated at different temperatures as indicated for 3–5 d. (C) Dnt1 is involved in rDNA silencing. Wild-type and dnt1Δ cells carrying ura4+ gene inserted at the rDNA repeats (rDNA::ura4+) and control wild-type cells carrying endogenous (ura4+) or deleted ura4+ gene (ura4-D18) were grown at 30°C and serially diluted and spotted onto the indicated plates. The plates were incubated at 30°C for 3–5 d before photography. (D) Dnt1 and Clp1 interact in the two-hybrid assay. Clp1 was fused with the DNA-binding domain of GAL4 (BD) and Dnt1 with the transcriptional activation domain of GAL4 (AD). Both fusion constructs were coexpressed and the growth on SD, −Leu, −Trp and SD, −Leu, −Trp, −His is shown. As controls, coexpressions of Clp1 and empty AD vector, and empty DB vector with Dnt1 (negative control) or an AD fusion with S. pombe Cdc25 (positive control) are shown. The two-hybrid interaction between Clp1 and Cdc25 has been described previously (Wolfe and Gould, 2004).
The similarity of Dnt1 to Net1/Cfi1 is intriguing, because both Dnt1 and Net1/Cfi1 were identified in similar screens for suppressors of the SIN and MEN pathways, respectively. As described above, we found that dnt1Δ rescues not only the cdc14-118 myo2-E1 double mutant but also either single mutant (Table 3). Further study showed that dnt1Δ can weakly suppress other SIN mutants (Table 3), consistent with Dnt1 being an inhibitor of the SIN. To examine functional conservation between Net1/Cfi1 and Dnt1, we tested whether Net1/Cfi1 could reverse the effects of dnt1Δ on the SIN. We had found that dnt1Δ partially rescues the growth defect of the cdc14-118 mutant, allowing it to grow at 33°C (Table 3). We expressed Net1/Cfi1 from plasmids (pREP41X-NET1-GFP, pREP3X-NET1-GFP) under the control of inducible nmt1 promoter in dnt1Δ cdc14-118 cells, and we observed that these cells are dead at 33°C, whereas cells with control plasmid grow well, showing that Net1/Cfi1 expression reversed the rescue phenotype of dnt1Δ at this temperature (Figure 5B). It is possible that this simply reflects toxicity associated with expression of NET1/CFI1 in S. pombe. However, Net1/Cfi1 expressed from either strong (pREP3X) or intermediate (pREP41X) nmt1 promoters gives a similar reversion phenotype; also, the reversion of suppression occurred even in the presence of thiamine when only low levels of Net1/Cfi1 are expressed. Furthermore, Net1/Cfi1 expression did not inhibit growth at lower temperatures, showing that at these expression levels it was not acting as a general growth inhibitor.
Table 3.
Summary of rescue of SIN mutants by dnt1 Δ
| 25°C | 30°C | 33°C | 36°C | |
|---|---|---|---|---|
| sid4-A1 | ++a | − | − | − |
| dnt1Δ sid4-A1 | ++ | + | − | − |
| cdc11-123 | ++ | ++ | ++ | − |
| dnt1Δ cdc11-123 | ++ | ++ | ++ | − |
| spg1-106 | ++ | − | − | − |
| dnt1Δ spg1-106 | ++ | − | − | − |
| cdc7-24 | ++ | ++ | ++ | + |
| dnt1Δ cdc7-24 | ++ | ++ | ++ | + |
| sid1-125 | ++ | − | − | − |
| dnt1Δ sid1-125 | ++ | + | − | − |
| sid1-239 | ++ | ++ | ++ | − |
| dnt1Δ sid1-239 | ++ | ++ | ++ | + |
| cdc14-118 | ++ | ++ | − | − |
| dnt1Δ cdc14-118 | ++ | ++ | ++ | − |
| sid2-250 | ++ | − | − | − |
| dnt1Δ sid2-250 | ++ | + | +/− | − |
| mob1-1 | ++ | + | − | − |
| dnt1Δ mob1-1 | ++ | + | − | − |
Growth was examined with serial dilution drop test at different temperatures.
a ++, good growth; +, weak growth; +/−, weak growth with variations in growth in different clones; −, no growth.
Other similarities between Dnt1 and Net1/Cfi1 include the fact that both proteins localize to the nucleolus (Figure 3), and like Net1/Cfi1 (Straight et al., 1999; Shou et al., 2001), we found that Dnt1 is required for rDNA silencing as judged by increased expression (i.e., derepression) of a reporter gene (ura4+) integrated into the rDNA repeats (Thon and Verhein-Hansen, 2000) (Figure 5C). Although we have not tested directly whether other suppressors have a similar effect on rDNA silencing, it would not be surprising if they did because RNA Pol I transcription activity is required for rDNA silencing in S. cerevisiae (Shou et al., 2001). Additionally, as with Net1/Cfi1 (Shou and Deshaies, 2002), Dnt1 is also involved in maintenance of minichromosomes. dnt1Δ cells showed an almost 100-fold increase in minichromosome loss rate compared with wild-type cells: with loss rate of 1.78 × 10−2 in dnt1Δ cells versus 2 × 10−4 in wild-type cells.
Although the genetic interactions we observe between dnt1Δ and mutants in the RNA polymerase I machinery suggest that Dnt1 may participate in Pol I transcription like Net1/Cfi1, we do not think that it plays a direct role, because unlike the net1Δ/cfi1Δ mutant, dnt1Δ cells do not have reduced growth rates compared with wild-type cells. Additionally, we did not observe cross-complementation between Dnt1 and Net1/Cfi1 for their putative roles in Pol I transcription. Specifically, we found that Net1/Cfi1 could not rescue the synthetic growth defects we observed in dnt1Δ rrn5-S6 strains (data not shown). We also tested whether Dnt1 could rescue the growth defects of net1Δ/cfi1Δ cells at high temperatures, which are thought to be due to defects in Pol I activity, because they can be rescued by overexpression by Pol I transcription factors (Shou et al., 2001). However, Dnt1 was not able to rescue the growth defects of net1Δ/cfi1Δ cells at high temperatures (data not shown). In summary, although we found some interesting similarities between Dnt1 and Net1/Cfi1, the proteins do not seem to be functionally interchangeable.
Does Dnt1 Act by Antagonizing Clp1?
We next tested whether Dnt1 and Net1/Cfi1 inhibit the SIN and MEN signaling pathways, respectively, through a common mechanism. It is known that Net1/Cfi1 inhibits MEN signaling by binding to the Cdc14 phosphatase, and both sequestering it in the nucleolus and inhibiting its phosphatase activity (Traverso et al., 2001). Although we were able to detect a modest interaction between Dnt1 and Clp1 in the yeast two-hybrid assay (Figure 5D), we have been unable to detect an interaction between endogenous or bacterially expressed Dnt1 and Clp1 by coimmunoprecipitation or in vitro binding assays (data not shown). In addition, bacterially expressed Dnt1 does not seem to inhibit Clp1 phosphatase activity in vitro (Ray and McCollum, unpublished data).
dnt1Δ and the Other Suppressors Do Not Cause Premature Release of Clp1 from Nucleolus
Because dnt1+ and the other suppressors we identified encode nucleolar proteins like Clp1, we thought that the suppressors might act by causing release of Clp1 from the nucleolus and allowing it to remain active and promote cytokinesis checkpoint signaling. To address whether the suppressors we isolated have effects on nucleolar localization of Clp1, we examined the localization of Clp1-GFP in dnt1Δ, rrn5-S6, sdc4-12, and nuc1-632 mutant strains (Figure 6A). All cells were first grown at 25°C, and then they were shifted to 30°C for 4 h, the temperature at which they showed suppression of cdc14-118 myo2-E1. Except for dnt1Δ mutant, all the other mutants show a characteristic ring-shaped DNA phenotype at permissive or restrictive temperature, with the nucleolus in the center of the nucleus. However, we did not find Clp1 to be released prematurely in interphase cells in any of the suppressor mutants, even at fully restrictive temperature (Figure 6A; data not shown). We also compared Clp1-GFP localization in cdc14-118 myo2-E1 and cdc14-118 myo2-E1 dnt1Δ cells to myo2-E1 cells after shift to 30°C. By comparing the ratio of the mean intensity of nucleolar verses cytoplasmic fluorescence in telophase cells we found, as expected, that Clp1 remains out of the nucleolus in myo2-E1 cells (Figure 6B) as normally occurs when cytokinesis is perturbed and the cytokinesis checkpoint is activated. However, Clp1-GFP returned to the nucleolus prematurely in both cdc14-118 myo2-E1 and cdc14-118 myo2-E1 dnt1Δ cells (Figure 6B). Together, these data indicate that the suppressors of the cdc14-118 myo2-E1 double mutant do not rescue by simply keeping Clp1 out of nucleolus.
Figure 6.
(A) Suppressors do not cause premature release of Clp1 from nucleolus. Wild-type and mutant cells with indicated genotypes were first grown at 25°C, and then they were shifted to 30 or 36°C for 4 h before being fixed and stained with DAPI. Arrows indicate the nuclei enlarged in insets. (B) Cells of the indicated genotypes were grown to log phase at 25°C, and then they were shifted to 30°C for 4 h. These cells were then methanol fixed and imaged for Clp1-GFP localization. The ratio of the mean average intensity of the Clp1-GFP signal in the nucleolus and cytoplasm is shown (n > 50). Representative images are shown. Only cells that were in telophase, as judged by nuclear positioning, were analyzed.
Dnt1 Can Regulate the SIN Independently of Clp1
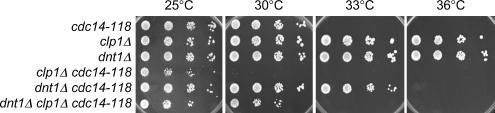
As described above, we found that dnt1Δ rescues not only the cdc14-118 myo2-E1 double mutant but also either single mutant as well as other SIN mutants. If the rescue of SIN mutants by dnt1Δ was through Clp1, then it should depend on clp1+. To test this, we compared the phenotypes of different combinations of single, double, and triple mutants between cdc14-118, clp1Δ, and dnt1Δ (Figure 7). We found that cdc14-118 cells can grow up to 30°C but that they die at the restrictive temperature of 33°C. As expected, deletion of Clp1 and Dnt1 have opposite effects on the cdc14-118 mutant, with clp1Δ reducing the restrictive temperature of cdc14-118 cells to 30°C and dnt1Δ raising the restrictive temperature of cdc14-118 cells to 36°C. Interestingly, deletion of dnt1+ raises the restrictive temperature of the cdc14-118 clp1Δ double mutant from 30 to 33°C, showing that Dnt1 can affect the SIN even in the absence of clp1+. We also found that dnt1Δ could promote growth of the myo2-E1 mutant in the absence of Clp1 (data not shown). Together, these results clearly show that dnt1Δ can promote SIN function independent of Clp1, and the simplest interpretation of our results is that Clp1 and Dnt1 act on the SIN independently.
Figure 7.
Dnt1 can regulate the SIN independently of Clp1. Wild-type and mutant cells with indicated genotypes were first grown in YE at 25°C, and then they were serially diluted and spotted on plates of YE. Plates were incubated at different temperatures as indicated for 3–5 d before photography.
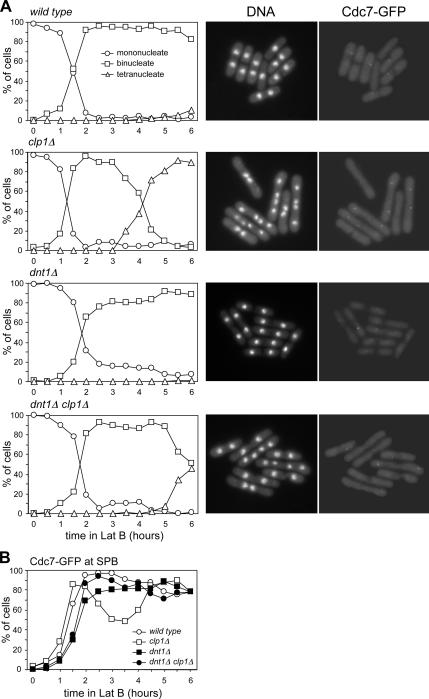
Another piece of evidence supporting the idea that Dnt1 can regulate SIN signaling and cytokinesis checkpoint in the absence of Clp1 came from our analysis of checkpoint activation and SIN signaling in dnt1Δ and clp1Δ single and double mutant cells. These cells also expressed Cdc7-GFP whose localization to the SPB can serve as a marker for SIN activation (for review, see McCollum and Gould, 2001). A low dosage of latrunculin, a drug that prevents actin polymerization (Ayscough et al., 1997), has been shown to be able to activate the cytokinesis checkpoint in S. pombe by slowing down actomyosin ring contraction (Mishra et al., 2004). We compared the kinetics of nuclear cycle progression in synchronized wild-type, dnt1Δ, clp1Δ, and clp1Δ dnt1Δ cells upon treatment with 4 μM latrunculin B in liquid cultures (Figure 8A). Wild-type and dntΔ cells, which have an intact checkpoint, remained binucleate with the SIN activated, and they slowly formed a septum (Figure 8, A and B). In contrast, clp1Δ cells, which lack the checkpoint, are unable to maintain SIN signaling, and they become multinucleate and are unable to form complete septa (Figure 8, A and B) (Mishra et al., 2004). Unlike clp1Δ single mutants, clp1Δ dnt1Δ cells maintain SIN signaling and remain blocked as binucleate cells for an extended period, although not as long as wild-type or dnt1Δ cells (Figure 8A). These data suggest that dnt1Δ allows cells to maintain SIN signaling and keep the cytokinesis checkpoint active even in the total absence of Clp1. Therefore, Dnt1 must be able to affect SIN signaling through an alternative pathway.
Figure 8.
dnt1Δ has a positive effect on SIN signaling even in the absence of Clp1. Wild-type and mutant cells with the indicated genotypes (all carrying Cdc7-GFP) were first grown in YE at 30°C, and then they were synchronized in early G2 by centrifugal elutriation and treated with 4 μM latrunculin B. Cells were withdrawn every 30 min for a period of 6 h, and then they were fixed and stained with DAPI. (A) Left, number of nuclei over time for each strain. Right, examples of DNA staining and Cdc7-GFP signal at the 4-h time point for each strain. (B) Quantification of Cdc7-GFP signals is shown. At least 200 cells were counted for each time point.
DISCUSSION
Our screen for suppressors of the cytokinesis checkpoint defect of SIN mutants identified several nucleolar proteins, Dnt1, Rrn5, and Nuc1, and one uncharacterized protein. Dnt1 seems to act as an inhibitor of SIN signaling, and it is also involved in rDNA silencing. Dnt1 has sequence similarity to two nucleolar proteins from budding yeast called Net1/Cfi1 and Tof2 that are both involved in rDNA silencing. Interestingly S. cerevisiae Net1/Cfi1, and a second nucleolar protein involved rDNA silencing called Fob1 also have roles in regulating the MEN pathway (Straight et al., 1999; Shou et al., 2001; Huang and Moazed, 2003; Stegmeier et al., 2004). It is not known whether Tof2 regulates MEN signaling. The effects of Net1/Cfi1 and Fob1 in mitotic exit control seem to be through the Cdc14 phosphatase. The key function of Net1 in controlling MEN in budding yeast is to bind to and inhibit the Cdc14 phosphatase in the nucleolus (Shou et al., 1999). However, our data suggest that unlike Net1/Cfi1, Dnt1 can regulate the SIN independently of the Clp1 phosphatase. We only found very weak interaction between Dnt1 and Clp1 by two-hybrid analysis but not in coimmunoprecipitation or in vitro binding assays. The weak interaction might suggest that Dnt1 is a substrate of Clp1, consistent with our observations that Dnt1 is phosphorylated on Cdk1 sites in vivo (Jin and McCollum, unpublished observations). Furthermore, the absence of Dnt1 does not lead to premature release of Clp1 into nucleoplasm or cytoplasm. Given the apparent differences between Net1/Cfi1 and Dnt1, it is surprising that Net1 could reverse the suppression of cdc14-118 cells by dnt1Δ. However, this could be explained if both proteins inhibit the SIN through different mechanisms. The simplest model to explain our data on the relationship between Dnt1 and Clp1 is that the two proteins independently regulate the SIN.
The mechanism by which the suppressors we identified promote SIN signaling is unclear. We do not think that the suppression is due to loss of rDNA silencing, because deletion of Sir2, which is required for rDNA silencing (Shankaranarayana et al., 2003), does not rescue cdc14-118 myo2-E1 mutants (data not shown). In principle, perturbation of RNA Pol I activity could rescue cdc14-118 myo2-E1 mutants through an indirect mechanism such as reduction of protein synthesis rates caused by Pol I inhibition, or changes in general transcription rates leading to increased levels of Cdc14 or Myo2 protein. We think this unlikely, primarily because dnt1Δ cells grow at wild-type growth rates, thus they are unlikely to be significantly impaired for Pol I activity; and second, graded reduction in overall protein synthesis rates by using tetracycline did not promote rescue of cdc14-118 myo2-E1 mutants, nor does deletion of dnt1 have a significant effect on Cdc14, Sid1, or Myo2 protein levels (data not shown). The other mutations in RNA Pol I factors might then rescue SIN signaling, because they all disrupt nucleolar localization of Dnt1, but the exact function of Dnt1 in SIN signaling remains to be determined.
What is the link between nucleolar proteins and SIN regulation? Definition of the nucleolus as the site of rDNA transcription and ribosome biogenesis is well established in both yeast and higher eukaryotes. Recent studies have expanded the functions of nucleolus to include roles in recruitment and exclusion of regulatory complexes (Garcia and Pillus, 1999; San-Segundo and Roeder, 1999; Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999). The synthesis of ribosomes consumes a vast amount of the resources in rapidly growing cells, and the nucleolus is emerging as a key control point for the regulation of cell growth and division, both in yeast and human cells (for review, see Dez and Tollervey, 2004). More generally, the high metabolic cost of ribosome synthesis may have selected for its close integration with cell growth and division. Together, these results suggest that the nucleolus may serve as a site to integrate signals governing cell growth and cell cycle progression, and this will serve as an exciting avenue for further research.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Drs. Kathy Gould (Vanderbilt University), Angelika Amon (M.I.T.), and Viesturs Simanis (ISREC) for providing strains and plasmids. We thank Rachel E. Lamson for providing wild-type budding yeast genomic DNA and Matthew J. Winters for advice on two-hybrid assay. We thank the McCollum laboratory members for discussions. This work was supported by National Institutes of Health grants GM-058406-08 and GM-068786-04 (to D.M.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0853) on May 30, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Ayscough K. R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D. G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam R., Chen S. L., Shou W., Mah A. S., Alexandru G., Nasmyth K., Annan R. S., Carr S. A., Deshaies R. J. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M. K., McCollum D., Gould K. L. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1997;283:494–506. doi: 10.1016/s0076-6879(97)83039-x. [DOI] [PubMed] [Google Scholar]
- Chen C.-T., Peli-Gulli M.-P., Simanis V., McCollum D. S.pombe orthologues of the FEAR proteins are not required for release of CDC14-family phosphatase Clp1/Flp1 from the nucleolus during mitosis. J. Cell Sci. 2006;119:4462–4466. doi: 10.1242/jcs.03220. [DOI] [PubMed] [Google Scholar]
- Cho H. P., Liu Y., Gomez M., Dunlap J., Tyers M., Wang Y. The dual-specificity phosphatase CDC14B bundles and stabilizes microtubules. Mol. Cell Biol. 2005;25:4541–4551. doi: 10.1128/MCB.25.11.4541-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cueille N., Salimova E., Esteban V., Blanco M., Moreno S., Bueno A., Simanis V. Flp1, a fission yeast orthologue of the S. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 2001;114:2649–2664. doi: 10.1242/jcs.114.14.2649. [DOI] [PubMed] [Google Scholar]
- Dez C., Tollervey D. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Garcia S. N., Pillus L. Net results of nucleolar dynamics. Cell. 1999;97:825–828. doi: 10.1016/s0092-8674(00)80794-1. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Guertin D. A., Trautmann S., McCollum D. Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Funahashi S. I., Uemura T., Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 1986;5:2973–2979. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Konoha G., Toda T., Yanagida M. Essential roles of the RNA polymerase I largest subunit and DNA topoisomerases in the formation of fission yeast nucleolus. J. Cell Biol. 1989;108:243–253. doi: 10.1083/jcb.108.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Brito I. L., Villen J., Gygi S. P., Amon A., Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser B. K., Zimmerman Z. A., Charbonneau H., Jackson P. K. Disruption of centrosome structure, chromosome segregation, and cytokinesis by misexpression of human Cdc14A phosphatase. Mol. Biol. Cell. 2002;13:2289–2300. doi: 10.1091/mbc.01-11-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang H., Balasubramanian M. K. A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J. Cell Sci. 2000;113:1223–1230. doi: 10.1242/jcs.113.7.1223. [DOI] [PubMed] [Google Scholar]
- Liu M., Guo A., Boukhgalter B., Van Den Heuvel K., Tripp M., Pape L. Characterization of the fission yeast ribosomal DNA binding factor: components share homology with Upstream Activating Factor and with SWI/SNF subunits. Nucleic Acids Res. 2002;30:5347–5359. doi: 10.1093/nar/gkf683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum D., Gould K. L. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- Mishra M., Karagiannis J., Trautmann S., Wang H., McCollum D., Balasubramanian M. K. The Clp1p/Flp1p phosphatase ensures completion of cytokinesis in response to minor perturbation of the cell division machinery in Schizosaccharomyces pombe. J. Cell Sci. 2004;117:3897–3910. doi: 10.1242/jcs.01244. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- San-Segundo P. A., Roeder G. S. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97:313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- Shankaranarayana G. D., Motamedi M. R., Moazed D., Grewal S. I. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 2003;13:1240–1246. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- Shou W., Deshaies R. J. Multiple telophase arrest bypassed (tab) mutants alleviate the essential requirement for Cdc15 in exit from mitosis in S. cerevisiae. BMC Genet. 2002;3:4. doi: 10.1186/1471-2156-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W., et al. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell. 2001;8:45–55. doi: 10.1016/s1097-2765(01)00291-x. [DOI] [PubMed] [Google Scholar]
- Shou W., Seol J. H., Shevchenko A., Baskerville C., Moazed D., Chen Z. W., Jang J., Shevchenko A., Charbonneau H., Deshaies R. J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Simanis V. Events at the end of mitosis in the budding and fission yeasts. J. Cell Sci. 2003;116:4263–4275. doi: 10.1242/jcs.00807. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Huang J., Rahal R., Zmolik J., Moazed D., Amon A. The replication fork block protein Fob1 functions as a negative regulator of the FEAR network. Curr. Biol. 2004;14:467–480. doi: 10.1016/j.cub.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Visintin R., Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Straight A. F., Shou W., Dowd G. J., Turck C. W., Deshaies R. J., Johnson A. D., Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Thon G., Verhein-Hansen J. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics. 2000;155:551–568. doi: 10.1093/genetics/155.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S., Wolfe B. A., Jorgensen P., Tyers M., Gould K. L., McCollum D. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 2001;11:931–940. doi: 10.1016/s0960-9822(01)00268-8. [DOI] [PubMed] [Google Scholar]
- Traverso E. E., Baskerville C., Liu Y., Shou W., James P., Deshaies R. J., Charbonneau H. Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J. Biol. Chem. 2001;276:21924–21931. doi: 10.1074/jbc.M011689200. [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang E. S., Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Wolfe B. A., Gould K. L. Fission yeast Clp1p phosphatase affects G2/M transition and mitotic exit through Cdc25p inactivation. EMBO J. 2004;23:919–929. doi: 10.1038/sj.emboj.7600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. A., McDonald W. H., Yates J. R., 3rd, Gould K. L. Phospho-regulation of the Cdc14/Clp1 phosphatase delays late mitotic events in S. pombe. Dev. Cell. 2006;11:423–430. doi: 10.1016/j.devcel.2006.07.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.