Abstract
Mutations in the parkin gene result in an autosomal recessive juvenile-onset form of Parkinson's disease. As an E3 ubiquitin-ligase, parkin promotes the attachment of ubiquitin onto specific substrate proteins. Defects in the ubiquitination of parkin substrates are therefore believed to lead to neurodegeneration in Parkinson's disease. Here, we identify the PSD-95/Discs-large/Zona Occludens-1 (PDZ) protein PICK1 as a novel parkin substrate. We find that parkin binds PICK1 via a PDZ-mediated interaction, which predominantly promotes PICK1 monoubiquitination rather than polyubiquitination. Consistent with monoubiquitination and recent work implicating parkin in proteasome-independent pathways, parkin does not promote PICK1 degradation. However, parkin regulates the effects of PICK1 on one of its other PDZ partners, the acid-sensing ion channel (ASIC). Overexpression of wild-type, but not PDZ binding– or E3 ubiquitin-ligase–defective parkin abolishes the previously described, protein kinase C-induced, PICK1-dependent potentiation of ASIC2a currents in non-neuronal cells. Conversely, the loss of parkin in hippocampal neurons from parkin knockout mice unmasks prominent potentiation of native ASIC currents, which is normally suppressed by endogenous parkin in wild-type neurons. Given that ASIC channels contribute to excitotoxicity, our work provides a mechanism explaining how defects in parkin-mediated PICK1 monoubiquitination could enhance ASIC activity and thereby promote neurodegeneration in Parkinson's disease.
INTRODUCTION
Parkinson's disease (PD) is characterized by the selective and progressive loss of midbrain dopamine neurons resulting in motor dysfunction and disability. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism, which accounts for a large proportion of genetically linked PD cases (Kitada et al., 1998). Parkin encodes a 465-amino acid protein (∼52 kDa) that is expressed in multiple tissues and functions in the ubiquitin (Ub) system as an E3 Ub-ligase (Shimura et al., 2000). Ubiquitination of substrate proteins is a tightly regulated process, requiring the combined activity of three enzymes: an E1 Ub-activating enzyme, E2 Ub-conjugating enzymes, and E3 Ub-ligases (Hershko and Ciechanover, 1998). E3 Ub-ligases bind substrate proteins and therefore regulate and confer specificity to the ubiquitination reaction. Typically, ubiquitination leads to the assembly of a K48-linked polyubiquitin chain on the substrate, which targets it for degradation by the 26S proteasome, a large multimeric proteolytic complex (Voges et al., 1999). Accordingly, defects in parkin-mediated ubiquitination have been proposed to result in the failure to target parkin substrates to the proteasome for degradation (Kahle et al., 2000; Feany and Pallanck, 2003; Giasson and Lee, 2003). The ensuing accumulation of parkin substrates is believed, in turn, to induce the cellular toxicity and dopamine neuron loss seen in PD. Although numerous parkin substrates have been identified (Zhang et al., 2000; Chung et al., 2001; Imai et al., 2001; Corti et al., 2003; Staropoli et al., 2003), there is still controversy as to which of these accumulate in the brains of parkin knockout (KO) mice (Goldberg et al., 2003; Itier et al., 2003; Von Coelln et al., 2004; Ko et al., 2005; Perez and Palmiter, 2005; Periquet et al., 2005) and as to their respective roles in the pathogenesis of PD (Cookson, 2005; Moore et al., 2005).
In addition to its traditional role, Ub can serve as a reversible posttranslational modification that regulates the function of tagged proteins without necessarily leading to their destruction by the proteasome (Hicke and Dunn, 2003; Mukhopadhyay and Riezman, 2007). Depending on the length and architecture of the Ub chain, ubiquitination has been implicated in a variety of cellular functions as diverse as signal transduction, transcription, and membrane trafficking (Mukhopadhyay and Riezman, 2007). Indeed, it was recently shown that, under certain circumstances, parkin can also mediate the assembly of K63-, rather that K48-linked, Ub chains, supporting a role for parkin in proteasome-independent ubiquitination pathways (Doss-Pepe et al., 2005; Lim et al., 2005). Additionally, conjugation of a single Ub moiety can function as a regulatory mechanism, influencing the interactions and trafficking of target proteins (Hicke and Dunn, 2003; Mukhopadhyay and Riezman, 2007). Parkin has recently been shown to monoubiquitinate and multi-monoubiquitinate itself in vitro (Hampe et al., 2006; Matsuda et al., 2006). Our own work has identified Eps15 as the first substrate of parkin-mediated monoubiquitination (Fallon et al., 2006), implicating parkin in a Ub-mediated growth-factor receptor trafficking and signaling pathway (Husnjak and Dikic, 2006). Thus, parkin-mediated modification of substrates with either K63-linked Ub chains or with monoubiquitin might help explain the lack of accumulation of most known parkin substrates in parkin mutant animals.
Parkin contains a ubiquitin-like domain (Ubl) at its N-terminus and two RING motifs, separated by an in-between-RINGs (IBR) domain, at its C-terminus. RING motifs are common in E3 Ub-ligases and serve to bind E2 enzymes (Lorick et al., 1999; Joazeiro and Weissman, 2000). Interestingly, many familial PD mutations in parkin are clustered within this E2-binding region (Kitada et al., 1998; Giasson and Lee, 2001), indicating a link between defects in parkin E3 Ub-ligase activity and neurodegeneration. Finally, we have recently shown that the last three amino acids (−FDV) at the extreme C-terminus of parkin function as a class II PDZ (PSD-95/discs large/ZO-1) binding motif (Fallon et al., 2002). PDZ domains bind the C-terminal tails of proteins in a sequence-specific manner. In general, two classes of interactions are distinguished. Class I PDZ domains are specific for the tripeptide sequence (S/T)XΦ (where Φ is any hydrophobic amino acid) and class II domains bind the C-terminal sequence ΦXΦ (Songyang et al., 1997). PDZ domain-containing proteins often function in trafficking or as scaffolds for the assembly of large protein complexes. Indeed, we showed that the parkin PDZ-binding motif binds the PDZ protein CASK; however, CASK itself is not ubiquitinated by parkin (Fallon et al., 2002). We reported that parkin colocalizes with CASK in postsynaptic densities (PSD), where many PDZ proteins are localized and that parkin is associated with the NMDA receptor-signaling complex. These findings suggest a role for parkin-mediated ubiquitination in synaptic transmission and plasticity. Identification of additional proteins that interact with the parkin PDZ-binding motif may therefore provide further insight into the molecular mechanisms that govern neuronal death in PD.
We report here that another PDZ protein, PICK1 (protein interacting with C-kinase 1), also binds parkin in a PDZ-dependent manner. PICK1 is a synaptic scaffolding protein known to functionally interact with an assortment of neurotransmitter receptors, transporters, and ion channels (Madsen et al., 2005). Unlike CASK, however, PICK1 is a substrate for parkin-mediated ubiquitination. Moreover, we find that parkin predominantly monoubiquitinates PICK1 and hence does not promote its degradation by the proteasome. As monoubiquitination influences the interactions, sorting, and trafficking of modified proteins (Hicke and Dunn, 2003; Mukhopadhyay and Riezman, 2007), parkin-mediated monoubiquitination of PICK1 might regulate its ability to interact with and regulate the function of its other synaptic PDZ-binding partners. Consistent with this hypothesis, we find that overexpression of parkin in non-neuronal cells abolishes the previously described PICK1-dependent, protein kinase C (PKC)-induced potentiation of acid-sensing ion channel subunit 2a (ASIC2a) currents (Baron et al., 2002a). Conversely, loss of endogenous parkin markedly enhances the PKC-induced potentiation of native ASIC currents in neurons. Taken together, our data show that parkin regulates ASIC function via PICK1 monoubiquitination. As excitatory ASIC currents have been implicated in synaptic plasticity (Wemmie et al., 2002, 2003) and neuronal injury (Xiong et al., 2004), we propose that defects in parkin-mediated monoubiquitination of PICK1 may result in aberrant ASIC signaling and contribute to dopamine neuron degeneration in PD.
MATERIALS AND METHODS
Constructs, Antibodies, and Reagents
Rat parkin was cloned as described previously (Fallon et al., 2002). GST-parkin constructs were prepared by subcloning various fragments into pGEX-4T2 or pGEX-5X-1 (Amersham Biosciences, Piscataway, NJ). The myc-PICK1 construct was a kind gift from Dr. Richard Huganir (Johns Hopkins University, Baltimore). GST-PICK1 constructs were generated by subcloning it into the SalI and NotI sites of the pGEX-4T-2 vector. The human ASIC2a clone in pcDNA3 was kindly provided by Dr. François Besnard (Sanofi-Synthelabo, Rueil-Malmaison, France). Flag-parkin was generated by PCR-amplifying the full-length rat cDNA using a 5′ oligonucleotide primer encoding an in-frame Flag epitope. The PCR product was then subcloned into the EcoRI and NotI sites of pcDNA3.1 (Invitrogen, Carlsbad, CA). The Flag-parkin (C431F, W453*, F463*, D464*) and myc-PICK1 (K27A/D28A) mutants were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). M2 Flag Agarose and Flag peptide were purchased from Sigma-Aldrich (St. Louis, MO). The antibodies used were: mouse monoclonal Flag and synaptophysin (Sigma-Aldrich), PSD95, NR1, CASK and synuclein (BD Bioscience, San Jose, CA), ubiquitin (Covance, Madison, WI), 9E10 myc, C2 actin, and PRK8 parkin (Santa Cruz Biotechnology, Santa Cruz, CA); rabbit polyclonal MALS-1/Veli-1 (Zymed, San Francisco, CA), ASIC2a (Alomone Laboratories, Jerusalem, Israel), PICK1 (kind gift from Dr. Richard Huganir, Johns Hopkins, Baltimore, MD), PICK1 (Affinity BioReagents, Golden, CO), and parkin (Cell Signaling, Beverly, MA); goat polyclonal N-18 PICK1 (Santa Cruz). Lactacystin was from Calbiochem (La Jolla, CA). Cycloheximide was from Sigma-Aldrich.
Mouse Brain Synaptosomes
Whole mouse brain was fractionated by differential centrifugation as previously described (Huttner et al., 1983; Fallon et al., 2002). Briefly, brain was homogenized in 0.32 M sucrose, 10 mM HEPES, pH 7.4 supplemented with protease inhibitors: 0.5 μg/ml leupeptin, 0.5 μg/ml aprotinin, 100 μg/ml benzamidine, 20 μg/ml phenylmethylsulfonyl fluoride. The homogenate was centrifuged for 10 min at 1000 × g, and the supernatant (S1) was collected and centrifuged again for 15 min at 12,000 × g to produce a synaptic pellet (P2). P2 was resuspended in the original volume of buffer and centrifuged for 15 min at 13,000 × g to produce the P2′ pellet. The soft, white component of P2′ was used as the crude synaptosome fraction. To further fractionate synaptosomes into subsynaptic components, P2′ was resuspended in 9 volumes of water and disrupted in a glass-Teflon homogenizer (three strokes). The water was adjusted to 10 mM HEPES and the sample was centrifuged for 20 min at 33,000 × g to yield the synaptic plasma membrane–enriched pellet (LP1). The supernatant (LS1) was centrifuged for 2 h at 260,000 × g to yield the synaptic vesicle–enriched pellet (LP2) and synaptic cytosol-enriched supernatant (LS2). Equal amounts of protein from each fraction were loaded for immunoblotting.
Cell Lines and Culture
COS-7 and HEK293 cells were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% fetal bovine serum (heat inactivated), 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. HEK293 cells were transfected with calcium phosphate, and COS-7 cells were transfected with Lipofectamine 2000 (Invitrogen). Stable Flag-parkin and control pcDNA3.1 HeLa cell lines were generated as described previously (Fallon et al., 2006) and maintained in the aforementioned medium supplemented with 100 μg/ml G418 sulfate.
In Vitro Transcription, Translation, and Expression of Bacterial Fusion Proteins
In vitro translation of myc-PICK1 was performed using the TNT rabbit reticulocyte lysate kit (Promega, Madison, WI.) according to the manufacturer's instructions. All glutathione S-transferase (GST) and His fusion proteins were expressed in Escherichia coli BL21 strain. GST fusion proteins were affinity-purified on glutathione Sepharose 4B beads (Amersham Biosciences) overnight at 4°C in 20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM dithiothreitol (DTT), and 0.1 mM ZnSO4. Untagged wild-type and mutant PICK1 were obtained by incubating GST-PICK1 with thrombin (10 U/mg of protein) for 2 h at room temperature. His-tagged proteins were affinity purified using Ni-NTA Agarose (Qiagen, Chatsworth, CA) according to the manufacturer's instructions.
GST-Binding Assays
Mouse brain synaptosomes were resuspended in 1% deoxycholic acid/50 mM Tris-HCl, pH 9.0, and protease inhibitors, solubilized on ice for 30 min, and cleared by centrifugation at 100,000 × g for 30 min. Triton X-100 was added to 1%, and the preparation was incubated overnight at 4°C with equimolar amounts GST fusion proteins and immobilized on glutathione-Sepharose beads in the following binding buffer: 50 mM Tris-HCl, 100 mM NaCl, 1 mM DTT, 0.1% Triton X-100, 10% glycerol, to pH 7.5 plus protease inhibitors. In vitro–translated Myc-PICK1 and HEK293 cell lysates were incubated for 3 h at 4°C with equimolar amounts of glutathione S-transferase fusion proteins, immobilized on glutathione-Sepharose beads in the following binding buffer: 50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1 mM DTT, 0.5% Triton X-100 plus protease inhibitors. In all cases, the beads were rinsed four times in the respective binding buffer, and bound proteins were eluted in SDS sample buffer at 65°C. Samples were subjected to SDS-PAGE followed by immunoblotting as described below.
Immunoprecipitation and Immunoblotting
For coimmunoprecipitation of PICK1 and parkin, cells were lysed 48 h after transfection in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% Triton X-100, plus protease inhibitors for 30 min on ice. Lysates were cleared by pelleting at 14,000 × g for 10 min, and the supernatant was incubated with primary antibody for 2 h at 4°C. The lysates were incubated with protein G-Sepharose beads for 1 h followed by washing of the immunoprecipitates four times with lysis buffer and elution of bound proteins in SDS sample buffer at 65°C. Samples were subjected to SDS-PAGE followed by electrotransfer to nitrocellulose membrane. Membranes were incubated with primary antibody overnight and secondary antibody for 1 h at room temperature. Proteins were detected using enhanced chemiluminescence (ECL) from Perkin Elmer-Cetus Life Sciences (Boston, MA). To examine the ubiquitination of PICK1, cells were preincubated in 2 μM lactacystin for 30 min and then lysed in RIPA buffer consisting of 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 0.1% SDS, 10 mM N-ethylmaleimide, and 1 mM EDTA plus protease inhibitors. Ub blots were performed as previously described (Avantaggiati et al., 1996). Briefly, after transferring protein to polyvinylidene difluoride (PVDF; instead of nitrocellulose), the membrane was denatured in guanidine-HCl for 30 min and blocked in 5% bovine serum albumin (BSA)/Tris-buffered saline–Tween (TBS-T) for 6 h before incubation with the Ub antibody in BSA overnight.
Parkin Solubility Assay
Transfected HEK293 cells were lysed on ice in 0.5% Triton X-100, 50 mM Tris-HCl, pH 7.4, and 100 mM NaCl plus protease inhibitors for 30 min. The lysates were centrifuged at 12,000 × g for 10 min, and the supernatant was collected as the soluble fraction. The pellet was rinsed twice with the same buffer and resuspended in 2% SDS, boiled for 10 min, and sheared with a 25-gauge needle. The solubilized pellet was then recentrifuged at 12,000 × g for 1 min, and the SDS-solubilized supernatant was collected as the insoluble pellet fraction. To compare the relative distribution of parkin, equal amounts of protein, as determined with the BCA protein assay kit (Pierce, Rockford, IL), of soluble and insoluble pellet fractions were processed for SDS-PAGE and analyzed by immunoblotting for parkin and CASK (as a loading control).
Cell Fractionation
Stable Flag-parkin and control HeLa cells were fractionated using either sequential detergent extraction or mechanical lysis. For detergent extraction, the cells were lysed with 0.5% Triton X-100 in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and protease inhibitors on ice for 30 min followed by centrifugation at 18,000 × g for 10 min. The supernatant was collected as the Triton-soluble fraction, whereas the pellet was sheared with a 27-gauge needle and re-extracted with 1% deoxycholic acid in 100 mM Tris-HCl, pH 8.8, and protease inhibitors on ice for 30 min followed by centrifugation at 79,000 × g for 30 min. The supernatant was collected as the deoxycholic acid–soluble fraction, whereas the pellet was re-extracted with 2% SDS, boiled for 10 min, and centrifuged at 79,000 × g for 30 min. The supernatant was collected as the SDS-soluble fraction, and the pellet was discarded. Equal volumes of each fraction were loaded and processed for SDS-PAGE followed by immunoblotting for PICK1 and parkin. For mechanical lysis, cells were collected in 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 2.6 mM 2-mercaptoethanol, 50 mM NaF, and 2 mM Na3VO4 sheared with 10 passages of a 27-gauge needle, and centrifuged at 100,000 × g for 30 min. The supernatant was collected as the cytosolic fraction, and the pellet was extracted in the same buffer with the addition of 1% Triton X-100 and 0.1% SDS, sheared with five passages of a 27-gauge needle, and collected as the membrane fraction. To load similar amounts of protein, 4:1 volume ratios of cytosol:membrane fraction were processed for SDS-PAGE followed by immunoblotting for PICK1 and parkin.
In Vitro Ubiquitination Assay
The assay was adapted from a previously described protocol (Matsuda et al., 2006). Briefly, purified GST fusion proteins (0.08 μg/μl GST, GST-parkin, or GST-parkinC431F), bound to glutathione-Sepharose beads, were resuspended in 50 μl of ubiquitination buffer (50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, and 2 mM DTT) containing 90 nM E1 enzyme (Boston Biochem, Cambridge, MA), 0.05 μg/μl either His6-Ubc7 (kind gift from Dr. Ryosuke Takahashi, RIKEN Brain Science Institute, Tokyo, Japan) or other E2 enzymes as indicated (Boston Biochem, Cambridge, MA), 4 mM ATP, 0.2 mM Ub (Boston Biochem), and 0.02 μg/μl wild-type or KD/AA mutant PICK1. The reactions were incubated for 1 h at 37°C and terminated by the addition of SDS sample buffer and boiling.
Genotyping
Genomic DNA from mouse tails was PCR-amplified from parkin wild-type, heterozygous, and knockout mice (Itier et al., 2003) using 35 cycles at 94°C for 1 min, 45°C (wild-type) or 42°C (knockout) for 1 min, and 72°C for 1 min with following primers: wild-type, forward 5′-tgctctggggttcgtc-3′, reverse 5′-tccactggcagagtaaatgt-3′; knockout, forward 5′-ttgttttgccaagttctaat-3′, reverse 5′-tccactggcagagtaaatgt-3′.
Primary Neuronal Culture
Cortical and hippocampal neurons were cultured from the brains of embryonic day 15 (E15)-E16 embryos obtained by mating parkin heterozygous mice. The cortex or hippocampi from individual embryos were dissected, incubated in 0.03% trypsin for 15 min at 37°C, and dissociated mechanically. Cells were plated in DMEM containing 10% inactivated horse serum (Sigma), 10% F-12 HAM nutrient mixture, 25 U/ml penicillin, and 25 μg/ml streptomycin. Hippocampal neurons were plated at a density of ∼50,000 cells per 35-mm poly-l-lysine–coated tissue culture plates (Falcon, Lincoln Park, NJ). Cortical neurons were plated at a density ∼25,000 cells per well on poly-l-lysine–coated 12-well plates. After 48 h, the medium was replaced with Neurobasal medium (Invitrogen) with 2% B27 supplement (Invitrogen) and kept in 95% air and 5% CO2 at 37°C.
Cycloheximide Pulse Chase
Ten days after plating, cortical neurons from parkin wild-type and knockout mice were incubated in 40 μg/ml cycloheximide in the presence or absence of 2 μM of the proteasome inhibitor lactacystin. After the indicated times, neurons were harvested, lysed in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.5% Triton X-100 plus protease inhibitors and 50 μg/time point were immunoblotted with the indicated antibodies.
Electrophysiological Recordings in Transfected COS-7 Cells and Hippocampal Neurons
COS-7 cells were transfected with Polyfect (Qiagen) according to the manufacturer's protocols and were used for electrophysiological recordings 24–48 h after transfection. Whole-cell patch-clamp recordings (Vh of −60 mV) were performed using pipettes filled with internal solution, pH 7.2, containing (in mM): K-gluconate 120, MgCl2 1, NaOH 4, and HEPES 10. Drug applications and rapid changes in extracellular pH were induced by shifting one out of three outlets in front of the cell using a fast microperfusion system at a rate of 1 ml/min (SF-77B, Warner Instruments, Morris Plains, NJ). The perfusion solution, pH 7.4, comprised (in mM): NaCl 145, NaOH 5, KCl 3, MgCl2 1, CaCl2 0.9, and HEPES 10. We used 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) instead of HEPES to buffer the extracellular solutions at pH 5.0. Membrane currents (DC, 200 Hz) were recorded using an Axopatch 200B amplifier and digitized at 500 Hz. Hippocampal neurons with typical triangular-shaped cell bodies, from parkin wild-type, heterozygous, and knockout mice, were selected for recording 11 d after plating. The standard external solution contained (in mM): NaCl 150, KCl 5, MgCl2 1, CaCl2 2, and glucose 10, buffered to various pH values with either 10 mM HEPES (pH 6.0–7.4) or 10 mM MES (pH <6.0), 300–330 mOsm/l. The patch pipette solution for whole-cell patch recording was (in mM): KCl 120, NaCl 30, MgCl2 1, CaCl2 0.5, EGTA 5, Mg-ATP 2, and HEPES 10. The internal solution was adjusted to pH 7.2 with Tris-base. All experiments were carried out at room temperature (20–23°C). Results were expressed as amplitude of peak currents evoked by pH 5.0 or as current density, defined as the ratio of peak amplitude over membrane capacitance (pA/pF).
RESULTS
Parkin Directly Binds the Synaptic PDZ Protein PICK1 via its C-Terminus
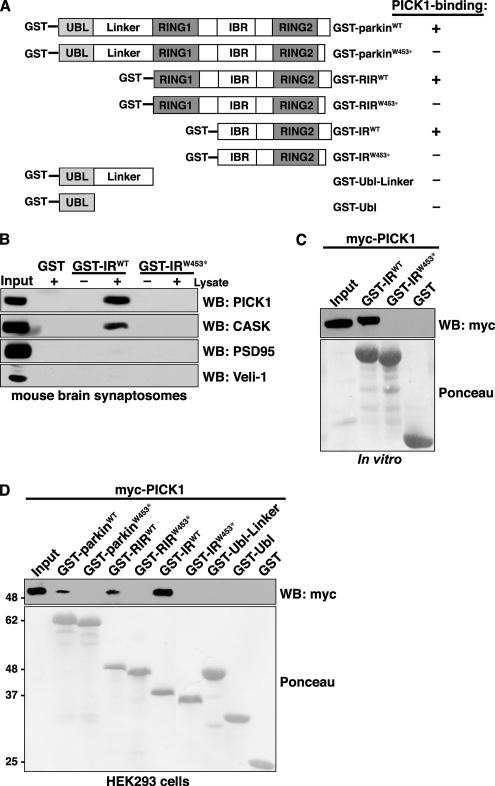
The last three amino acids of parkin (−FDV) encode a type II PDZ-binding motif. We reported previously that the PDZ protein CASK binds to parkin via this C-terminal motif (Fallon et al., 2002). As proteins with PDZ-binding motifs often interact with several distinct PDZ proteins (Srivastava et al., 1998; Dev et al., 1999), we used a candidate approach to identify additional PDZ proteins that interact with the C-terminus of parkin. We carried out pulldowns from mouse brain synaptosomes using GST fused to various parkin domains (Figure 1A). In particular, we compared binding of several endogenous PDZ proteins from mouse brain to GST-IRWT, encoding the parkin C-terminal IBR, RING2, and PDZ-binding motif. We find that, in addition to CASK, GST-IRWT binds to the PDZ protein PICK1 (Figure 1B). In contrast, neither CASK nor PICK1 binds to a GST-IR construct containing the familial PD-linked mutation, W453stop (GST-IRW453*). This mutation is particularly interesting because it truncates only the last 13 amino acids of parkin, eliminating its PDZ-binding motif, but leaves the critical E2-binding RING domain intact (Lucking et al., 2000; Zhang et al., 2000). Interestingly, although CASK contains a well-characterized class II PDZ domain, the PDZ domain of PICK1 can interact with both type I and type II as well as with atypical PDZ ligands (Staudinger et al., 1997; Dev et al., 1999; Williams et al., 2003; Madsen et al., 2005). In contrast, both PSD95 and Veli-1 contain distinct type I PDZ domains and, accordingly, these do not bind to the GST-IRWT construct. Thus, the C-terminus of parkin interacts not only with the type II PDZ protein CASK but also interacts with the atypical PDZ protein PICK1. To determine whether the interaction observed between the parkin C-terminus and PICK1 is direct, we performed pulldowns using in vitro–translated myc-tagged PICK1 (myc-PICK1) and the bacterially expressed and purified parkin GST-IR fusion constructs. We find that PICK1 interacts directly with the parkin C-terminus (Figure 1C). Further, this interaction requires the presence of the parkin PDZ-binding motif as the GST-IRW453* construct fails to bind. Next, we asked whether PICK1 could interact with other parkin domains, not present in the N-terminally truncated GST-IR constructs. To this end, we performed pulldowns from HEK293 cells expressing myc-PICK1 using GST fusion constructs encoding various parkin domains (Figure 1A). Mapping experiments demonstrate that PICK1 fails to bind all parkin constructs that lack the PDZ-binding motif (Figure 1D). Importantly, these data also indicate that PICK1 is able to interact with full-length parkin (GST-parkinWT), in addition to the C-terminal PDZ-binding motif in isolation.
Figure 1.
PDZ-dependent, direct interaction between parkin and PICK1. (A) GST-parkin fusion constructs are shown with the following functional domains: UBL, ubiquitin-like domain; RING, RING finger motif; IBR, in-between-RINGs motif. Constructs containing the parkin C-terminus encode both the wild-type and truncated PD-linked mutant, W453*. The ability of each GST-parkin construct to interact with wild-type PICK1 is indicated in the right column. (B) The C-terminus of parkin pulls down endogenous PICK1 from mouse brain synaptosomes. Solubilized synaptosomes were incubated with wild-type and mutant parkin C-terminal fusion proteins, GST-IRWT and GST-IRW453*. Binding was assayed by SDS-PAGE and Western blotting with antibodies against candidate PDZ proteins. As shown previously, CASK as well as the PDZ protein PICK1 associate specifically with the wild-type parkin construct and not with the truncated construct or GST alone. (C) PICK1 directly binds the extreme C-terminal residues of parkin in pulldown assays. In vitro–translated myc-PICK1 is selectively pulled down by the wild-type parkin carboxyl-terminal construct, GST-IRWT, and not by the PDZ-binding site mutant, GST-IRW453*, or by GST alone. (D) Domain mapping of interaction of PICK1 with parkin. Myc-PICK1 was transiently expressed in HEK293 cells and pulled down with the indicated GST-parkin fusion proteins or by GST alone. Western blots show that myc-PICK1 is only retained by fusion constructs containing the extreme C-terminal residues of parkin. The ponceau indicates comparable levels of fusion proteins were used in each binding assay.
Identification of Critical Residues Involved in the Parkin-PICK1 PDZ-mediated Interaction
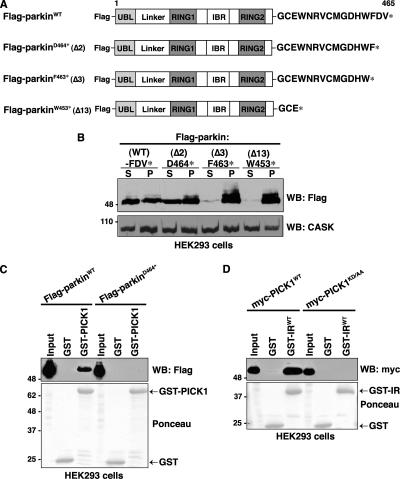
We anticipated that parkin constructs lacking the PDZ-binding motif (i.e., W453*) would be unable to bind PICK1. However, a recent report suggested that C-terminal truncations of parkin tend to induce misfolding and cause aggregation of the protein in cells (Winklhofer et al., 2003). Deleting of as few as four carboxyl-terminal amino acids completely interferes with parkin folding in transfected N2a cells; however, loss of the final one or two residues has minimal impact on folding compared with the wild-type protein (Winklhofer et al., 2003). The lack of association between PICK1 and the parkinW453* mutant might therefore result from improper parkin folding rather than the absence of the PDZ-binding motif per se. To investigate the PDZ-dependent association between parkin and PICK1 in cells, we first determined the effects of systematic C-terminal amino acid deletions on parkin solubility in HEK293 cells (Figure 2A). We find that deletion of as little as the final three residues of Flag-tagged parkin (Flag-parkinF463*) renders the protein nearly completely insoluble, as measured by the amount of parkin remaining in the supernatant after extraction with Triton X-100 (Figure 2B). In agreement with previous work (Winklhofer et al., 2003), the W453* truncation is also found exclusively in the Triton-insoluble pellet fraction, whereas deletion of the last two amino acids results in levels of soluble parkin comparable to those of the wild-type protein. We therefore used Flag-parkinD464* to further investigate the role of the parkin C-terminus in PICK1 binding. To confirm that the parkin C-terminus is required for PICK1 binding, we performed GST-PICK1 pulldowns from HEK293 cells transiently expressing either Flag-parkinWT or Flag-parkinD464* (Figure 2C). GST-PICK1 shows selective binding to wild-type but not parkinD464*, confirming that the interaction is PDZ-dependent.
Figure 2.
Identification of critical residues involved in the parkin-PICK1 PDZ-mediated interaction. (A) Schematic representation of Flag-parkin constructs. (B) C-terminal truncations exceeding two amino acids render parkin insoluble. HEK293 cells were transiently transfected with the indicated wild-type and C-terminal deletion constructs of parkin. Cells were harvested, lysed in detergent buffer (0.5% Triton X-100 in TBS), and fractionated by centrifugation, and parkin present in the detergent-soluble (S) and -insoluble (P) fractions was analyzed by Western blotting with anti-Flag antibodies. Deletion of the last two residues of parkin (D464*) does not affect parkin solubility compared with the wild-type protein. However, deletion of the final 3 (F463*) or 13 (W453*) amino acids of parkin renders the protein insoluble to mild detergent. Endogenous CASK levels are shown in the bottom panel as a loading control. (C) Deleting the final two amino acids of the PDZ-binding motif (Flag-parkinD464*) of parkin eliminates binding with PICK1 in vitro. Extracts from HEK293 cells transiently transfected with wild-type or PDZ mutant Flag-parkin were incubated with either GST-PICK1 or GST alone. Binding was assayed by SDS-PAGE and Western blotting with the indicated antibodies. (D) Mutation of the PDZ domain of PICK1 disrupts the interaction with parkin. Extracts from HEK293 cells transiently transfected with wild-type or PDZ mutant myc-PICK1 (myc-PICK1KD/AA) were incubated with the indicated GST fusion proteins. Binding was assayed by SDS-PAGE and Western blotting with the indicated antibodies.
PICK1 contains a single PDZ domain at its N-terminus. To test the hypothesis that the PDZ domain of PICK1 is responsible for the interaction with parkin, we mutated PICK1 residues Lys-27 and Asp-28 to alanine residues (myc-PICK1KD/AA). Mutations of these two residues have been well characterized to interfere specifically with PDZ-interactions between PICK1 and its ligands (Staudinger et al., 1997; Xia et al., 1999) because they form part of the carboxylate-binding loop of the PDZ domain, which binds to the last residue at the C-terminus of interacting proteins (Doyle et al., 1996; Songyang et al., 1997). As shown in Figure 2D, the myc-PICK1KD/AA mutant does not interact with the parkin GST-IRWT fusion construct, confirming that the PDZ domain of PICK1 is responsible for the interaction with parkin.
Parkin and PICK1 Interact in Cells
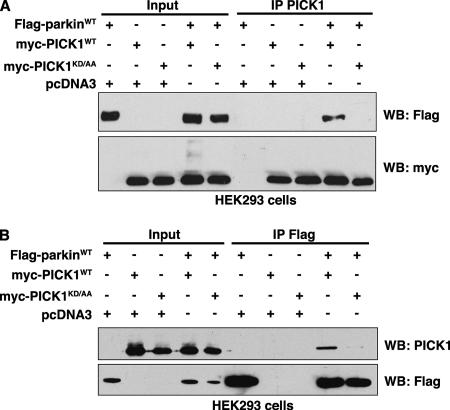
To determine whether parkin and PICK1 interact in cells as well as in vitro, we carried out coimmunoprecipitation experiments using HEK293 cells overexpressing full-length Flag-parkin and either wild-type or PDZ mutant myc-PICK1. As shown in Figure 3A, parkin coimmunoprecipitates with PICK1 only when the PDZ domain of PICK1 is intact. Conversely, PICK1 is coimmunoprecipitated by a Flag antibody only when both Flag-parkin and wild-type myc-PICK1 are expressed together (Figure 3B). These results demonstrate that PICK1 and full-length parkin associate in mammalian cells in a PDZ-dependent manner. However, as both parkin and PICK1 comigrate with the immunoglobulin heavy chain at 45–50 kDa, we were unable to detect coimmunoprecipitation of endogenous PICK1 with parkin using currently available antibodies.
Figure 3.
Parkin and PICK1 associate in cells via a PDZ-mediated interaction. HEK293 cells were transfected with expression vectors encoding Flag-parkin and wild-type myc-PICK1 or myc-PICK1 PDZ mutant K27A,D28A (KD/AA). Extracts were prepared from transfected cells and were immunoprecipitated with (A) anti-PICK1 or (B) anti-Flag antibodies. After separation of immunoprecipitates by SDS-PAGE, Western blotting was performed with the indicated antibodies. In both A and B, parkin associates only with wild-type PICK1. Mutations in the carboxylate-binding loop of PICK1 (KD/AA) inhibit these interactions.
Parkin Promotes PDZ-dependent PICK1 Monoubiquitination
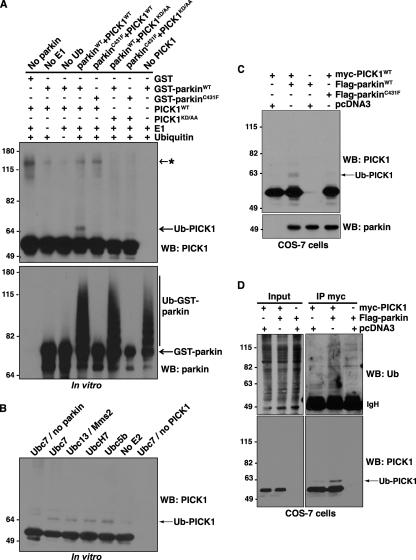
Given that parkin functions as an E3 Ub-ligase, we asked whether parkin could ubiquitinate PICK1 directly using an in vitro ubiquitination reaction containing purified recombinant E1, E2 (Ubc7), Ub, PICK1, and GST-parkin bound to glutathione-Sepharose beads. The reactions were immunoblotted with antibodies against PICK1 and parkin to assess PICK1 ubiquitination and parkin self-ubiquitination, respectively (Figure 4A). We find that PICK1 is ubiquitinated by parkin, as shown by a characteristic 8-kDa shift in the molecular weight of PICK1 (Figure 4A). In contrast, PICK1 ubiquitination is not observed in reactions lacking parkin, E1, or Ub. As expected, the PD-linked parkin mutant, C431F, which is E3 Ub-ligase inactive (Fallon et al., 2006; Matsuda et al., 2006), does not support parkin self-ubiquitination and failed to ubiquitinate PICK1 (Figure 4A). Moreover, neither wild-type nor C431F mutant parkin could ubiquitinate the PICK1 PDZ domain mutant, PICK1KD/AA (Figure 4A). Because PICK1KD/AA does not bind parkin (Figure 3, A and B), our findings indicate that parkin-mediated PICK1 ubiquitination requires both functional parkin E3 Ub-ligase activity and the parkin-PICK1 PDZ interaction. Interestingly, we could reliably detect only a single PICK1-immunoreactive band, 8 kDa above the unmodified protein, indicating that PICK1 is predominantly monoubiquitinated. The lack of PICK1 polyubiquitination is unlikely to stem from inherent limitations of our assay, because GST-parkin in the same reaction displayed a typical smear characteristic of polyubiquitination or multi-monoubiquitination, as reported previously (Hampe et al., 2006; Matsuda et al., 2006). It is also unlikely that the lack of PICK1 polyubiquitination or multi-monoubiquitination can be explained by the specific E2 (Ubc7) used in our assay. Indeed, reactions carried out with a panel of E2s that have been shown to support parkin-mediated ubiquitination (Matsuda et al., 2006) yielded very similar patterns of PICK1 ubiquitination (Figure 4B). Taken together, the findings indicate that PICK1 is predominantly monoubiquitinated by parkin in a PDZ-dependent manner.
Figure 4.
Parkin promotes PDZ-dependent PICK1 monoubiquitination. (A) In vitro ubiquitination assays were performed using purified recombinant E1s, E2 (Ubc7), GST-parkin (wild-type or C431F E3 Ub-ligase inactive mutant), PICK1 (wild-type or KD/AA PDZ-mutant), Ub, and ATP. Reagents were combined as indicated at 37°C for 1 h. Reactions were immunoblotted to detect Ub-modified PICK1 species. Monoubiquitination of PICK1 by parkin is dependent on both an intact PDZ domain in PICK1 and functional E3 Ub-ligase activity of parkin. Asterisk (*) indicates nonspecific band, present with or without functional parkin. (B) Multiple E2 enzymes can support parkin-mediated PICK1-monoubiquitination. Ubiquitination reactions were carried out as above with wild-type GST-parkin and PICK1 in the presence of the indicated E2s. (C and D) Parkin enhances the monoubiquitination of PICK1 in cells. (C) Extracts were prepared from COS-7 cells transfected with expression vectors encoding myc-PICK1 and wild-type Flag-parkin or the E3 Ub-ligase inactive parkin mutant Flag-parkinC431F. Proteins were separated by SDS-PAGE, and Western blotting was performed with the indicated antibodies. (D) COS-7 cells were transfected with expression vectors encoding myc-PICK1 and wild-type Flag-parkin. Cells were preincubated with proteasome inhibitor (2 μM lactacystin) for 30 min to allow ubiquitinated species to accumulate. Cells were then lysed in RIPA buffer containing NEM, and soluble proteins were immunoprecipitated with anti-myc antibody. After separation of immunoprecipitates by SDS-PAGE, immunoblotting was used to detect ubiquitinated PICK1.
Considering that parkin ubiquitinates PICK1 in vitro, we asked whether parkin could also promote PICK1 ubiquitination in cells. Examination of transfected COS-7 cell lysates reveals an 8-kDa upward shift in the mobility of myc-PICK1 in the presence of wild-type Flag-parkin but not the E3 Ub-ligase–inactive Flag-parkinC431F (Figure 4C). To further substantiate that PICK1 is mono- rather than polyubiquitinated, we also carried out anti-myc immunoprecipitation experiments in cells expressing myc-PICK1 in the presence or absence of Flag-parkin (Figure 4D). PICK1 immunoblots reveal prominent PICK1-monoubiquitin, rather than higher molecular weight PICK1-polyubiquitin conjugates, despite preincubation of the cells with lactacystin to prevent the degradation of polyubiquitinated proteins by the proteasome (Figure 4D, bottom panel). Further, it is unlikely that the monoubiquitinated species results from PICK1 deubiquitination after cell lysis as N-ethylmaleimide, an inhibitor of deubiquitinating enzymes, was included in all steps after lysis. Indeed, probing of the immunoprecipitates with an antibody against Ub reveals a clear enhancement in the amount of polyubiquitinated species associated with PICK1 in cells overexpressing parkin (Figure 4D, top panel). Unfortunately, it was not possible to clearly distinguish the monoubiquitinated PICK1 band, visible in the PICK1 blot (Figure 4D, bottom panel), from among this smear of ubiquitinated species. Considering that these bands are not immunoreactive for PICK1, they likely correspond to distinct parkin-ubiquitinated proteins that coimmunoprecipitate with PICK1 in cells. However, we cannot exclude that our PICK1 antibody recognizes only monoubiquitinated and not more heavily ubiquitinated forms of PICK1. Nonetheless, taken together, these results demonstrate that PICK1 can serve as a substrate for parkin-mediated ubiquitination both in vitro and in cells and that parkin predominantly promotes PICK1 monoubiquitination.
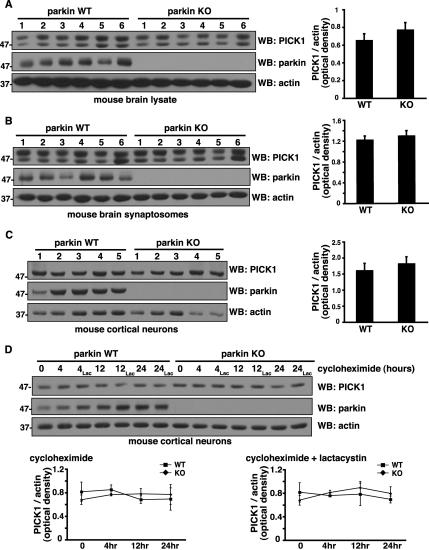
Parkin Does Not Promote PICK1 Degradation by the Proteasome
If parkin promotes polyubiquitination and proteasome-dependent degradation of PICK1, then we should see an increase in PICK1 protein levels in the absence of parkin. Conversely, if parkin primarily promotes monoubiquitination, then we predict that PICK1 protein levels would not be altered in the absence of parkin. Consistent with the latter possibility, we find that steady state PICK1 levels are similar in parkin wild-type and knockout mouse whole brain lysates and synaptic fractions (Figure 5, A and B). Similarly, we could detect no differences in PICK1 levels in cortical neurons cultured from parkin wild-type and knockout mouse brains (Figure 5C). Thus, steady-state PICK1 levels do not increase in the absence of parkin in neurons, consistent with parkin-mediated PICK1 monoubiquitination, rather than polyubiquitination. To examine directly whether parkin promotes PICK1 degradation, we treated parkin wild-type and knockout cortical neurons with the protein synthesis inhibitor cycloheximide and monitored endogenous PICK1 turnover during 24 h (Figure 5D). Remarkably, levels remained stable over this time period, suggesting that PICK1 is quite a long-lived protein in neurons. Endogenous parkin levels also remained stable over the time course, consistent with recent reports that it is predominantly multi-monoubiquitinated (Hampe et al., 2006; Matsuda et al., 2006) and functions in proteasome-independent pathways (Doss-Pepe et al., 2005; Lim et al., 2005; Fallon et al., 2006). Indeed, incubation of the neurons with lactacystin did not affect PICK1 levels, further indicating that its turnover is not regulated by the proteasome. More importantly, we did not observe a more rapid rate of PICK1 degradation in wild-type compared with parkin knockout neurons, either with or without lactacystin, indicating that parkin does not promote endogenous PICK1 degradation in neurons. Taken together with our ubiquitination (Figure 4) and steady state (Figure 5, A–C) data, these findings indicate that parkin predominantly promotes PICK1 monoubiquitination, which does not target it for degradation by the proteasome.
Figure 5.
Parkin does not promote PICK1 degradation by the proteasome. (A–C) Steady state endogenous PICK1 levels in mouse whole brain lysates (A), synaptosomes (B), and cortical neurons (C) from parkin wild-type and knockout mice. Equal amounts of protein were loaded from the indicated number of mice and immunoblotted with the antibodies shown. Densitometric analyses of endogenous PICK1 band intensities, normalized to actin, used as a loading control, are presented (right) as mean ± SEM. Student's t test reveals no significant differences between parkin wild-type and knockout samples. (D) Degradation rate of endogenous PICK1 in cortical neurons cultured from parkin wild-type and knockout mice. New protein synthesis was blocked with cycloheximide (40 μg/ml). In parallel cultures, the proteasomal inhibitor lactacystin (2 μM) was applied in addition to cycloheximide. Cells were harvested at the indicated time points after treatment, and protein levels were assessed by immunoblotting with the indicated antibodies. Densitometric analyses of PICK1 band intensities, normalized to actin, are presented (below) as mean ± SEM of five experiments. ANOVA reveals no significant differences in endogenous PICK1 turnover between parkin wild-type and knockout samples regardless of lactacystin treatment.
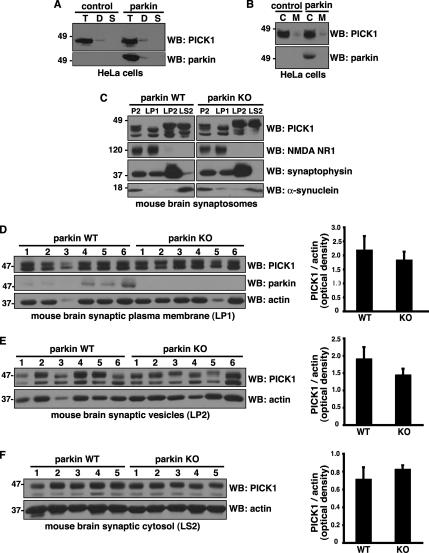
Parkin Does Not Affect PICK1 Subcellular Localization
Given that parkin does not promote PICK1 turnover and that monoubiquitination controls protein trafficking, we tested whether parkin could regulate PICK1 subcellular localization. Using a sequential detergent extraction procedure in HeLa cell lines stably expressing either Flag-parkin or pcDNA3.1 as a negative control (Fallon et al., 2006), we found that the majority of endogenous PICK1 cofractionated with parkin in the Triton X-100–soluble fraction (Figure 6A). A small amount of PICK1 and parkin could also be detected in the Triton X-100–resistant, deoxycholate soluble fraction, suggesting their localization in lipid raft or caveolar compartments. Similarly, using mechanical lysis followed by differential centrifugation, we found that the majority of parkin and PICK cofractionated in the cytosolic fraction (Figure 6B). Most importantly, the distribution of PICK1 was similar in both the parkin and control HeLa cell lines, indicating that parkin expression does not play a major role in the subcellular localization of endogenous PICK1 in non-neuronal cells. Next, to test whether parkin affects PICK1 localization in brain, we used a well-established differential centrifugation procedure to prepare synaptic fractions from parkin wild-type and knockout mice (Huttner et al., 1983; Fallon et al., 2002). The procedure yielded crude synaptosomes (P2) and synaptic plasma membrane-(LP1), synaptic vesicle- (LP2), and synaptic cytosol (LS2)-enriched fractions. As expected, the NR1 subunit of the NMDA receptor was enriched in LP1, whereas the synaptic vesicle protein synaptophysin was enriched in LP2 and the cytosolic protein α-synuclein was enriched in LS2 (Figure 6C). We found that PICK1 was present in all fractions, but particularly enriched in the synaptic vesicle (LP2) and synaptic cytosol (LS2) fractions. However, no differences were apparent in the distribution of PICK1 between the parkin wild-type and knockout fractions (Figure 6C). Even when normalized relative to actin, careful densitometric quantification revealed no statistically significant difference in PICK1 levels in synaptic plasma membrane (Figure 6D), synaptic vesicles (Figure 6E), and synaptic cytosol (Figure 6F) between parkin wild-type and knockout brain. Thus, parkin does not regulate the steady-state subsynaptic distribution of PICK1 in brain.
Figure 6.
Parkin does not affect PICK1 subcellular localization. (A and B) Subcellular distribution of endogenous PICK1 in HeLa cells stably expressing Flag-parkin or empty vector control. (A) Cells were sequentially extracted with 0.5% Triton X-100 (T), 1% deoxycholic acid (D), and 2% SDS (S). Equal volumes were loaded and immunoblotted with the indicated antibodies. Both PICK1 and parkin were predominantly found in the Triton X-100–soluble fraction with a small amount in the Triton X-100–resistant, deoxycholic acid–soluble fraction. (B) Cells were lysed mechanically without detergent, and the cytosolic (C) and membrane fractions (M) were separated by centrifugation. Similar amounts of protein (volume ratio of 4:1 cytosol:membrane fraction) were loaded and immunoblotted with the indicated antibodies. Both PICK1 and parkin were predominantly found in the cytosolic fraction. Importantly, parkin expression did not change endogenous PICK1 distribution using either the detergent (A) or mechanical (B) fractionation procedure. (C) Distribution of PICK1 in subsynaptic fractions prepared from parkin wild-type and knockout mouse brain synaptosomes (P2). Immunoblotting showed a similar distribution pattern of PICK1 in parkin wild-type and knockout fractions. Antibodies against NMDA NR1, synaptophysin, and α-synuclein were used as markers of synaptic plasma membrane (LP1), synaptic vesicle (LP2), and synaptic cytosolic (LS2) fractions, respectively. (D–F) Quantification of PICK1 levels in synaptic plasma membrane (D), synaptic vesicle (E), and synaptic cytosol (F) from parkin wild-type and knockout mice. Equal amounts of protein were loaded from the indicated number of mice and immunoblotted with the antibodies shown. Densitometric analyses of endogenous PICK1 band intensities, normalized to actin, used as a loading control, are presented (right) as mean ± SEM. Student's t test reveals no significant differences between parkin wild-type and knockout samples.
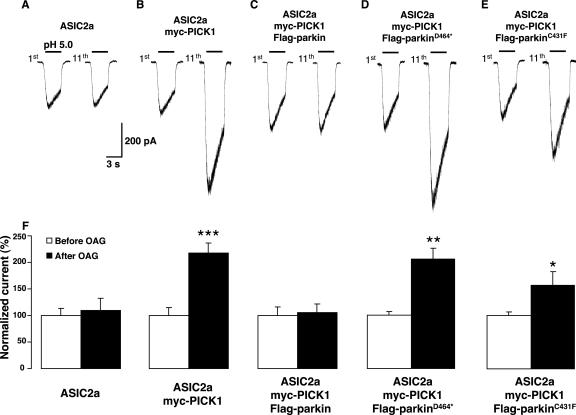
Parkin Suppresses PICK1-mediated Potentiation of ASIC2a Currents
Through its single PDZ domain, PICK1 binds numerous synaptic proteins, including several neurotransmitter receptors, transporters, and ion channels (Madsen et al., 2005). Furthermore, PICK1 can dimerize via its central coiled-coil motif and thereby promote protein complex formation between its binding partners. One such PICK1-associated protein is the proton-gated ion channel ASIC2a, a mammalian member of the degenerin/epithelial Na+ channel (DEG/ENaC) superfamily (Duggan et al., 2002; Hruska-Hageman et al., 2002). Interestingly, Baron et al. (2002a) reported that activation of PKC strongly potentiates ASIC2a currents in a PICK1-dependent manner. Considering that parkin monoubiquitinates PICK1, we asked whether parkin could functionally affect PICK1-mediated potentiation of ASIC2a currents. We therefore recorded proton-gated ASIC2a inward currents after repeated application of pH 5.0, 2-min apart, from COS-7 cells transfected with either ASIC2a alone, ASIC2a with myc-PICK1, or ASIC2a, myc-PICK1 and Flag-parkin together. In cells expressing ASIC2a alone, peak-current channel responses are similar before and after sustained application of the diacylglycerol analog OAG (1-oleyl-2-acetyl-sn-glycerol; Figure 7, A and F). Consistent with the findings of Baron et al. (2002a), when cells expressing ASIC2a channels are cotransfected with myc-PICK1, pH 5.0-evoked ASIC2a currents are strongly potentiated by 50 μM OAG treatment (Figure 7, B and F). The effect was significant within 8 min of OAG application and continued to increase until a plateau was reached. Further, the potentiation of ASIC2a currents persisted even 6 min after the OAG application was stopped. Importantly, we find that coexpression of Flag-parkin with myc-PICK1 and ASIC2a completely abolishes the PICK1-mediated potentiation of ASIC2a currents induced by OAG, without affecting basal ASIC2a current amplitudes or kinetics (Figure 7, C and F). Cotransfection of Flag-parkin and ASIC2a without myc-PICK1 had no effect on ASIC2a currents (normalized current ratio of 11th response/1st response = 100 ± 12 vs. 106 ± 13%, n = 3), suggesting that parkin blocks potentiation of ASIC2a currents via its effect on PICK1 rather than via a direct effect on ASIC2a. To further elucidate the mechanisms involved, we tested the ability of different parkin mutants to modify ASIC2a function. Deletion of the final two C-terminal residues of parkin (Flag-parkin464*) abolishes binding to PICK1 in our GST pulldown assays (Figure 2C). In contrast to wild-type Flag-parkin, coexpression of Flag-parkin464* with ASIC2a and myc-PICK1 failed to suppress the OAG-induced potentiation of ASIC2a currents (Figure 7, D and F). Therefore, the PDZ-dependent interaction between parkin and PICK1 is required to suppress the OAG-induced potentiation of ASIC2a currents. We further tested a point mutation in the E2-binding RING region (Flag-parkinC431F) that interferes with parkin self-ubiquitination (Figure 4A) and parkin-mediated ubiquitination of PICK1 both in vitro and in cells (Figure 4, A and C). Importantly, Flag-parkinC431F also failed to suppress the OAG-induced PICK1-dependent potentiation of ASIC2a currents (Figure 7, E and F). Thus, the ability of parkin to suppress PICK1-dependent, OAG-induced ASIC2a current potentiation requires functional parkin E3 Ub-ligase activity. Together, the findings indicate that PDZ-dependent monoubiquitination of PICK1 regulates ASIC2a channel function.
Figure 7.
Parkin suppresses PICK1-mediated potentiation of ASIC2a currents. (A–E) Representative ASIC2a inward currents (1st response and 11th response) evoked by consecutive application of extracellular solution at pH 5.0, 2 min apart, before and after OAG (50 μM, 10 min) from COS-7 cells transfected with ASIC2a alone (A), ASIC2a + myc-PICK1 (B), ASIC2a + myc-PICK1 and either wild-type Flag-parkin (C), Flag-parkinD464* (D), or Flag-parkinC431F (E). (F) Quantitative analysis of PICK1-induced potentiation of ASIC2a responses (n = 9) and of the blockade of the PICK1-mediated effect by wild-type, but not by D464* nor C431F mutant parkin coexpression (n = 4–7), expressed as mean ± SEM of normalized currents. *p < 0.05; **p < 0.01; ***p < 0.001, Student's t test.
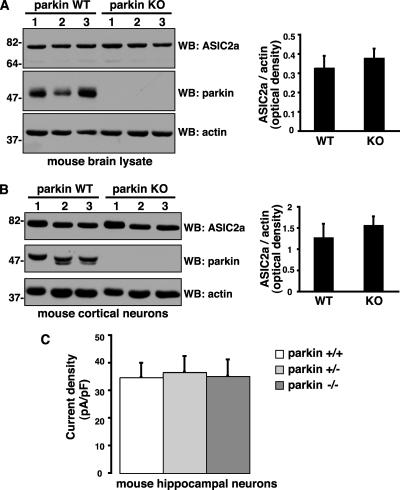
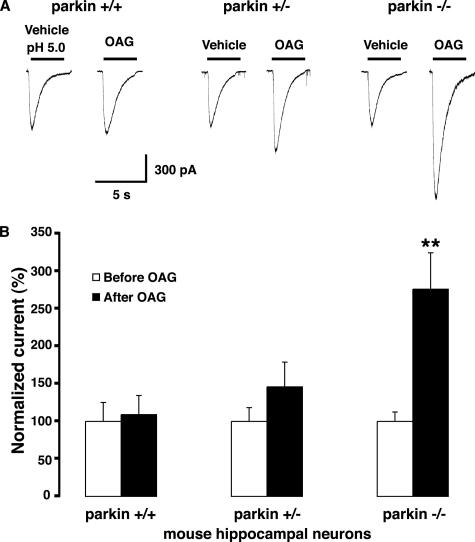
Endogenous Parkin Suppresses Potentiation of Native ASIC Currents in Neurons
To examine whether endogenous parkin could also affect native ASIC channel levels, localization and function in neurons, we used parkin knockout mice, which mimic the loss of parkin protein observed in patients with parkin gene deletions (Itier et al., 2003). PICK1 regulates the stability and trafficking of several of its PDZ domain–binding partners (Xia et al., 1999; Torres et al., 2001; Williams et al., 2003; Hanley, 2006). We therefore tested whether parkin-mediated PICK1 monoubiquitination affected ASIC2a levels or surface targeting. We found no differences in endogenous steady state ASIC2a levels in the brain or in cultured cortical neurons from wild-type and parkin knockout mice (Figure 8, A and B). In hippocampal neurons, proton-evoked ASIC currents are mediated predominantly by homomeric ASIC1a channels and heteromeric ASIC1a/2a and ASIC1a/2b channels (Baron et al., 2002b; Askwith et al., 2004). Therefore, ASIC-like current densities are likely to reflect total surface expression of endogenous functional ASIC channels in these neurons. We found that ASIC current densities were similar in parkin wild-type, heterozygous, and knockout hippocampal neurons (Figure 8C). Thus, the loss of parkin does not affect the steady state levels or the surface expression of endogenous ASIC channels under basal conditions. Because overexpression of parkin suppresses PICK1-dependent, PKC-induced ASIC2a current potentiation in non-neuronal cells (Figure 7), we speculated that, conversely, the loss of parkin might enhance the potentiation of native ASIC current in neurons. Indeed, native ASIC currents in hippocampal neurons from parkin knockout mice showed strong potentiation after OAG treatment (50 μM for 30 min), whereas heterozygotes showed an intermediate albeit nonsignificant trend toward potentiation (Figure 9). The results suggest a gene-dosage effect of parkin in modulating ASIC channels in central neurons. In contrast, OAG treatment did not potentiate ASIC currents in wild-type neurons (Figure 9), suggesting that endogenous levels of parkin are sufficient to completely block the PKC-induced potentiation of native ASIC currents. Thus, parkin may normally function to suppress ASIC current potentiation in neurons, whereas, in the absence of parkin, an enhanced propensity to ASIC channel overactivation with the associated vulnerability to cell damage and excitotoxicity may contribute to neurodegeneration in PD.
Figure 8.
Parkin does not affect ASIC2a levels or native ASIC current densities. (A and B) Steady state endogenous ASIC2a levels in mouse whole brain lysates (A) and cortical neurons (B) from parkin wild-type and knockout mice. Equal amounts of protein were loaded and immunoblotted with the antibodies shown. Densitometric analyses of endogenous ASIC2a band intensities, normalized to actin, used as a loading control, are presented (right) as mean ± SEM, (n = 6). Student's t test reveals no significant differences between parkin wild-type and knockout samples. (C) Native ASIC current densities were similar in hippocampal neurons cultured from parkin wild-type (34.68 ± 5.33 pA/pF, n = 7), heterozygous (36.58 ± 5.83 pA/pF, n = 9), and knockout (35.13 ± 6.1 pA/pF, n = 10) mice.
Figure 9.
Loss of parkin unmasks native ASIC current potentiation in hippocampal neurons. (A) Representative native ASIC inward currents evoked by consecutive application of extracellular solution at pH 5.0, 2 min apart, before and after OAG (50 μM, 30 min) in hippocampal neurons cultured from parkin wild-type, heterozygous, and knockout mice. (B) ASIC currents in parkin knockout neurons mice showed strong OAG-induced potentiation (+176%), whereas heterozygotes showed an intermediate albeit nonsignificant trend toward potentiation (+46%). In contrast, OAG treatment did not potentiate ASIC currents in wild-type neurons (peak currents evoked by pH 5.0, OAG: 906 ± 211 pA; vehicle: 830 ± 208 pA). The data are expressed as mean ± SEM of normalized currents (n = 5–10). **p < 0.01, Student's t test.
DISCUSSION
Our previous study established that the parkin carboxyl-terminus encodes a type II PDZ-binding motif that interacts with, but does not ubiquitinate, the PDZ scaffolding protein CASK. The present work demonstrates that CASK is not unique in its ability to associate with the C-terminal tail of parkin but that the PDZ protein PICK1, a PKCα-binding protein, also associates with parkin via a PDZ-mediated interaction. In contrast to CASK, we find that parkin ubiquitinates PICK1 both in vitro and in cells. Moreover, ubiquitination of PICK1 requires a PDZ-mediated interaction with parkin. PDZ-dependent ubiquitination of substrates has been reported previously for other E3 Ub-ligases. For example, the RING-type E3 Ub-ligase LNX requires its first PDZ domain to ubiquitinate its substrate, mNumb (Nie et al., 2004). Likewise, the human papillomavirus E6 protein contains a PDZ-binding motif, which targets the PDZ protein hScrib to the HECT domain E3 Ub-ligase E6AP for ubiquitination (Nakagawa and Huibregtse, 2000). We find that parkin conjugates only a single Ub moiety onto PICK1 in vitro and promotes PICK1 monoubiquitination in cells. Nonetheless, we cannot completely rule out that a given cellular context or a cofactor not present in our assay might tilt the balance toward PICK1 polyubiquitination under certain circumstances. For instance, the E4 activity mediated by CHIP (carboxy-terminus of the Hsp70-interacting protein) has been previously shown to enhance polyubiquitin chain formation on the parkin substrate Pael-R (Imai et al., 2002). Further supporting a role for parkin in mono-, rather than polyubiquitination of PICK1, we find that steady state levels of PICK1 are not increased in whole brain, synaptic fractions, or cultured cortical neuron lysates prepared from parkin knockout mice compared with wild-type littermates. Moreover, cycloheximide pulse-chase experiments show that the rate of PICK1 degradation is similar in cortical neurons prepared from parkin knockout mice and wild-type mice. We therefore propose that parkin predominantly functions in PICK1 monoubiquitination and does not target PICK1 for proteasome-mediated degradation. The results are consistent with recent work showing that parkin predominantly monoubiquitinates and multi-monoubiquitinates itself (Hampe et al., 2006; Matsuda et al., 2006) and that it monoubiquitinates Eps15 (Fallon et al., 2006). Despite considerable effort, we were unable to detect ubiquitination of endogenous PICK1 in cultured neurons or in brain. This can be notoriously difficult considering the low abundance and short half-life of ubiquitinated proteins and the lack of sensitive antibodies and methods to detect them. Accordingly, we speculate that PICK1 ubiquitination is very rapid and transient in neurons. Nonetheless, we provide strong evidence that parkin ubiquitinates PICK1 in vitro and in cells and that parkin E3 Ub-ligase activity is required to suppress the PKC-induced, PICK1-dependent potentiation of ASIC2a currents. Moreover, we find that the potentiation of native ASIC currents is markedly enhanced in parkin knockout neurons, consistent with parkin-mediated ubiquitination of PICK1 in vivo.
PICK1 was originally identified in a yeast two-hybrid screen with the catalytic domain of PKCα as bait (Staudinger et al., 1995). It has since been shown to bind numerous transmembrane receptors, transporters, and ion channels via its single N-terminal PDZ domain (Madsen et al., 2005). In contrast to most PDZ proteins, it is capable of interacting with both class I and class II PDZ ligands, as well as with various atypical motifs (Staudinger et al., 1997; Dev et al., 1999; Madsen et al., 2005). PICK1 also contains a coiled-coil motif that has been proposed to form a BAR (Bin/amphiphysin/Rvs.) domain, which mediates protein–protein interactions, lipid-binding, and endocytic processes (Peter et al., 2004; Lu and Ziff, 2005; Jin et al., 2006). The coiled-coil/BAR domain also promotes PICK1 dimerization (Staudinger et al., 1997), which may allow the pairing of multiple combinations of PDZ ligands. Indeed, PICK1 is known to target the activated form of PKCα to several transmembrane proteins for phosphorylation in a PDZ-dependent manner, which, in turn, alters their trafficking and functional regulation (Xia et al., 1999; Baron et al., 2002a; Williams et al., 2003). For instance, PICK1 binds to the C-termini of several isoforms of the ASIC family of proton-gated ion channels (ASIC1a, 2a, and 2b) via its PDZ domain (Duggan et al., 2002; Hruska-Hageman et al., 2002). Further, PICK1 is believed to target activated PKCα to ASIC2a, thereby leading to the phosphorylation of the channel in its cytoplasmic tail and to the potentiation of ASIC2a currents (Baron et al., 2002a). Considering that parkin promotes PICK1 monoubiquitination, we hypothesized that it might influence the ability of PICK1 to potentiate ASIC2a activity. Indeed, we find that parkin abolishes the ability of PICK1 to potentiate PKC-stimulated ASIC2a currents. In contrast to wild-type parkin, the parkinD464 mutant, which cannot bind PICK1, fails to suppress potentiation, demonstrating that the effect is PDZ-dependent. Similarly, the E3 Ub-ligase inactive, parkinC431F mutant fails to suppress potentiation, indicating that the effect also requires parkin-mediated ubiquitination. Interestingly, the magnitude of potentiation with parkinC431F was somewhat lower than with parkinD464* or than in the absence of parkin. Therefore, we cannot exclude that parkinC431F may have some residual low-level E3 Ub-ligase activity toward PICK1 in vivo, despite the complete absence of function in vitro or that part of the inhibitory effect on ASIC2a potentiation is independent of parkin's E3 Ub-ligase activity. Consistent with our findings using overexpression in non-neuronal cells, we also observed an inverse relationship between endogenous parkin levels and PKC-induced potentiation of native ASIC channels in hippocampal neurons from parkin wild-type, heterozygous, and knockout mice. Although potentiation was prominent in the absence of parkin, it was undetectable in neurons in the presence of wild-type levels of endogenous parkin. The finding may explain why PICK1-dependent, PKC-induced ASIC current potentiation has only been reported in heterologous systems, because the phenomenon may have been masked by endogenous parkin in neurons.
Parkin has been implicated previously in the regulation of cell surface receptors, channels and transporters. Parkin was shown to modulate P2X receptor function in PC12 cells. However, in contrast to our findings, ATP-induced currents were potentiated rather than suppressed by overexpression of parkin, and the effect was independent of PKC (Sato et al., 2006). Further, ubiquitination by parkin of P2X or other potential substrates in this process was not reported. Parkin was also found to ubiquitinate the dopamine transporter (DAT), which like ASIC2a, is a PDZ ligand of PICK1 (Torres et al., 2001; Jiang et al., 2004). By ubiquitinating misfolded DAT in the endoplasmic reticulum, parkin appears to enhance the expression of functional DAT at the cell surface. However, the role of PICK1 in this process was not examined. It was also recently reported that cell surface DAT is ubiquitinated and internalized in response to PKC activation (Miranda et al., 2005, 2007), a process that involves the HECT-family E3 Ub-ligase Nedd4–2 rather than parkin (Sorkina et al., 2006). Nedd4 also ubiquitinates and controls the cell surface expression of the Liddle's syndrome-associated epithelial sodium channel (ENaC; Staub et al., 1997), which like ASIC2a is a member of the degenerin/ENaC superfamily of ion channels. In contrast, we did not observe Ub-modified forms of ASIC2a on immunoblots (not shown), suggesting that parkin acts on PICK1 rather than by directly ubiquitinating ASIC2a itself. Moreover, we did not detect differences in steady state ASIC2a levels or native ASIC current densities in neurons cultured from parkin knockout and wild-type mice, suggesting that parkin does not regulate constitutive ASIC channel turnover or surface targeting. Thus, in contrast to Nedd4, which can ubiquitinate cell surface receptors, channels, and transporters, parkin appears to act predominantly on downstream adaptor proteins involved in endocytosis and trafficking. In addition to PICK1, parkin monoubiquitinates Eps15, a Ub-binding protein involved in epidermal growth factor (EGF) receptor (EGFR) trafficking and signaling (Fallon et al., 2006). Similar to our findings with ASIC channels, parkin does not appear to directly ubiquitinate EGFR nor does it affect constitutive EGFR turnover or surface targeting. However, parkin regulates EGFR internalization and PI3K-Akt signaling indirectly by ubiquitinating Eps15, reminiscent of the indirect effects of PICK1 ubiquitination on ASIC channels. In the future, it will be interesting to determine whether parkin-mediated ubiquitination also regulates other PICK1 ligands, such as the AMPA receptor and UNC5H1 (Xia et al., 1999; Williams et al., 2003; Madsen et al., 2005) and to further characterize the mechanisms involved. Importantly, central ASIC channels are involved in synaptic plasticity (Wemmie et al., 2002) and mediate a significant component of calcium-dependent excitotoxicity after ischemic injury (Xiong et al., 2004). Thus, the current work showing that endogenous parkin masks PICK1-dependent ASIC potentiation in hippocampal neurons suggests that it might also normally protect neurons from channel overactivity leading to excitotoxicity. Because ASICs have recently been implicated in midbrain dopamine neuron injury (Pidoplichko and Dani, 2006), the unmasking of ASIC current potentiation in parkin knockout neurons may reflect an increased vulnerability of these neurons to injury in patients with PD-associated parkin mutations.
ACKNOWLEDGMENTS
We thank Dominique Blais for her expert technical assistance. We thank Drs. Richard Huganir (Johns Hopkins University, Baltimore), François Besnard (Sanofi-Synthelabo, Rueil-Malmaison, France), and Ryosuke Takahashi (RIKEN Brain Science Institute, Tokyo) for kindly providing reagents. E.F. and P.S. are supported by the Canadian Institutes for Health Research and the Parkinson's Society of Canada. P.S. and E.F. are Killam Scholars of the Montreal Neurological Institute.
Abbreviations used:
- ASIC
acid-sensing ion channel
- CHX
cycloheximide
- ENaC
epithelial sodium channel
- GST
glutathione S-transferase
- NEM
N-ethylmaleimide
- OAG
1-oleyl-2-acetyl-sn-glycerol
- PD
Parkinson's disease
- PKC
protein kinase C
- PDZ
PSD-95/Discs-large/Zona Occludens-1
- PICK1
protein interacting with C-kinase 1
- PSD
postsynaptic density
- Ub
ubiquitin.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-11-1027) on June 6, 2007.
REFERENCES
- Askwith C. C., Wemmie J. A., Price M. P., Rokhlina T., Welsh M. J. Acid-sensing ion channel 2 (ASIC2) modulates ASIC1 H+-activated currents in hippocampal neurons. J. Biol. Chem. 2004;279:18296–18305. doi: 10.1074/jbc.M312145200. [DOI] [PubMed] [Google Scholar]
- Avantaggiati M. L., Carbone M., Graessmann A., Nakatani Y., Howard B., Levine A. S. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 1996;15:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- Baron A., Deval E., Salinas M., Lingueglia E., Voilley N., Lazdunski M. Protein kinase C stimulates the acid-sensing ion channel ASIC2a via the PDZ domain-containing protein PICK1. J. Biol. Chem. 2002a;277:50463–50468. doi: 10.1074/jbc.M208848200. [DOI] [PubMed] [Google Scholar]
- Baron A., Waldmann R., Lazdunski M. ASIC-like, proton-activated currents in rat hippocampal neurons. J. Physiol. 2002b;539:485–494. doi: 10.1113/jphysiol.2001.014837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K. K., Zhang Y., Lim K. L., Tanaka Y., Huang H., Gao J., Ross C. A., Dawson V. L., Dawson T. M. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1, implications for Lewy-body formation in Parkinson disease. Nat. Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- Cookson M. R. The biochemistry of Parkinson's disease. Annu. Rev. Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- Corti O., et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum. Mol. Genet. 2003;12:1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- Dev K. K., Nishimune A., Henley J. M., Nakanishi S. The protein kinase C alpha binding protein PICK1 interacts with short but not long form alternative splice variants of AMPA receptor subunits. Neuropharmacology. 1999;38:635–644. doi: 10.1016/s0028-3908(98)00230-5. [DOI] [PubMed] [Google Scholar]
- Doss-Pepe E. W., Chen L., Madura K. Alpha-synuclein and parkin contribute to the assembly of ubiquitin lysine 63-linked multiubiquitin chains. J. Biol. Chem. 2005;280:16619–16624. doi: 10.1074/jbc.M413591200. [DOI] [PubMed] [Google Scholar]
- Doyle D. A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- Duggan A., Garcia-Anoveros J., Corey D. P. The PDZ domain protein PICK1 and the sodium channel BNaC1 interact and localize at mechanosensory terminals of dorsal root ganglion neurons and dendrites of central neurons. J. Biol. Chem. 2002;277:5203–5208. doi: 10.1074/jbc.M104748200. [DOI] [PubMed] [Google Scholar]
- Fallon L., et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat. Cell Biol. 2006;8:834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- Fallon L., Moreau F., Croft B. G., Labib N., Gu W. J., Fon E. A. Parkin and CASK/LIN-2 associate via a PDZ-mediated interaction and are co-localized in lipid rafts and postsynaptic densities in brain. J. Biol. Chem. 2002;277:486–491. doi: 10.1074/jbc.M109806200. [DOI] [PubMed] [Google Scholar]
- Feany M. B., Pallanck L. J. Parkin: a multipurpose neuroprotective agent? Neuron. 2003;38:13–16. doi: 10.1016/s0896-6273(03)00201-0. [DOI] [PubMed] [Google Scholar]
- Giasson B. I., Lee V. M. Parkin and the molecular pathways of Parkinson's disease. Neuron. 2001;31:885–888. doi: 10.1016/s0896-6273(01)00439-1. [DOI] [PubMed] [Google Scholar]
- Giasson B. I., Lee V. M. Are ubiquitination pathways central to Parkinson's disease? Cell. 2003;114:1–8. doi: 10.1016/s0092-8674(03)00509-9. [DOI] [PubMed] [Google Scholar]
- Goldberg M. S., et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- Hampe C., Ardila-Osorio H., Fournier M., Brice A., Corti O. Biochemical analysis of Parkinson's disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum. Mol. Genet. 2006;15:2059–2075. doi: 10.1093/hmg/ddl131. [DOI] [PubMed] [Google Scholar]
- Hanley J. G. Molecular mechanisms for regulation of AMPAR trafficking by PICK1. Biochem. Soc. Trans. 2006;34:931–935. doi: 10.1042/BST0340931. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hicke L., Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Hruska-Hageman A. M., Wemmie J. A., Price M. P., Welsh M. J. Interaction of the synaptic protein PICK1 (protein interacting with C kinase 1) with the non-voltage gated sodium channels BNC1 (brain Na+ channel 1) and ASIC (acid-sensing ion channel) Biochem. J. 2002;361:443–450. doi: 10.1042/0264-6021:3610443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K., Dikic I. EGFR trafficking: parkin' in a jam. Nat. Cell Biol. 2006;8:787–788. doi: 10.1038/ncb0806-787. [DOI] [PubMed] [Google Scholar]
- Huttner W. B., Schiebler W., Greengard P., De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J. Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Soda M., Hatakeyama S., Akagi T., Hashikawa T., Nakayama K. I., Takahashi R. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol. Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- Imai Y., Soda M., Inoue H., Hattori N., Mizuno Y., Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Itier J. M., et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum. Mol. Genet. 2003;12:2277–2291. doi: 10.1093/hmg/ddg239. [DOI] [PubMed] [Google Scholar]
- Jiang H., Jiang Q., Feng J. Parkin increases dopamine uptake by enhancing the cell surface expression of dopamine transporter. J. Biol. Chem. 2004;279:54380–54386. doi: 10.1074/jbc.M409282200. [DOI] [PubMed] [Google Scholar]
- Jin W., Ge W. P., Xu J., Cao M., Peng L., Yung W., Liao D., Duan S., Zhang M., Xia J. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J. Neurosci. 2006;26:2380–2390. doi: 10.1523/JNEUROSCI.3503-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C. A., Weissman A. M. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Kahle P. J., Leimer U., Haass C. Does failure of parkin-mediated ubiquitination cause juvenile parkinsonism? Trends Biochem. Sci. 2000;25:524–527. doi: 10.1016/s0968-0004(00)01682-0. [DOI] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism [see comments] Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Ko H. S., et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J. Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. L., et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1, implications for Lewy body formation. J. Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Ziff E. B. PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron. 2005;47:407–421. doi: 10.1016/j.neuron.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Lucking C. B., et al. Association between early-onset Parkinson's disease and mutations in the parkin gene. French Parkinson's Disease Genetics Study Group. N. Engl. J. Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- Madsen K. L., Beuming T., Niv M. Y., Chang C. W., Dev K. K., Weinstein H., Gether U. Molecular determinants for the complex binding specificity of the PDZ domain in PICK1. J. Biol. Chem. 2005;280:20539–20548. doi: 10.1074/jbc.M500577200. [DOI] [PubMed] [Google Scholar]
- Matsuda N., Kitami T., Suzuki T., Mizuno Y., Hattori N., Tanaka K. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J. Biol. Chem. 2006;281:3204–3209. doi: 10.1074/jbc.M510393200. [DOI] [PubMed] [Google Scholar]
- Miranda M., Dionne K. R., Sorkina T., Sorkin A. Three ubiquitin conjugation sites in the amino terminus of the dopamine transporter mediate protein kinase C-dependent endocytosis of the transporter. Mol. Biol. Cell. 2007;18:313–323. doi: 10.1091/mbc.E06-08-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M., Wu C. C., Sorkina T., Korstjens D., Sorkin A. Enhanced ubiquitylation and accelerated degradation of the dopamine transporter mediated by protein kinase C. J. Biol. Chem. 2005;280:35617–35624. doi: 10.1074/jbc.M506618200. [DOI] [PubMed] [Google Scholar]
- Moore D. J., West A. B., Dawson V. L., Dawson T. M. Molecular pathophysiology of Parkinson's disease. Annu. Rev. Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Huibregtse J. M. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 2000;20:8244–8253. doi: 10.1128/mcb.20.21.8244-8253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li S. S., McGlade C. J. A novel PTB-PDZ domain interaction mediates isoform-specific ubiquitylation of mammalian Numb. J. Biol. Chem. 2004;279:20807–20815. doi: 10.1074/jbc.M311396200. [DOI] [PubMed] [Google Scholar]
- Perez F. A., Palmiter R. D. Parkin-deficient mice are not a robust model of parkinsonism. Proc. Natl. Acad. Sci. USA. 2005;102:2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periquet M., Corti O., Jacquier S., Brice A. Proteomic analysis of parkin knockout mice: alterations in energy metabolism, protein handling and synaptic function. J. Neurochem. 2005;95:1259–1276. doi: 10.1111/j.1471-4159.2005.03442.x. [DOI] [PubMed] [Google Scholar]
- Peter B. J., Kent H. M., Mills I. G., Vallis Y., Butler P. J., Evans P. R., McMahon H. T. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Pidoplichko V. I., Dani J. A. Acid-sensitive ionic channels in midbrain dopamine neurons are sensitive to ammonium, which may contribute to hyperammonemia damage. Proc. Natl. Acad. Sci. USA. 2006;103:11376–11380. doi: 10.1073/pnas.0600768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., et al. Parkin potentiates ATP-induced currents due to activation of P2X receptors in PC12 cells. J. Cell. Physiol. 2006;209:172–182. doi: 10.1002/jcp.20719. [DOI] [PubMed] [Google Scholar]
- Shimura H., et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Crompton A., Chan A. C., Anderson J. M., Cantley L. C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Sorkina T., Miranda M., Dionne K. R., Hoover B. R., Zahniser N. R., Sorkin A. RNA interference screen reveals an essential role of Nedd4-2 in dopamine transporter ubiquitination and endocytosis. J. Neurosci. 2006;26:8195–8205. doi: 10.1523/JNEUROSCI.1301-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., et al. Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- Staropoli J. F., McDermott C., Martinat C., Schulman B., Demireva E., Abeliovich A. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735–749. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L., Rotin D. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J., Lu J., Olson E. N. Specific interaction of the PDZ domain protein PICK1 with the COOH terminus of protein kinase C-alpha. J. Biol. Chem. 1997;272:32019–32024. doi: 10.1074/jbc.272.51.32019. [DOI] [PubMed] [Google Scholar]
- Staudinger J., Zhou J., Burgess R., Elledge S. J., Olson E. N. PICK1, a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J. Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres G. E., Yao W. D., Mohn A. R., Quan H., Kim K. M., Levey A. I., Staudinger J., Caron M. G. Functional interaction between monoamine plasma membrane transporters and the synaptic PDZ domain-containing protein PICK1. Neuron. 2001;30:121–134. doi: 10.1016/s0896-6273(01)00267-7. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Von Coelln R., Thomas B., Savitt J. M., Lim K. L., Sasaki M., Hess E. J., Dawson V. L., Dawson T. M. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc. Natl. Acad. Sci. USA. 2004;101:10744–10749. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie J. A., Askwith C. C., Lamani E., Cassell M. D., Freeman J. H., Jr, Welsh M. J. Acid-sensing ion channel 1 is localized in brain regions with high synaptic density and contributes to fear conditioning. J. Neurosci. 2003;23:5496–5502. doi: 10.1523/JNEUROSCI.23-13-05496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie J. A., et al. The acid-activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron. 2002;34:463–477. doi: 10.1016/s0896-6273(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Williams M. E., Wu S. C., McKenna W. L., Hinck L. Surface expression of the netrin receptor UNC5H1 is regulated through a protein kinase C-interacting protein/protein kinase-dependent mechanism. J. Neurosci. 2003;23:11279–11288. doi: 10.1523/JNEUROSCI.23-36-11279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winklhofer K. F., Henn I. H., Kay-Jackson P. C., Heller U., Tatzelt J. Inactivation of parkin by oxidative stress and C-terminal truncations: a protective role of molecular chaperones. J. Biol. Chem. 2003;278:47199–47208. doi: 10.1074/jbc.M306769200. [DOI] [PubMed] [Google Scholar]
- Xia J., Zhang X., Staudinger J., Huganir R. L. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- Xiong Z. G., et al. Neuroprotection in ischemia: blocking calcium-permeable acid-sensing ion channels. Cell. 2004;118:687–698. doi: 10.1016/j.cell.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gao J., Chung K. K., Huang H., Dawson V. L., Dawson T. M. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]