Abstract
Mono-ADP-ribosylation is emerging as an important posttranslational modification that modulates a variety of cell signaling pathways. Here, we present evidence that mono-ADP-ribosylation of the transcriptional corepressor C terminal binding protein, brefeldin A (BFA)-induced ADP-ribosylated substrate (CtBP1/BARS) regulates neutral lipid storage in droplets that are surrounded by a monolayer of phospholipid and associated proteins. CtBP1/BARS is an NAD-binding protein that becomes ribosylated when cells are exposed to BFA. Both endogenous lipid droplets and droplets enlarged by oleate treatment are lost after 12-h exposure to BFA. Lipid loss requires new protein synthesis, and it is blocked by multiple ribosylation inhibitors, but it is not stimulated by disruption of the Golgi apparatus or the endoplasmic reticulum unfolded protein response. Small interfering RNA knockdown of CtBP1/BARS mimics the effect of BFA, and mouse embryonic fibroblasts derived from embryos that are deficient in CtBP1/BARS seem to be defective in lipid accumulation. We conclude that mono-ADP-ribosylation of CtBP1/BARS inactivates its repressor function, which leads to the activation of genes that regulate neutral lipid storage.
INTRODUCTION
Lipid droplets are metabolically active organelles rich in proteins that regulate lipid synthesis, storage, and degradation as well as membrane traffic (Brasaemle et al., 2004, Liu et al., 2004, Mlickova et al., 2004, Pol et al., 2004, Umlauf et al., 2004). Lipid droplets or lipid bodies (Martin and Parton, 2005) are common to most cells, and they seem to originate from the endoplasmic reticulum in eukaryotic cells (van Meer, 2001) or the plasma membrane in bacterial cells (Waltermann et al., 2005). We have proposed that the core machinery responsible for lipid accumulation deserves special status as an organelle that packages and distributes lipids in cells, and we have suggested the name adiposome to designate this cellular compartment (Liu et al., 2004). Thus, in response to an increase in cellular fatty acid or cholesterol, adiposomes package esterified lipids into droplets surrounded by a monolayer of phospholipid. Virtually any eukaryotic or bacterial cell can be induced to make lipid droplets, which suggests all cells contain the adiposome core machinery. Among the proteins present on lipid droplets are caveolin-1 and -2 (Fujimoto et al., 2001, Liu et al., 2004, Pol et al., 2004). Caveolin-1 (Cav-1) was originally discovered as the principal protein component of the filamentous caveolae coat (Rothberg et al., 1992). Droplet-associated Cav-1 markedly increases when cells are grown overnight in the presence of oleic acid or exposed for short times to the fungal metabolite brefeldin A (BFA) (Pol et al., 2004). BFA also inhibits caveolae internalization (Richards et al., 2002). BFA is best known for its ability to block membrane traffic between the endoplasmic reticulum (ER) and Golgi by interfering with COPI-coated vesicle assembly. BFA blocks membrane traffic by preventing GDP-GTP exchange of Arf1, a key regulator of COPI coatomer recruitment, thereby maintaining Arf1 in the GDP-bound state (Donaldson et al., 1992, Helms and Rothman, 1992, Palmer et al., 1993, Jackson and Casanova, 2000). Arf1-GTP can also regulate phospholipase D1, which converts phosphatidylcholine to phosphatidic acid (PA). PA is a key regulatory lipid in a variety of signaling pathways, and it is a modulator of membrane curvature (Freyberg et al., 2003). Another effect of BFA is to stimulate mono-ADP-ribosylation of C terminal binding protein, BFA-induced ADP-ribosylated substrate (CtBP1/BARS) (Di Girolamo et al., 1995, Feng et al., 2003). CtBP1/BARS functions as both a transcriptional corepressor (Zhang et al., 2000) and as a modulator of Golgi structure (Nardini et al., 2003). CtBP1/BARS is an NAD-regulated dehydrogenase that is homologous to 2-hydroxyacid dehydrogenases (Kumar et al., 2002). It functions as a transcriptional corepressor by binding to PXDLS sequences present in target transcription factors (Nardini et al., 2003). Here, we present evidence that CtBP1/BARS regulates lipid accumulation in lipid droplets and that BFA-stimulated mono-ADP-ribosylation of CtBP1/BARS results in a dramatic increase in fatty acid efflux and the concomitant loss of lipid droplets from cells.
MATERIALS AND METHODS
Materials
Monoclonal antibody (mAb) α-GM130, mAb α-CtBP1/BARS, mAb α-CtBP2, and mAb α-GRP78 (BIP) were purchased from BD Biosciences (San Jose, CA); mAb α-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (clone 6C5) was from Millipore (Billerica, MA); polyclonal antibody (pAb) α-green fluorescent protein (GFP) was from J. Seemann; mAb α-adipocyte differentiation-related protein (ADRP) was from Research Diagnostics (Flanders, NJ); pAb α-Rab18 was from Calbiochem (La Jolla, CA); pAb α-cyclophilin A was from ABR–Affinity BioReagents (Golden, CO); and mAb α-tubulin, mAb α-actin, and oil red O were from Sigma-Aldrich (St. Louis, MO). pAb against εCOP and βCOP were used as described previously (Lowe and Kreis, 1996). BFA was from Epicenter Technologies (Madison, WI); Mowiol and protease inhibitor cocktail 3 were obtained from Calbiochem. Alexa Fluor 488 and Alexa Fluor 568 fluorescent secondary antibodies were purchased from Invitrogen (Carlsbad, CA). [3H]Oleate was from PerkinElmer-Cetus (Wellesley, MA). Fortified cosmic calf serum and fetal bovine serum were from Hyclone (Logan, UT); Si-gel G plates were from Whatman (Brentford Middlesex, United Kingdom). Nicotinamide and oleate were from Sigma-Aldrich. m-Iodobenzylguanidine (MIBG) was from Calbiochem. All other reagents were obtained from Calbiochem, Invitrogen, and Sigma-Aldrich.
Cells and Tissue Culture
Normal rat kidney (NRK), Chinese hamster ovary (CHO), and immortalized human fibroblasts (SV589) were maintained in DMEM with high glucose (4.5 g/l) supplemented with 10% fortified cosmic calf serum (CHO and NRK) or 10% fetal bovine serum (SV589) plus 100 U/ml penicillin, 100 μg/ml streptomycin at 37°C in a standard tissue culture incubator with 5% CO2. CHO cells were additionally supplemented with 40 μg/ml l-proline. CHO SRD-12 A cells were grown as described previously (Rawson et al., 1998). Mouse embryo fibroblasts (wild type [WT] and CtBP1/2−/−) were kindly provided by Dr. Hildebrand (University of Pittsburgh, Pittsburgh, PA), and they were cultured as described previously (Hildebrand and Soriano, 2002).
Neutral Lipid Staining
For oil red O staining, cells grown on glass coverslips were washed twice with phosphate-buffered saline (PBS), and then they were fixed with 4% formaldehyde in PBS for 20 min at room temperature (RT). Cells were then washed 3 × 5 min with PBS, and they were incubated with a filtered (0.2 μm) 3:2 dilution (with H2O) of a 1% stock solution of oil red O in isopropanol. Cells were stained for 10 min at RT, washed 3 × 5 min with PBS, and mounted on a glass slide with Mowiol.
Immunofluorescence
For immunofluorescence, cells were either fixed immediately and permeabilized for 30 min with methanol (−20°C), or they were first fixed with 4% formaldehyde in PBS as described above and then permeabilized with methanol (−20°C for 30 min). Alternatively, cells were permeabilized with 0.1% Triton X-100 (in PBS) for 5 min on ice. After fixation and permeabilization, cells were washed 3 × 5 min with PBS, and then they were incubated with primary antibodies for 30 min at 37°C in a humidified chamber. Cells were then washed 3 × 5 min with PBS and incubated with Alexa Fluor 488- or Alexa Fluor 568-conjugated fluorescent secondary antibodies for 30 min at 37°C in a humidified chamber. After incubation, cells were washed 3 × 5 min with PBS, and then they were mounted on a glass slide with Mowiol. Cells (for oil red O staining and immunofluorescence) were observed with a Zeiss Axioplan 2E microscope (Carl Zeiss, Thornwood, NY) by using Plan-Neofluar 40×/1.3 oil differential interference contrast (DIC) and Plan-Neofluar 63×/1.4 oil DIC objectives. Pictures were taken with a monochrome digital camera (ORCA-II; Hamamatsu, Hamamatsu City, Japan), analyzed using Openlab (Improvision, Lexington, MA), and colorized with Adobe Photoshop (Adobe Systems, San Jose, CA).
Cell Fractionation
For some experiments, we used purified droplets (designated purified droplets) prepared by the method of Liu et al. (2004). In other experiments, we used droplet-enriched fractions (designated enriched droplets). These fractions were isolated from cells grown to confluence in 100-mm dishes. Cells were washed twice with PBS on ice, and then they were scraped into 5 ml of ice-cold PBS. Cells were pelleted (500 × g for 5 min at 4°C), and then they were resuspended in 1 ml of buffer A (250 mM sucrose and 20 mM Tricine, pH 7.8, plus proteinase inhibitors). After incubation for 20 min on ice, cells were homogenized with a dounce homogenizer (20 strokes), and then they were transferred to a 1.5-ml tube. The lysate was centrifuged (1020 × g for 7 min at 4°C), and the postnuclear supernatant (PNS) fraction was transferred to a new 1.5-ml tube and centrifuged for 40 min at 264,499 × g at 4°C. The top 500 μl (containing lipid droplet-enriched phase) was collected and transferred to a new 1.5-ml tube. The 500 μl was reduced to 30 μl by repeated centrifugation at 20,000 × g for 2 min at 4°C and sequential removal of the liquid from the bottom of the centrifuge tube. One milliliter of acetone was added to the 30 μl to precipitate the protein. After incubation for 10 min on ice, precipitated proteins were centrifuged at 20,000 × g for 20 min at 4°C. The supernatant fraction was removed, the pellet was dried, and the proteins were dissolved in SDS sample buffer containing 2% β-mercaptoethanol, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and processed for immunoblotting.
Lipid Extraction and Thin Layer Chromatography (TLC)
Lipids were extracted by the method of Bligh and Dyer (1959). Briefly, lipids were extracted by adding CHCl3:methanol (2:1), mixed, and centrifuged at 1000 × g for 10 min. The lower CHCl3 phase was removed, and it was transferred to a fresh tube. The sample was dried under nitrogen and resuspended in CHCl3. Extracted lipids were separated on Si-gel G plates by using a hexane:diethyl ether:acetic acid (80:20:1, vol/vol/vol) mixture for 50 min. Lipids were visualized by exposure to iodine vapor, and images were quantified using National Institutes of Health ImageJ software (http://rsb.info.nih.gov/ij/). Lipids were identified by relative migration to known standards.
Ribosylation Assay
Rat brain membranes were prepared, and BFA-dependent ADP-ribosylation was carried out by the method of Valente et al. (2005). Briefly, ADP-ribsoylation was carried out by combining solution A (1.5 mg/ml rat brain membranes, 60 μg/ml BFA or dimethyl sulfoxide [DMSO] and 5 mM dithiothreitol [DTT]) with solution B (250 μM NAD [Sigma-Aldrich] and 480 μCi/ml [32P]NAD [GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom] and 0.3–0.5 mg/ml cytosol from cultured cells). Both solutions were prepared in ribosylation buffer (50 mM potassium phosphate buffer, pH 7.5, 1.25 mM MgCl2, 0.5 mM ATP, 0.5 mM GTP, and 5 mM thymidine), and incubation was carried out at 37°C for 2 h. The sample was then centrifuged for 10 min at 18,000 × g, and the supernatant was recovered, processed for SDS-PAGE, and transferred onto polyvinylidene difluoride (PVDF)-membrane. Radioactivity was either visualized using Typhoon 9410 Image analysis (GE Healthcare) or by exposure to x-ray film. After exposure, membranes were processed for immunoblotting.
Knockdown of CtBP1/BARS and Quantification of Lipid Droplets
SV589 (65,000 cells/dish) were seeded into 35-mm dishes the day before each experiment. At day 0, cells were transfected with small interfering RNA (siRNA) oligonucleotides against either CtBP1/BARS (target sequence AACCACCACCTCATCAACGACTT or AAGGAGCATTTGGAAGTCAATTT) or as a control microsomal triglyceride transfer protein (target sequence AAACAGAAGCAGGCTTGGAGTTT) using Oligofectamine (Invitrogen) according to the manufacturer's protocol. On day 1, the medium was changed to regular medium without antibiotics, and on day 3 the cells were split 1:3, and they were plated onto glass coverslips. On day 4, cells were washed three times with PBS, and they were fixed with formaldehyde as described above and processed for oil red O staining and immunofluorescence. Acquired images were used to quantify the area of the lipid droplets by using ImageJ. Images from control cells were randomly chosen, and the lipid droplet size was measured in 120 cells for each point. For cells transfected with CtBP1/BARS siRNA, the fluorescence intensity of the cell in the CtBP1/BARS channel was measured and compared with that of control cells. Cells exhibiting a >70% decrease in the CtBP1/BARS signal were selected, and the droplet area was quantified as described above. The result is presented as the average droplet area ± SE of three independent experiments. In other experiments, siRNA-treated cells were assayed for the amount of CtBP1/BARS protein by immunoblotting.
Other Methods
Living NRK cells were observed with a Zeiss Axiovert 200M microscope equipped with a temperature-controlled incubation chamber. Cells were cultured in CO2 independent medium (Invitrogen) containing 10% cosmic calf serum for up to 18 h, and then they were viewed by DIC with a Plan-Neofluar 40×/1.3 oil DIC objective. Images were taken (1 frame/min) for up to 14 h by using an ORCA-285 monochrome digital camera (Hamamatsu) and Openlab software (Improvision). Immunoblotting was carried out by separating proteins on either 12 or 10% polyacrylamide gels in SDS buffer containing 2% β-mercaptoethanol, transferred to PVDF membranes, and processed for detection of proteins by using enhanced chemiluminescence. Transfections with cDNA were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
RESULTS
BFA Stimulates Lipid Efflux from Cells
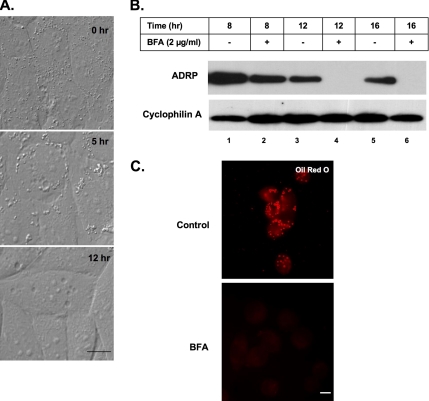
Previous studies suggested that BFA inhibits fatty acid uptake by modulating caveolae function (Pol et al., 2004, Richards et al., 2002). We used time-lapse microscopy to monitor the effects of BFA on lipid accumulation in cells grown in the presence of oleate for various times (Figure 1A; see movie in Supplemental Material). For this experiment, we used NRK cells, which have relatively few lipid droplets. During the first 5 h of incubation, lipid accumulated in cells as evidenced by an increase in the size (compare 0 with 5 h) of the droplets. After 5 h, however, the size and the number of droplets began to decline. By 12 h, virtually no droplets remained, even though we did not remove oleate from the medium. Therefore, rather than blocking the formation of lipid droplets, BFA dramatically stimulates their loss.
Figure 1.
Effect of BFA on uptake and storage of oleate. (A) NRK cells were continuously grown in the presence of 2 μg/ml BFA plus 100 μM oleate for the indicated time. These images are taken from the movie supplied in supplementary data. (B) CHO K2 cells were incubated in the presence of either 2 μg/ml BFA or EtOH carrier for the indicated time. Droplet-enriched fractions were prepared, equal amounts of protein separated on 12% SDS-PAGE and immunoblotted with α-ADRP and α-cyclophilin A IgG (loading control). (C) CHO cells were incubated in the presence of 2 μg/ml BFA (BFA) or EtOH carrier (control) for 12 h. Cells were fixed and processed to visualize neutral lipids with oil red O. Bar, 10 μm.
We used both visual and biochemical assays to confirm that BFA effected lipid retention. CHO K2 cells, which naturally have a large amount of droplets, were incubated for various times in the presence or absence of BFA, and the droplet fraction was prepared at the indicated time points (Figure 1B). In control cells, the droplet fraction is a milky white cake at the top of the centrifuge tube after the PNS is centrifuged at 250,000 × g. The amount of white cake markedly declined after a 12-h incubation in BFA, indicating a loss in lipid content. To quantify these results, a 500-μl sample from the top of each tube was processed to measure by immunoblotting the amount of ADRP, which is a well-accepted lipid droplet marker (van Meer, 2001). BFA caused a marked loss of ADRP (lanes 4 and 6). These results were confirmed by immunofluorescence (Figure 1C), which showed that a 12-h incubation in the presence of BFA caused a marked loss of oil red O-positive droplets from CHO cells.
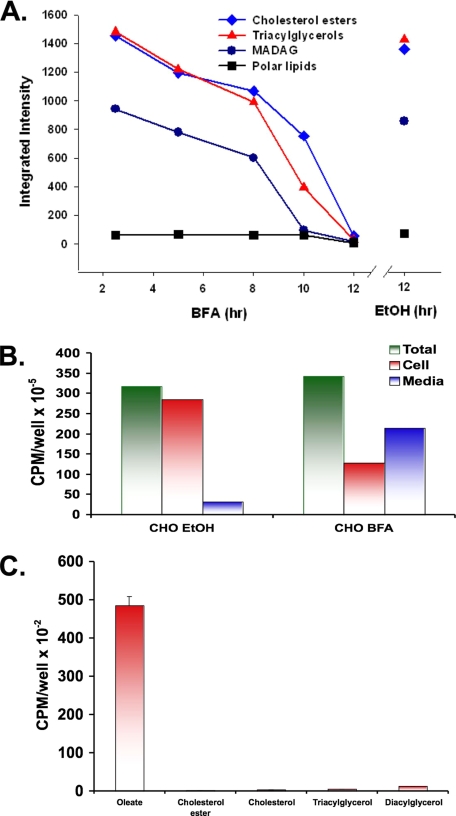
We next used TLC to separate lipids extracted from the droplet fraction isolated from CHO cells exposed to BFA for various times (Figure 2A). After a 5-h incubation in the presence of BFA, the level of stored cholesterol ester (♦), and monoalk(en)yl diacylglycerol (•) and triacylglycerol (▴) began to decline. Incubation in the presence of the ethanol carrier for 12 h had no effect. To better understand the fate of the lost lipids, CHO cells were allowed to incorporate [3H]oleate for 24 h, and then they were washed and incubated in the presence of either BFA or the ethanol carrier for 12 h. At the end of the incubation, the amount of radiolabeled lipid in the cells and the medium was determined (Figure 2B). Similar amounts of oleate were incorporated into both sets of cells. BFA caused a 63% loss of radioactivity from the cells, and most of it occurred in the medium. In a separate experiment performed under similar conditions (Figure 2C), the radiolabeled lipid in the medium of BFA-treated cells was identified as oleate. Therefore, BFA promotes the breakdown of stored neutral lipid and the subsequent release of free fatty acid into the medium.
Figure 2.
BFA stimulates release of free fatty acids from stored neutral lipid droplets. (A) CHO K2 cells were incubated in the presence of 2 μg/ml BFA for the indicated time at 37°C or EtOH carrier for 12 h. Purified droplets were prepared and the lipids extracted, separated by TLC, stained with iodine, and quantified using ImageJ software. (B) CHO K2 cells were incubated in the presence of 100 μM oleate plus 12 μCi of [3H]oleate for 24 h. Cells were then washed and incubated in the presence of 2 μg/ml BFA or EtOH carrier for 12 h at 37°C. Media were collected, the cells washed and homogenized, and the radioactivity in both samples was measured. (C) The 3H lipids in the medium from the BFA-treated cells was extracted and separated by TLC along with the standards for the indicated lipids. The area in the TLC next to the indicated standard was scraped, and the amount of radioactivity was measured.
BFA Effect Is Not Due to Golgi disruption
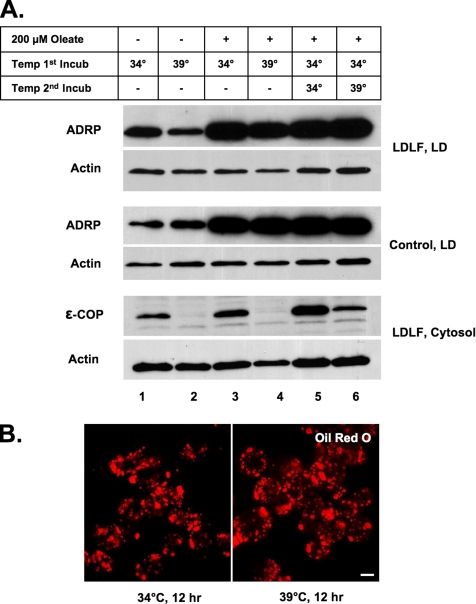
As little as 1 μg/ml (3.5 μM) BFA caused lipid loss from CHO cells, NRK cells, differentiated 3T3 L1 adipocytes, and both normal and immortalized human fibroblasts (data not shown). To determine whether lipid loss is linked to the well-characterized effect that BFA has on membrane traffic, we looked at the affects of Golgi disruption on droplet loss using LDLF cells (Figure 3). This temperature-sensitive CHO cell line exhibits a defect at the restrictive temperature in ER-to-Golgi traffic owing to a mutation in the εCOP protein (Guo et al., 1996). The Golgi disassembles at the restrictive temperature of 39°C due to the degradation of εCOP, thereby mimicking the effects of BFA on Golgi structure. Cells were incubated for 12 h at either 34°C (no effect on εCOP) or 39°C (εCOP degraded) in the presence or absence of 200 μM oleate. The droplet fraction was prepared, and the level of ADRP was measured by immunoblotting (Figure 3A, LD). Initially, there was a significant amount of ADRP in the droplet fraction of both LDLF and wild-type cells (lanes 1 and 2), indicating that these cells normally have droplets. Oleate addition markedly increased the amount of ADRP in both LDLF and wild-type cells (lanes 3–6). Regardless of the temperature or exposure to oleate, however, the droplet fraction in both LDLF and wild-type cells contained similar amounts of ADRP. Likewise, the restrictive temperature had a negligible effect on the number of oil red O-positive droplets (Figure 3B). A 12-h incubation at the restrictive temperature caused a marked loss of εCOP from the cytosol (Figure 3A, lanes 2 and 4), and these conditions also disrupted the Golgi apparatus (data not shown). We also compared the effects of the restrictive temperature on the loss of accumulated lipids from both LDLF and wild-type cells (Figure 3A, lanes 5 and 6). Cells were incubated at the nonrestrictive temperature in the presence of oleate for 12 h, oleate was removed, and the cells were incubated further at the indicated temperature for 6 h. Although the restrictive temperature caused an ∼70% reduction in εCOP (cytosol, Figure 3A, lane 6), the ADRP levels in the droplet fraction (Figure 3A, lane 6) and the number of oil red O-positive droplets in both sets of cells remained unchanged. We also found that disrupting the Golgi apparatus in NRK cells with the drug Exo2 (Feng et al., 2004), which has a similar effect to BFA on the Golgi structure, had no effect on the number or size of the droplets (Supplemental Figure 1). Together, these results indicate that neither the assembly of COPI coats, the function of εCOP, nor protein secretion are involved in BFA-stimulated lipid loss.
Figure 3.
BFA-stimulated loss of lipid droplets is not dependent on Golgi fragmentation (A and B). (A) Normal (control) and temperature-sensitive CHO LDLF cells were grown either in the presence or absence of 200 μM oleate at the permissive (34°C) or restrictive (39°C) temperature for 12 h, or they were incubated in the presence of 200 μM oleate at the permissive temperature before shifting to the restrictive temperature for 6 h (lanes 5 and 6). Either droplet-enriched (LD) or cytosol fractions were prepared, and equal amounts of protein were separated by 12% SDS-PAGE and immunoblotted with α-ADRP and α-actin IgG (loading control). (B) LDLF cells were grown in the presence of 200 μM oleate for 12 h. The oleate containing medium was removed, and the cells incubated at either the permissive or restrictive temperature for an additional 12 h before staining with oil red O.
BFA Effect Requires New Protein Synthesis
Since BFA is not stimulating the loss of droplets through its ability to disrupt membrane traffic to and from the Golgi apparatus, we explored the possible that either protein synthesis, ER stress, or genes regulated by sterol regulatory element-binding protein (SREBP) might be involved (Figure 4).
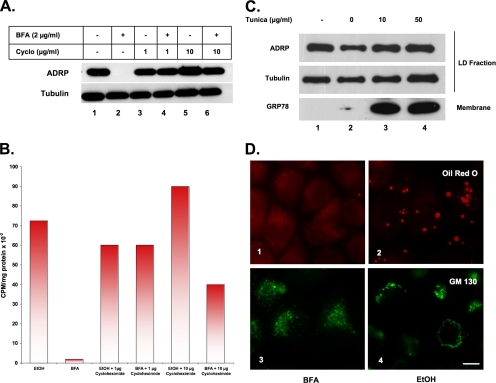
Figure 4.
BFA-stimulated lipid loss requires protein synthesis (A and B), and it is not regulated by UPR (C) or SREBP (D). (A) CHO K2 cells were incubated in the presence of BFA or EtOH carrier plus the indicated concentration of cycloheximide for 12 h. Droplet-enriched fractions were prepared, and equal amounts of protein were separated by 12% SDS-PAGE and immunoblotted with α-ADRP and α-tubulin IgG (load control). (B) CHO cells were incubated in the presence of 5 μCi of [3H]oleate for 24 h to label endogenous neutral lipids, washed, and then incubated further in the presence of BFA or EtOH carrier plus the indicated concentration of cycloheximide. Lipids were extracted from the droplet-enriched fraction with acetone, and the specific radioactivity was determined. The figure shown is representative of three independent experiments. (C) CHO K2 cells were incubated in the presence of the indicated concentration of tunicamycin or DMSO carrier for 12 h at 37°C. Droplet-enriched fraction and total membranes were isolated, and equal amounts of protein separated by 12% SDS-PAGE and immunoblotted with α-ADRP and α-tubulin IgG (loading control). Total membranes from the same preparation were separated the same way, but they were immunoblotted with α-GRP78 (BIP) IgG. (D) CHO cells deficient in SREBP S1P (SRD-12A) were incubated in the presence of either 2 μg/ml BFA (1 and 3) or EtOH carrier (2 and 4) for 12 h. Cells were fixed and processed for immunofluorescence analysis to detect GM130 (3 and 4) or oil red O staining (lanes 1 and 2). Bar, 10 μm.
We first looked at a requirement for protein synthesis (Figure 4, A and B). CHO cells containing numerous endogenous lipid droplets were incubated for 12 h in the presence or absence of BFA plus either nothing (lanes 1 and 2), 1 μg/ml cycloheximide (lanes 3 and 4), or 10 μg/ml cycloheximide (lanes 5 and 6). We prepared the droplet fraction and immunoblotted for ADRP. Both concentrations of cycloheximide blocked the loss of ADRP, indicating that lipid loss was prevented. In a separate experiment (Figure 4B), we found that cycloheximide markedly inhibited BFA-stimulated loss of [3H]oleate stored in neutral lipids. Protein synthesis inhibitors also blocked the loss of oil red O-positive droplets, but it did not block BFA-stimulated disruption of the Golgi apparatus in the same cell (data not shown). Similar results were obtained using the protein synthesis inhibitor emetine (data not shown). These results suggest that BFA-stimulated loss of lipid requires either new protein synthesis from existing RNA or the activation of specific genes.
Two sets of candidate genes that might be activated by BFA are those that regulate either the unfolded protein response (UPR) or SREBP. BFA is commonly used to induce ER stress (Misumi et al., 1986), presumably because disrupting ER-to-Golgi traffic causes the accumulation of unfolded proteins in the ER. UPR can be stimulated without disrupting the Golgi apparatus by blocking protein glycosylation with tunicamycin (Travers et al., 2000). When we incubated CHO K2 cells for 12 h in the presence of either 10 μg/ml or 50 μg/ml tunicamycin, we did not see any loss of ADRP from the droplet fraction (Figure 4C, lanes 3 and 4), even though this stress condition causes a marked increase in the amount of the ER chaperone GRP78 (Figure 4C), which is known to be up-regulated in response to UPR (Kaufman, 1999). Similar results were obtained with the UPR-inducing agent DTT (data not shown). Therefore, genes activated by the UPR do not seem to stimulate the loss of droplets.
SREBP regulates (Horton et al., 2003) a number of proteins associated with droplets (Liu et al., 2004), and BFA induces the inappropriate release of SREBP from the ER by causing the relocation of critical processing proteases from the Golgi apparatus to the ER (DeBose-Boyd et al., 1999). Therefore, SREBP-regulated genes might be up-regulated by BFA. If SREBP were involved, however, any condition that caused relocation of Golgi proteases to the ER would activate lipid loss, yet relocation of Golgi elements to the ER in LDLF cells did not promote lipid loss (Figure 3A). A more specific test, however, is the ability of BFA to stimulate loss of droplets in CHO cells deficient in the SREBP site-1 cleavage enzyme S1P (Figure 4D). Incubation with BFA for 12 h was just as effective at stimulating loss of droplets in these cells as in wild-type cells (compare 1 and 2). GM130 staining showed that the Golgi apparatus was disrupted by BFA, so these cells are not resistant to this drug (compare 3 and 4).
CtBP1/BARS Is a Target for BFA-stimulated Lipid Loss
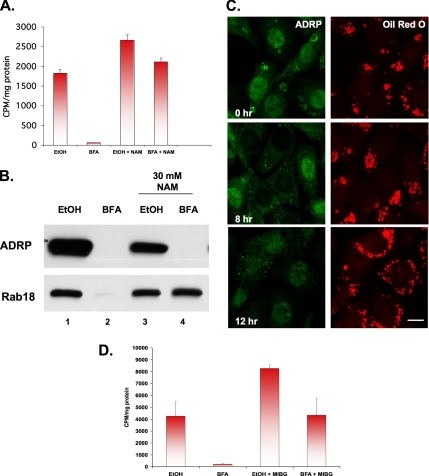
A poorly characterized effect of BFA is its ability to stimulate the mono-ADP-ribosylation of CtBP1/BARS and GAPDH (Spano et al., 1999). CtBP1/BARS interacts with the nuclear hormone receptor corepressor RIP140 (Vo et al., 2001), and RIP140 has been implicated in regulating fat accumulation in animals (Leonardsson et al., 2004). To explore a possible role for ribosylation in BFA-induced lipid droplet loss (Figure 5A), we incubated CHO K2 cells in the presence of trace amounts of [3H]oleate for 24 h to label the endogenous droplet fatty acid pool. The label was removed, and the cells were exposed to BFA for 12 h in the presence or absence of the ribosylation inhibitor nicotinamide (NAM). Exposure to BFA alone (Figure 5A, BFA) stimulated a marked loss of [3H]oleate-labeled lipids from the droplet fraction. The presence of 30 mM NAM (Figure 5A, BFA+NAM) blocked this effect, whereas NAM with the ethanol carrier (EtOH+NAM) caused a slight increase in radioactivity. We used immunoblotting of droplet marker proteins to confirm these results (Figure 5B). Unexpectedly, NAM did not block BFA-stimulated loss of ADRP from the droplet fraction (lane 4), even though another droplet-associated protein, Rab18, remained unchanged (compare lane 2 with lane 4). Immunofluorescence analysis confirmed that NAM blocks BFA-stimulated lipid loss (Figure 5C, oil red O) but that it causes a decrease in the amount of ADRP staining around each droplet (Figure 5C, ADRP). Two other ribosylation inhibitors (N-methylnicotinamide and 3-aminobenzamide) also blocked the ability of BFA to stimulate loss of droplets (data not shown). Finally, we tested MIBG, which is considered to be a more specific inhibitor of mono-ADP-ribosylation (Yau et al., 2004). As little as 100 μM MIBG blocked BFA-stimulated loss of [3H]oleate-labeled lipids from the droplet fraction (Figure 5D, BFA+MIBG). As we found with NAM, MIBG with the ethanol carrier markedly increased the amount of [3H]oleate lipid in the cell (Figure 5D, EtOH+MIBG). These results suggest that BFA-stimulated secretion of oleate depends on protein mono-ADP-ribosylation.
Figure 5.
BFA-stimulated loss of lipid droplets is blocked by inhibitors of mono-ADP ribosylation. (A) CHO K2 cells were incubated in the presence of 5 μCi of [3H]oleate for 24 h to label neutral lipids, washed, and then incubated further in the presence of 2 μg/ml BFA or EtOH carrier for 12 h at 37°C in the presence or absence of 30 mM NAM. Droplet-enriched fractions were prepared, and the lipid was extracted with acetone and counted. Each bar is the average of three experiments ± SE. (B) CHO K2 cells were incubated in the presence of 2 μg/ml BFA or EtOH carrier plus or minus 30 mM NAM. Purified droplets were prepared, and equal-volume fractions were separated by 12% SDS-PAGE and immunoblotted with α-ADRP and α-Rab18 IgG. (C) NRK cells were processed as described in Figure 1B to increase the number of droplets before incubating the cells in the presence of 2 μg/ml BFA plus 30 mM NAM for the indicated time. Cells were stained with α-ADRP IgG and oil red O. Bar, 10 μm. (D) The experimental design is the same as described in A, except that 100 μM MIBG was used instead of NAM. Each bar is the average of triplicate measurements ± SE.
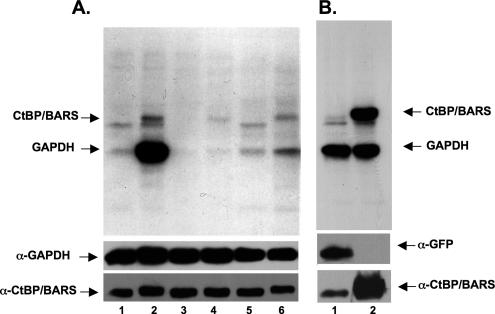
Because both NAM and MIBG block the effects of BFA on lipid loss, we next determined whether BFA stimulates protein ribosylation in these cells (Figure 6). To test for protein ribosylation, rat brain membranes were mixed with [32P]-NAD plus cytosol extracted from the cell of interest and incubated in the presence or absence of BFA for 2 h at 37°C (Valente et al., 2005) before separating the proteins on SDS-PAGE and processing for autoradiography. BFA induced the appearance of two bands, one band with the molecular weight of CtBP1/BARS, and the other band with the molecular weight of GAPDH (Figure 6A, compare lane 2 with lane 1). We confirmed that CtBP1/BARS is the top band by assaying for ribosylation in fibroblasts expressing a cDNA for either GFP (Figure 6B, lane 1) or CtBP1/BARS (Figure 6B, lane 2). Only the CtBP/BARS-expressing cells displayed a band of the correct molecular weight. BFA-dependent ribosylation could be observed in all cell lines we tested (Supplemental Figure 2). Finally, 30 mM NAM (Figure 6A, lane 4) reduced the incorporation of [32P]-NAD into both CtBP1/BARS and GAPDH relative to no treatment (Figure 6A, lane 2). It also reduced ribosylation of two bands that seem not to be sensitive to BFA (Figure 6A, lane 3). Measurement of each band indicated that NAM caused a reduction of >75% in the amount of incorporated [32P]-NAD into CtBP1/BARS. A similar result was obtained with 100 μM MIBG, which caused >50% reduction (Figure 6A, compare lane 6 with lane 2, and Supplemental Figure 3). Unlike NAM, MIBG alone did not reduce the ribosylation of the BFA insensitive bands (compare lane 5 with lane 1). Finally, Exo2 did not stimulate the mono-ADP ribosylation of either CtBP1/BARS or GAPDH (Supplemental Figure 1B).
Figure 6.
BFA stimulates ribosylation of CtBP/BARS and GAPDH. (A) Cytosol from CHO cells containing [32P]-NAD was either not treated (lane 1), incubated in the presence of BFA (lane 2), incubated in the presence of ribosylaton inhibitors NAM (lane 3) or MIBG (lane 5), incubated in the presence of BFA plus 30 mM NAM (lane 4) or BFA plus 100 μM MIBG (lane 6) for 2 h at 37°C. The reaction was stopped and membranes and cytosol were separated by centrifugation. The supernatant fraction was separated by SDS-PAGE, transferred to PVDF-membrane and processed for autoradiography. The PVDF-membrane was then processed for detection of GAPDH and CtBP1/BARS by immunoblotting. (B) Post-nuclear supernatant from Cos-7 cells expressing the cDNA for either GFP (lane 1) or CtBP1/BARS (lane 2) was incubated in the presence of BFA plus [32P]-NAD as described. The reaction was stopped, and the supernatant fraction processed as described above. The PVDF-membrane was then processed for immunoblotting to detect CtBP1/BARS or GFP.
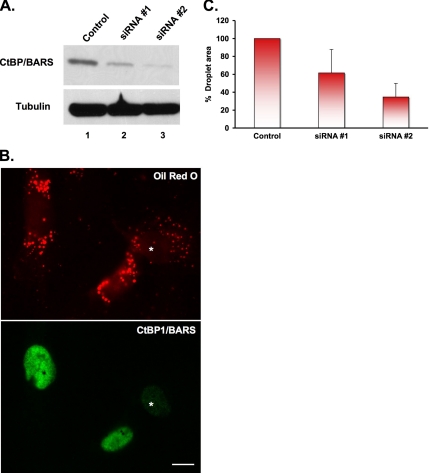
Of the two BFA-dependent ribosylated proteins, we focused on CtBP1/BARS because of its known interaction with the transcriptional corepressor RIP140 (Vo et al., 2001). We first used RNA interference to reduce the level of CtBP1/BARS in SV589 cells, and then we measured the size of the droplets (Figure 7). Immunoblotting shows that two different siRNAs against CtBP1/BARS markedly reduce the amount of CtBP1/BARS protein (Figure 7A). Oil red O staining (Figure 7B) shows that SV589 cells naturally have lipid droplets. Because the majority of the CtBP1/BARS staining is detected in the nucleus (7B), we used the intensity of the nuclear staining to gauge the degree of CtBP1/BARS knockdown. Cells with a reduced nuclear staining had significantly smaller droplets (Figure 7B, *) compared with surrounding cells with normal levels of nuclear staining. We quantified these results by comparing the total droplet area in cells with normal nuclear staining intensity (control cells) with those that had a >70% reduction in staining intensity (Figure 7C). Setting control transfected cells as 100%, we found that siRNA 1 reduced the lipid droplets by 40% and siRNA 2 by 60%. We conclude that down-regulation of CtBP1/BARS mimics the effect of BFA.
Figure 7.
Knockdown of CtBP1/BARS mimics the effects of BFA. (A) SV589 cells transfected with two different siRNA oligonucleotides for CtBP1/BARS (lanes 2 and 3) or an siRNA against microsomal transfer protein (control, lane 1) were cultured 4 d posttransfection before processing for immunoblotting using an α-CtBP1/BARS and α-tubulin IgG (loading control). (B) SV589 cells were processed similar to as described in A, except at day 3 posttransfection cells were seeded onto coverslips and incubated an additional day. Cells were washed and processed to localize CtBP1/BARS by immunofluorescence and droplets with oil red O. The asterisk marks a transfected cell that has a reduced level of CtBP1/BARS staining in the nucleus and decreased amount of oil red O staining compared with surrounding cells. Bar, 10 μm. (C) The area of the lipid droplets in the cells processed in B was then evaluated: the total droplet area in cells (120 cells/data point) exhibiting a >70% reduction in nuclear staining of CtBP1/BARS was analyzed using ImageJ and compared with control transfected cells. Each bar is the average of three experiments ± SE.
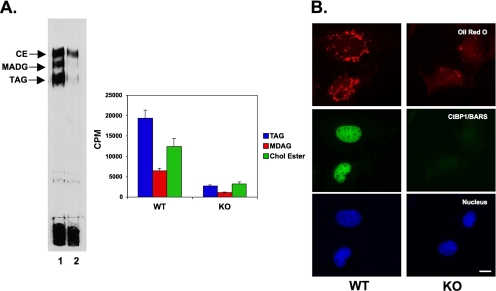
The effect of the CtBP1/BARS siRNA suggests that BFA may work by stimulating the ribosylation and inactivation of the transcriptional suppressor activity of CtBP1/BARS, thereby allowing the up-regulation of genes that govern lipid storage. This predicts that cells lacking CtBP/BARS will be defective in lipid storage. We obtained mouse embryo fibroblasts (MEFs) from CtBP/BARS knockout (KO) and matched wild-type mice (Hildebrand and Soriano, 2002), and we measured the amount of endogenous lipids (Figure 8A). We first measured the neutral lipid composition of these cells by using TLC (Figure 8). The two sets of cells were incubated in the presence of 10 μCi of [3H]oleate (300 nM oleate) for 24 h to label cellular lipids. The lipids were extracted, the neutral lipids were separated, and the bands were either visualized by autoradiography or scraped and counted in a scintillation counter (Figure 8A). As described previously (Bartz et al., 2007), the wild-type cells contained three major neutral lipids (Figure 8A, lane 1): cholesterol ester, monoalk(en)yl diacylglycerol, and triacylglycerol. The amount of all three lipids in CtBP1/BARS KO cells was markedly reduced (Figure 8A, lane 2). For example, KO cells contained nearly 10-fold less triacylglycerol (Figure 8A). To confirm these results, we used oil red O staining to observe the number of lipid droplets in the two sets of cells (Figure 8B). The KO cells clearly had many fewer droplets than wild-type cells. In summary, these results indicate that cells lacking CtBP/BARS display a reduced amount of lipid storage.
Figure 8.
CtBP/BARS-deficient MEFs are defective in lipid storage. Mouse embryo fibroblasts (WT and CtBP1/BARS1/2−/−) were cultured as described in text. (A) One set of cells was incubated in the presence of 50 μCi of [3H]oleate] for 24 h, washed, and processed to extract lipids as described in text. An equal volume of lipid extract from the two cells was separated by TLC, and then it was processed for autoradiography (left). In a separate experiment (right), equal volumes of lipid extract from the two cell lines were separated, scraped, and counted in a scintillation counter. The result is the average of three samples plus the SD. (B) WT and KO fibroblasts were grown as described in text. Cells were fixed and stained with α-CtBP1/BARS IgG, and the nucleus was visualized with Hoechst dye and the neutral lipids with oil red O. Bar, 10 μm.
DISCUSSION
BFA stimulates lipid loss independently of its well-characterized ability to disrupt ER/Golgi traffic. When we exposed cells to both oleate and BFA, we did not observe any detectable initial effect on fatty acid uptake or its subsequent esterification and storage. After a 5-h exposure, however, a set of genes become active that code for proteins involved in lipid secretion. Activation of fatty acid loss did not seem to depend on disruption of the Golgi apparatus, the unfolded protein response, or inappropriate activation of SREBP. By contrast, four different ribosylation inhibitors block the effect of BFA. The knockdown of the BFA-stimulated ribosylation substrate CtBP1/BARS causes droplet loss, and MEFs derived from CtBP1/BARS embryos are impaired in lipid storage. Because CtBP1/BARS is well known as a NAD-binding transcriptional corepressor (Zhang et al., 2002), we speculate that ribosylation inactivates its repressor activity, leading to the up-regulation of genes that code for proteins involved in the breakdown of neutral lipids and the export of fatty acids.
CtBP1/BARS belongs to a group of NAD/NADH binding proteins that has been implicated in regulating activities ranging from apoptosis to aging. Dimeric CtBP1/BARS is the active form of the transcriptional corepressor. Zhang et al. (2002) have proposed that CtBP1/BARS detects NAD/NADH ratios in the cell (NAD suppresses and NADH activates), thereby functioning as a redox sensor that regulates transcription. This model is particularly relevant to our studies, because it suggests a mechanism for tightly coupling the regulatory activity of CtBP1/BARS to glycolysis and fatty acid oxidation through the requirement for NAD as an electron acceptor in these metabolic pathways (Agarwal and Auchus, 2005). A potential target for CtBP1/BARS is the nuclear receptor corepressor RIP140 (Steel et al., 2005). Mice deficient in RIP140 exhibit a 70% reduction in total body fat and females are infertile. CtBP1/BARS is known to interact with RIP140, and it contribute to its repressor activity (Tazawa et al., 2003). Because ribosylation is known to inhibit the activity of enzymes such as glutamate dehydrogenase (Haigis et al., 2006, Herrero-Yraola et al., 2001), BFA-stimulated ribosylation may inactivate CtBP1/BARS. Inactive CtBP1/BARS, in turn, is unable to interact with transcriptional corepressors such as RIP140, and nuclear hormone target genes such as the glucocorticoid receptor become active. We speculate that ribosylation normally regulates the transcriptional corepressor activity of CtBP1/BARS because we found that the neutral lipid content of cells always increased when cells were exposed to ribosylation inhibitors alone.
The most thoroughly studied mono-ADP-ribosyltransferases are bacterial toxins that inactivate specific signaling pathways by ribosylating key intermediates (Corda and Di Girolamo, 2003). Much less is known about the function of endogenous mono-ADP-ribosyltransferases or enzymes such as ADP-ribosylarginine hydrolase that can deribosylate proteins. This raises the question of what intracellular enzyme might be responsible for ribosylating CtBP1/BARS. It is unlikely to be a member of the ART family of ectoribosyltransferases because all of these enzymes are either GPI-anchored to surface membranes or secreted. We are intrigued by the possibility that CtBP1/BARS itself is a ribosyltransferase capable of catalyzing either intra- or intermolecular ribosylation. The basis for this speculation is the recent evidence that purified SIRT6, an NAD-binding member of the Sir2-like protein (sirtuin) family of gene silencing factors, can ribosylate itself (Liszt et al., 2005). Moreover, multiple members of the sirtuin family have been found to exhibit weak mono-ADP-ribosyltransferase activity (Frye, 1999). Therefore, one model for how ribosylation might control the transcriptional repressor activity of CtBP1/BARS is that Arf1-GDP in a complex with an unknown Arf1 GEF stimulates the autoribosylation of CtBP1/BARS. Ribosylation, in turn, interferes with NAD binding, which blocks the interaction of CtBP1/BARS with PXDLS motifs on transcription repressors such as RIP140. As a consequence, target genes for these repressors become activated.
CtBP1/BARS has also been implicated in regulating the organization of the Golgi apparatus as an integral component of membrane fission machinery that converts lysophosphatidic acid to phosphatidic acid (Weigert et al., 1999). We do not think that modulation of Golgi organization by CtBP1/BARS is linked to lipid loss. Not only were we able to show experimentally that disrupting the Golgi by itself has no effect, but in the cells we used for these studies, the immunofluorescence only detected CtBP1/BARS in the nucleus. In addition, none of the ribosylation inhibitors we used had any effect on the organization of the Golgi, nor did they block the effect of BFA on Golgi structure. Most likely, the function of CtBP1/BARS in the Golgi apparatus is suppressed in cells containing many lipid droplets.
Nakamura et al. (2004) reported that 5 μg/ml BFA by itself stimulates the loss of ADRP from droplets. They also found that BFA caused an increase in the amount Rab18 on the droplet and that expressing dominant-negative Arf1T31N mimicked the effect of BFA by binding ADRP and dissociating it from the droplet. We, in contrast, did not see any effect of BFA alone on the level of ADRP on droplets nor did we observe an increase in Rab18. Instead, we saw that ADRP was lost from droplets only when cells were exposed to both BFA and NAM. Nevertheless, the two sets of observations may be related. BFA-stimulated ribosylation of CtBP1/BARS is probably dependent on Arf1-GDP, and Arf1-GDP is known to bind ADRP. Therefore, we speculate that in our system, the ribosylation inhibitors may stabilize the interaction between the Arf1-GDP generated by the BFA and any ADRP that has dissociated from the droplet, thereby favoring the accumulation of Arf1-GDP/ADRP in the cytoplasm.
Even though we have identified a mechanism that can explain how BFA turns on genes involved in lipid secretion, the identity of the genes remains unknown. We speculate these genes code for proteins involved in the hydrolysis of triacylglycerol and the transport of the released fatty acids to sites where they are secreted into the media. An interesting mechanistic clue comes from mice lacking RIP140, which exhibit a lipid retention phenotype (Leonardsson et al., 2004). Adipocytes from RIP140 knockout animals display a striking up-regulation of carnitine palmitoyltransferase 1b (>20-fold) and mitochondrial UCP1 (>100-fold), so the underlying mechanism of poor retention in these cells seems to be increased mitochondrial energy dissipation. BFA, by contrast, stimulates fatty acid secretion, not consumption. Tissue culture cells obtain most of their energy by glycolysis, so up regulation of UCP may not increase fatty acid oxidation in these cells. Nevertheless, it will be interesting to see whether ribosylation of CtBP1/BARS turns on different sets of genes in different cells with each set causing the loss of cellular neutral lipids.
Supplementary Material
ACKNOWLEDGMENTS
We thank Meifang Zhu, Tracy Diaz, and Charles Jason Hall for valuable technical assistance and Brenda Pallares for administrative assistance. We are grateful to Drs. Monty Krieger for the LDLF cells, Jeffrey D. Hildebrand for the CtBP/BARS-deficient mouse fibroblasts, Thomas Kirchhausen for the Exo2, and James E. Casanova for critical insights about CtBP/BARS. This work was supported by National Institutes of Health grants HL 20948 and GM-52016; the Perot Family Foundation; and the Cecil H. Green Distinguished Chair in Cellular and Molecular Biology.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0869) on May 30, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Agarwal A. K., Auchus R. J. Minireview: cellular redox state regulates hydroxysteroid dehydrogenase activity and intracellular hormone potency. Endocrinology. 2005;146:2531–2538. doi: 10.1210/en.2005-0061. [DOI] [PubMed] [Google Scholar]
- Bartz R., Li W. H., Venables B., Zehmer J. K., Roth M. R., Welti R., Anderson R. G., Liu P., Chapman K. D. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- Bligh E. G., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brasaemle D. L., Dolios G., Shapiro L., Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3–L1 adipocytes. J. Biol. Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- Corda D., Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBose-Boyd R. A., Brown M. S., Li W. P., Nohturfft A., Goldstein J. L., Espenshade P. J. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- Di Girolamo M., Silletta M. G., De Matteis M. A., Braca A., Colanzi A., Pawlak D., Rasenick M. M., Luini A., Corda D. Evidence that the 50-kDa substrate of brefeldin A-dependent ADP-ribosylation binds GTP and is modulated by the G-protein beta gamma subunit complex. Proc. Natl. Acad. Sci. USA. 1995;92:7065–7069. doi: 10.1073/pnas.92.15.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G., Finazzi D., Klausner R. D. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- Feng Y., Jadhav A. P., Rodighiero C., Fujinaga Y., Kirchhausen T., Lencer W. I. Retrograde transport of cholera toxin from the plasma membrane to the endoplasmic reticulum requires the trans-Golgi network but not the Golgi apparatus in Exo2-treated cells. EMBO Rep. 2004;5:596–601. doi: 10.1038/sj.embor.7400152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., et al. Exo 1, a new chemical inhibitor of the exocytic pathway. Proc. Natl. Acad. Sci. USA. 2003;100:6469–6474. doi: 10.1073/pnas.0631766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyberg Z., Siddhanta A., Shields D. “Slip, sliding away”: phospholipase D and the Golgi apparatus. Trends Cell Biol. 2003;13:540–546. doi: 10.1016/j.tcb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Frye R. A. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- Fujimoto T., Kogo H., Ishiguro K., Tauchi K., Nomura R. Caveolin-2 is targeted to lipid droplets, a new “membrane domain” in the cell. J. Cell Biol. 2001;152:1079–1085. doi: 10.1083/jcb.152.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Penman M., Trigatti B. L., Krieger M. A single point mutation in epsilon-COP results in temperature-sensitive, lethal defects in membrane transport in a Chinese hamster ovary cell mutant. J. Biol. Chem. 1996;271:11191–11196. doi: 10.1074/jbc.271.19.11191. [DOI] [PubMed] [Google Scholar]
- Haigis M. C. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Helms J. B., Rothman J. E. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- Herrero-Yraola A., Bakhit S. M., Franke P., Weise C., Schweiger M., Jorcke D., Ziegler M. Regulation of glutamate dehydrogenase by reversible ADP-ribosylation in mitochondria. EMBO J. 2001;20:2404–2412. doi: 10.1093/emboj/20.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand J. D., Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C. L., Casanova J. E. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Kaufman R. J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Kumar V., Carlson J. E., Ohgi K. A., Edwards T. A., Rose D. W., Escalante C. R., Rosenfeld M. G., Aggarwal A. K. Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol. Cell. 2002;10:857–869. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- Leonardsson G., et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. USA. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt G., Ford E., Kurtev M., Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., Anderson R. G. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- Lowe M., Kreis T. E. In vivo assembly of coatomer, the COP-I coat precursor. J. Biol. Chem. 1996;271:30725–30730. doi: 10.1074/jbc.271.48.30725. [DOI] [PubMed] [Google Scholar]
- Martin S., Parton R. G. Caveolin, cholesterol, and lipid bodies. Semin. Cell Dev. Biol. 2005;16:163–174. doi: 10.1016/j.semcdb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Misumi Y., Misumi Y., Miki K., Takatsuki A., Tamura G., Ikehara Y. Novel blockade by brefeldin A of intracellular transport of secretory proteins in cultured rat hepatocytes. J. Biol. Chem. 1986;261:11398–11403. [PubMed] [Google Scholar]
- Mlickova K., Roux E., Athenstaedt K., d'Andrea S., Daum G., Chardot T., Nicaud J. M. Lipid accumulation, lipid body formation, and acyl coenzyme A oxidases of the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2004;70:3918–3924. doi: 10.1128/AEM.70.7.3918-3924.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Akashi T., Taneda T., Kogo H., Kikuchi A., Fujimoto T. ADRP is dissociated from lipid droplets by ARF1-dependent mechanism. Biochem. Biophys. Res. Commun. 2004;322:957–965. doi: 10.1016/j.bbrc.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Nardini M., Spano S., Cericola C., Pesce A., Massaro A., Millo E., Luini A., D., Bolognesi M. CtBP/BARS: a dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 2003;22:3122–3130. doi: 10.1093/emboj/cdg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D. J., Helms J. B., Beckers C. J., Orci L., Rothman J. E. Binding of coatomer to Golgi membranes requires ADP-ribosylation factor. J. Biol. Chem. 1993;268:12083–12089. [PubMed] [Google Scholar]
- Pol A., Martin S., Fernandez M. A., Ferguson C., Carozzi A., Luetterforst R., Enrich C., Parton R. G. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol. Biol. Cell. 2004;15:99–110. doi: 10.1091/mbc.E03-06-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson R. B., Cheng D., Brown M. S., Goldstein J. L. Isolation of cholesterol-requiring mutant Chinese hamster ovary cells with defects in cleavage of sterol regulatory element-binding proteins at site 1. J. Biol. Chem. 1998;273:28261–28269. doi: 10.1074/jbc.273.43.28261. [DOI] [PubMed] [Google Scholar]
- Richards A. A., Stang E., Pepperkok R., Parton R. G. Inhibitors of COP-mediated transport and cholera toxin action inhibit simian virus 40 infection. Mol. Biol. Cell. 2002;13:1750–1764. doi: 10.1091/mbc.01-12-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg K. G., Heuser J. E., Donzell W. C., Ying Y. S., Glenney J. R., Anderson R. G. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- Spano S., et al. Molecular cloning and functional characterization of brefeldin A-ADP-ribosylated substrate. A novel protein involved in the maintenance of the Golgi structure. J. Biol. Chem. 1999;274:17705–17710. doi: 10.1074/jbc.274.25.17705. [DOI] [PubMed] [Google Scholar]
- Steel J. H., White R., Parker M. G. Role of the RIP140 corepressor in ovulation and adipose biology. J. Endocrinol. 2005;185:1–9. doi: 10.1677/joe.1.05896. [DOI] [PubMed] [Google Scholar]
- Tazawa H., Osman W., Shoji Y., Treuter E., Gustafsson J. A., Zilliacus J. Regulation of subnuclear localization is associated with a mechanism for nuclear receptor corepression by RIP140. Mol. Cell. Biol. 2003;23:4187–4198. doi: 10.1128/MCB.23.12.4187-4198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Umlauf E., Csaszar E., Moertelmaier M., Schuetz G. J., Parton R. G., Prohaska R. Association of stomatin with lipid bodies. J. Biol. Chem. 2004;279:23699–23709. doi: 10.1074/jbc.M310546200. [DOI] [PubMed] [Google Scholar]
- Valente C., Spano S., Luini A., Corda D. Purification and functional properties of the membrane fissioning protein CtBP3/BARS. Methods Enzymol. 2005;404:296–316. doi: 10.1016/S0076-6879(05)04027-9. [DOI] [PubMed] [Google Scholar]
- van Meer G. Caveolin, cholesterol, and lipid droplets? J. Cell Biol. 2001;152:F29–F34. doi: 10.1083/jcb.152.5.f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N., Fjeld C, Goodman R. H. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol. Cell. Biol. 2001;21:6181–6188. doi: 10.1128/MCB.21.18.6181-6188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltermann M., et al. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol. Microbiol. 2005;55:750–763. doi: 10.1111/j.1365-2958.2004.04441.x. [DOI] [PubMed] [Google Scholar]
- Weigert R., et al. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999;402:429–433. doi: 10.1038/46587. [DOI] [PubMed] [Google Scholar]
- Yau L., Litchie B, Zahradka P. MIBG, an inhibitor of arginine-dependent mono(ADP-ribosyl)ation, prevents differentiation of L6 skeletal myoblasts by inhibiting expression of myogenin and p21(cip1) Exp. Cell Res. 2004;301:320–330. doi: 10.1016/j.yexcr.2004.08.036. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Piston D. W, Goodman R. H. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Yao H, Vo N., Goodman R. H. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc. Natl. Acad. Sci USA. 2000;97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.