Abstract
The TOR kinases are regulators of growth in eukaryotic cells that assemble into two distinct protein complexes, TORC1 and TORC2, where TORC1 is inhibited by the antibiotic rapamycin. Present models favor a view wherein TORC1 regulates cell mass accumulation, and TORC2 regulates spatial aspects of growth, including organization of the actin cytoskeleton. Here, we demonstrate that in yeast both TORC1 and TORC2 fractionate with a novel form of detergent-resistant membranes that are distinct from detergent-resistant plasma membrane “rafts.” Proteomic analysis of these TOR-associated membranes revealed the presence of regulators of endocytosis and the actin cytoskeleton. Genetic analyses revealed a significant number of interactions between these components and TORC1, demonstrating a functional link between TORC1 and actin/endocytosis-related genes. Moreover, we found that inhibition of TORC1 by rapamycin 1) disrupted actin polarization, 2) delayed actin repolarization after glucose starvation, and 3) delayed accumulation of lucifer yellow within the vacuole. By combining our genetic results with database mining, we constructed a map of interactions that led to the identification of additional genetic interactions between TORC1 and components involved in membrane trafficking. Together, these results reveal the broad scope of cellular processes influenced by TORC1, and they underscore the functional overlap between TORC1 and TORC2.
INTRODUCTION
The rapamycin-sensitive TOR kinase is a highly conserved mediator of growth in eukaryotic cells (Wullschleger et al., 2006). There are two TOR kinases in budding yeast, Tor1p and Tor2p, that independently assemble with a distinct set of proteins to form what is termed TOR complex I (TORC1), the form of TOR that is inhibited by rapamycin (Wullschleger et al., 2006). Treating cells with rapamycin elicits widespread and characteristic changes in cellular behavior that mimic nutrient deprivation, including cessation of ribosome biogenesis and protein synthesis, induction of autophagy, cell cycle arrest at the G1/S boundary, and, ultimately, entry into stationary phase (Crespo and Hall, 2002; Rohde and Cardenas, 2004; Wullschleger et al., 2006). Rapamycin affects the expression of a wide array of nutrient-responsive genes as well as intracellular sorting of distinct amino acid permeases, results that are consistent with proposals that TORC1 conveys one or more nutrient-related signals within cells (Crespo and Hall, 2002; Rohde and Cardenas, 2004). Many events downstream from TORC1 are regulated by a phosphatase regulatory module that is composed of the type 2A phosphatases Pph21p and Pph22p, the related type 2A phosphatase Sit4p, and the two regulatory proteins Tap42p and Tip41p (Di Como and Arndt, 1996; Jiang and Broach, 1999; Jacinto et al., 2001; Düvel et al., 2003). TORC1 activity also intersects with several other nutrient-responsive kinases, including cAMP-regulated protein kinase A (PKA) and Sch9p (Jorgensen et al., 2004; Marion et al., 2004; Schmelzle et al., 2004; Zurita-Martinez and Cardenas, 2005; Chen and Powers, 2006). Precisely how TORC1 activity is integrated with these kinases and phosphatases remains poorly understood, and it is likely to be very complex.
Independently of its role in TORC1, Tor2p also assembles with an overlapping yet distinct set of proteins to form TORC2 (Loewith et al., 2002; Wedaman et al., 2003; Wullschleger et al., 2006), which plays a role in polarized cell growth and actin cytoskeletal organization (Loewith et al., 2002; Wullschleger et al., 2006). This Tor2p-unique activity involves signaling to components required for proper remodeling of actin, in the form of cortical patches, at the site of bud emergence (Schmidt et al., 1996; Schmidt et al., 1997; Helliwell et al., 1998; Crespo and Hall, 2002). These components include the Rho1p GTPase and its associated regulatory partners Rom2p and Sac7p, which function in part by signaling to protein kinase C (Pkc1p), an upstream activator of the mitogen-activated protein kinase (MAPK) Slt2p/Mpk1p (Helliwell et al., 1998; Pruyne and Bretscher, 2000; Crespo and Hall, 2002). More recently, the AGC kinase Ypk2p and two novel effectors, Slm1p and Slm2p, have been identified that also function downstream of TORC2 and that are required for proper actin polarization and cell viability (Audhya et al., 2004; Kamada et al., 2005). In addition, TORC2 has now been linked to a number of other cellular processes, including receptor-mediated endocytosis, phosphoinositide signaling, and calcineurin regulation as well as cell integrity maintenance (Schmelzle et al., 2002; deHart et al., 2003; Levin, 2005; Mulet et al., 2006; Tabuchi et al., 2006). Whether each of these different processes are linked to TORC2 via the Rho1p GTPase module and/or the newly discovered Slm proteins has not been determined. Interestingly, rapamycin treatment and/or inhibition of the Tap42p/Sit4p phosphatase system has also been shown to affect both actin polarization as well as cell integrity signaling, suggesting an unexpected degree of overlap with respect to functions carried out by TORC1 versus TORC2 (Torres et al., 2002; Wang and Jiang, 2003). In this regard, a recent chemical genetic screen has identified a large number of yeast gene deletion mutants that possess altered sensitivities to rapamycin (Xie et al., 2005). Intriguingly, mutants identified in this study represent several processes that have not been so far linked to TORC1 (Xie et al., 2005). Thus, the scope of cellular activities influenced by TORC1, although already extensive, could be even greater than is presently recognized.
Another important unresolved question concerns the site(s) of action of TORC1 and TORC2 within the cell. Previous studies have shown, using a variety of experimental approaches, that a significant portion of Tor1p and Tor2p are associated with an internal set of membranes that are adjacent to, yet apparently distinct from, the plasma membrane (Cardenas and Heitman, 1995; Kunz et al., 2000; Wedaman et al., 2003). In particular, using immunoelectron microscopy, we have localized both Tor1p and Tor2p as well as their common binding partner Lst8p to an intracellular membrane population that, at the ultrastructural level, closely resembles characteristic tracks and/or tubules that have been attributed to membranes of the endocytic pathway (Wedaman et al., 2003). By contrast, two components specific to TORC1, Tco89p and Kog1p, are also localized to vacuolar membranes, suggesting a more complex intracellular distribution of TORC1 components (Huh et al., 2003; Reinke et al., 2004; Araki et al., 2005). Moreover, it has recently been reported that a portion of Tor1p is localized within the nucleus where it specifically regulates 35S rDNA expression (Li et al., 2006). Thus, a full description of the intracellular environment(s) of the TOR kinases and their partners remains an important goal, and it is likely to be essential for a complete understanding of the functions of TORC1 and TORC2.
Here, we report findings that bear on several of these issues by characterizing further the membrane environment of TOR. We find that Tor1p and Tor2p associate with a novel form of detergent-resistant membranes that are enriched for proteins involved in actin cytoskeleton organization as well as endocytosis. Genetic analyses reveal a remarkable number of functional interactions between these proteins and components of TORC1; moreover, we demonstrate that, in addition to previously described effects on actin depolarization, rapamycin treatment severely impairs fluid phase endocytosis. Combining our genetic analyses with database mining, we constructed a network of functional interactions that identifies cellular pathways that are likely to be influenced by both TORC1 and actin/endocytosis-related components. This systems-based approach allowed us to establish a novel connection between TORC1 and a number of additional genes involved in vesicular trafficking.
MATERIALS AND METHODS
Strains, Media, and Genetic Methods
Yeast strains used in this study are listed in Table 1. Cells were grown in rich YPD (2% yeast extract, 1% peptone, and 2% dextrose) unless otherwise indicated. For tetrad dissection analysis, diploid strains were grown in sporulation medium (1% potassium acetate, 0.1% yeast extract, and 0.05% dextrose) for 3–6 d. Dissected tetrads were grown on YPD medium, except for mutants containing an SRV2 deletion, where SCD medium was used instead. Colonies derived from spores were grown at 30°C, except for mutants containing deletions of SLA2, TPD3, or SLT2, which were grown at 20°C, and deletions of SRV2, which were grown at 28°C. In all cases, deletion strains were constructed by replacement of corresponding open reading frame with a selectable marker as described previously (Brachmann et al., 1998; Longtine et al., 1998; Reinke et al., 2004). RVS161, SLA2, SEC8, KOG1, TCO89, LST8, and AVO3 were tagged at their carboxy termini with multiple copies of the Myc epitope as described previously (Brachmann et al., 1998; Longtine et al., 1998; Reinke et al., 2004). TOR1, TOR2, and RSP5 were tagged at their amino termini with three copies of the hemagglutinin (HA) epitope as described previously (Brachmann et al., 1998; Longtine et al., 1998; Wedaman et al., 2003; Reinke et al., 2004). Rapamycin (LC Labs, Woburn, MA) was added to YPD medium when indicated at a final concentration of 0.2 μg/ml.
Table 1.
Strains of S. cerevisiae used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303a | MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ | Nasmyth et al. (1990) |
| W303α | MATα ade2-1 trp1-1 can1-100 leu2-3 112 his3-11, 15 ura3 GAL+ | Nasmyth et al. (1990) |
| PLY293 | Same as W303a, except SEC8-MYC:TRP1 | This study |
| PLY391 | Same as W303a, except SLA2-MYC:TRP1 3HA-TOR2:His3MX6 | This study |
| PLY306 | Same as W303a, except KOG1-HA3:TRP1 | Reinke et al. (2004) |
| PLY307 | Same as W303a, except LST8-HA3:TRP1 | Reinke et al. (2004) |
| PLY323 | Same as W303a, except 3HA-RSP5:His3MX6 | This study |
| PLY327 | Same as W303a, except AVO3-MYC:TRP1 3HA-TOR2:His3MX6 | This study |
| PLY122 | Same as W303a, except 3HA-TOR2:His3MX6 | Wedaman et al. (2003) |
| PLY380 | Same as W303a, except TCO89-HA3:TRP | This study |
| Jk9-3da | MATa leu2-3, 112 trp1 ura3 rem1 his4 | Kunz et al. (1993) |
| JH1-11c | Same as Jk9-3da, except TOR1–1 | Helliwell et al. (1994) |
| LHY291 | MATa trp1 leu2 his3 ura3 lys2 bar1 | Stamenova et al. (2004) |
| PLY330 | Same as W303 α, except tor1::TRP1 | Reinke et al. (2004) |
| PLY254 | Same as W303a, except tor1::His3MX6 | Reinke et al. (2004) |
| PLY332 | Same as W303a, except tco89::His3MX6 | Reinke et al. (2004) |
| PLY438 | Same as W303α, except sla2::TRP1 | This study |
| PLY436 | Same as W303α, except rvs161::TRP1 | This study |
| PLY331 | Same as W303a, except avo2::His3MX6 | This study |
| PLY517 | Same as W303α, except slt2::KanMX6 | This study |
| PLY519 | Same as W303α, except srv2::KanMX6 | This study |
| PLY505 | Same as W303α, except bem2::KanMX6 | This study |
| PLY521 | Same as W303α, except ypk1::TRP1 | This study |
| PLY523 | Same as W303α, except tpd3::KanMX6 | This study |
| PLY525 | Same as W303α, except sac6::TRP1 | This study |
| PLY531 | Same as W303α, except she4::KanMX6 | This study |
| PLY533 | Same as W303α, except sro7::KanMX6 | This study |
| PLY576 | Same as W303α, except avo3-132-MYC:TRP1 | This study |
| PLY629 | Same as W303a, except ypt6::His3MX6 | This study |
| PLY644 | Same as W303a, except fen1::KanMX6 AVO3-MYC:TRP1 3HA-TOR2:His3MX6 | This study |
| PLY498 | Same as W303α, except pGAL-STE3-HA:URA3 | This study |
| PLY503 | Same as W303α, except pGAL-STE3-HA:URA3 sla2::TRP1 | This study |
| PLY348 | Same as W303a, except [pRS416-TLG1-MYC] | This study |
Antibodies and Other Reagents
Western blot analysis and immunoprecipitations were performed using monoclonal antibodies for HA (12CA5; Roche Diagnostics, Indianapolis, IN), c-Myc (9E10; Covance, Princeton, NJ), alkaline phosphatase (ALPp, 1D3; Invitrogen, Carlsbad, CA), Vps10p (18C8; Invitrogen), Pep12p (2C3; Invitrogen), carboxypeptidase Y (CPY) (10A5; Invitrogen), and Vph1p (V-ATPase subunit) (10D7; Invitrogen), and polyclonal antibodies for Tor1p (Santa Cruz Biotechnology, Santa Cruz, CA), Zwf1p (G6PDH) (Sigma-Aldrich, St. Louis, MO), Sec61p (a gift from Peter Walter, University of California, San Francisco, CA), and Pma1p and Chs3p (kindly provided by Randy Schekman, University of California, Berkeley, CA). Anti-goat immunoglobulin (Ig)G (Santa Cruz Biotechnology) and anti-mouse IgG and anti-rabbit IgG (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) secondary antibodies conjugated to horseradish peroxidase were used where appropriate. Rhodamine-phalloidin and FM4–64 were from Invitrogen. Lucifer yellow (LY) was purchased from Sigma-Aldrich.
Triton X-100 (TX-100) Supernatant/Pellet Assay
For each strain tested, 320 ml of cells was grown overnight at 30°C to 0.5 OD600/ml in YPD (160 OD cells). Cells were pelleted in 50-ml conical tubes, washed in H2O, pelleted again, and resuspended at 40 OD cells/ml in TNE buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5 mM EDTA) containing protease inhibitors (cocktail tablet; Roche Diagnostics) and 1 mM phenylmethylsulfonyl fluoride (PMSF). Yeast cells were lysed by bead beating for 2 min. The lysates were cleared by centrifugation at 500 × g, for 5 min at 4°C. Cleared lysates were then spun at 20,000 × g for 20 min at 4°C, yielding ∼2 ml of supernatant to be used for detergent treatment. After Bradford assays, the resulting supernatants were divided into 500-μl aliquots and treated 1:1 with TNE, TNE 1% Triton X-100, or TNE 1% Triton X-100 and 1 M NaCl (final concentrations). Samples were incubated on ice for 30 min during which time they were passed through a Hamilton syringe two times. Samples were spun at 100,000 × g for 1 h at 4°C. Supernatants were collected and trichloroacetic acid (TCA) precipitated, dried, and resuspended in 100 μl of sample buffer. Pellets were resuspended in 100 μl of sample buffer. Supernatants and pellets were analyzed by SDS-polyacrylamide gel electrophoresis followed by Western blot analysis.
OptiPrep Floatation Assay
OptiPrep floatation gradients and isolation of the Triton X-100–insoluble membranes were performed essentially as described previously (Bagnat et al., 2000) with minor modifications. Briefly, yeast cells were lysed by vortexing with glass beads in TNE buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5 mM EDTA) supplied with protease inhibitors tablet (Roche Applied Science, Indianapolis, IN) and 1 mM PMSF (Sigma-Aldrich). The lysates were cleared by centrifugation at 500 × g for 5 min at 4°C, but they were not subjected to the 20,000 × g centrifugation step described for the Triton X-100 supernatant/pellet assay. Cleared lysates were incubated either with 1% TX-100 or with an equal volume of TNEX buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, and 0.1% Triton X-100) for 30 min on ice. The lysates were adjusted to 40% OptiPrep (Nycomed, Oslo, Norway), and 4.2 ml of resulting mixture was sequentially overlaid with 6.7 ml of 30% OptiPrep in TNEX buffer and 1.1 ml of TNEX buffer. The samples were centrifuged at 100,000 × g in SW41 Ti rotor for 2.5 h, and 1.2-ml fractions were collected from the top of the gradient and subjected to Western blot analysis.
Proteomic Analysis
TX-100–treated cell extracts were subjected to OptiPrep gradient as described above, and equivalent groups of fractions, corresponding to 0, 30, and 40% of OptiPrep, collected from four sets of the identical gradients, were combined. TOR-containing (30% OptiPrep) and plasma membrane raft (0% OptiPrep) fractions were repeatedly subjected to incubation with 1% TX-100 followed by floatation on an OptiPrep gradient as described above. Proteins from fractions corresponding to 0–30% OptiPrep of the second gradient as well as those corresponding to 40% of the first gradient were TCA precipitated and identified by tandem mass spectrometry as described previously (Carroll et al., 1998; Link et al., 1999).
Fluorescence Microscopy
Rhodamine-phalloidin staining of polymerized actin was performed as described previously (Pringle et al., 1989). For quantification of cells with depolarized actin cytoskeleton, ∼100 small-budded or midsize-budded cells were counted for each condition. Cells were considered as having depolarized actin patches if six or more patches were found within the mother cell. Lucifer yellow uptake assay was performed essentially as described previously (Dulic et al., 1991). Fluorescence microscopy was performed using Nikon E600 fluorescence microscope and Orca-ER charge-coupled device camera (Hamamatsu, Bridgewater, NJ) controlled by Simple PCI software (Compix, Cranberry, PA). All actin-staining images are z-series projections of optical sections.
Ste3-HA Endocytosis Assay
Monitoring of Ste3p-HA endocytosis was performed as described previously (Walther et al., 2006). Samples were diluted to give equal protein concentration determined using bicinchoninic acid assay and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting using anti-HA antibody.
CPY Trafficking Assay
Metabolic cell labeling and immunoprecipitations were performed essentially as described previously (Vashist et al., 2001). Briefly, 12 A600 OD units of early log phase cells were harvested and resuspended in 3.6 ml of SCD medium lacking methionine and cysteine. After 30 min of incubation at 30°C, cells were labeled with 500 μCi of [35S]methionine/cysteine (Promix; GE Healthcare), and chase was initiated by adding cold solution of methionine and cysteine (2 mM each in a final concentration) and terminated by the addition trichloroacetic acid to 10%. CPY was immunoprecipitated from cell lysates with the mAb specific to CPY and resolved by 8.5% SDS-PAGE. Quantification of CPY band images was performed using ImageQuant 5.0 software (GE Healthcare).
Subcellular Fractionation of Spheroplasts: Sorbitol Overlay Assay
Conversion of 250 OD cells to spheroplasts and cell lysis were performed as described previously (Harsay and Bretscher, 1995; Harsay and Schekman, 2002; Nunnari et al., 2002). For sorbitol density gradient centrifugation, 1 ml of cleared extract was overlaid directly onto sorbitol step gradients prepared in 12.5-ml tubes (SW41; Beckman Coulter, Fullerton, CA) as follows: 1 ml, 80%; 2 ml, 70%; 2 ml, 60%; 2 ml, 50%; 2 ml, 40%, and 1 ml, 30% sorbitol/1× YEB. The gradients were spun for 40 h at 200,000 × g, and 0.5-ml fractions were collected from the top. A portion of the material collected from each of the fractions was analyzed by SDS-PAGE and Western blot analysis.
RESULTS
Exploring the Detergent-resistant Properties of TORC1 and TORC2
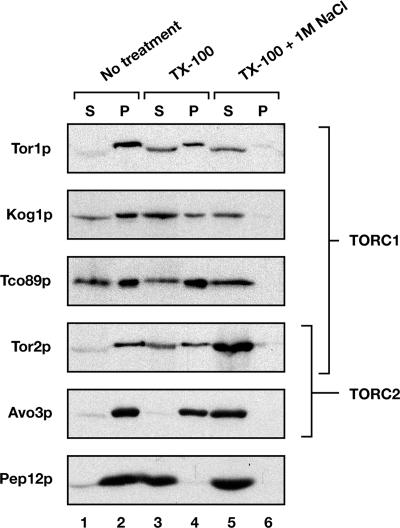
Results from previous studies have shown that Tor1p and Tor2p associate with membranes that are partially resistant to extraction with the nonionic detergent TX-100 (Kunz et al., 2000; Wedaman et al., 2003). We asked whether this was a general property of TORC1 and TORC2 by examining the behavior of representative components of each complex after detergent extraction and centrifugation of cell-free lysates. Indeed, for three members examined, Kog1p, Tco89p, and Avo3p, each behaved similarly to Tor1p and Tor2p in that they were efficiently converted to a soluble form only upon treatment with both 1% TX-100 and with 1 M NaCl (Figure 1). By contrast, a control membrane protein, the late endosomal marker Pep12p, was efficiently solubilized by detergent extraction alone (Figure 1).
Figure 1.
TORC1 and TORC2 components are resistant to extraction with Triton X-100. Cell extracts were prepared from W303a-derived strains, and they were treated with 1 M NaCl and/or Triton X-100, as indicated. After centrifugation, supernatants (S) and pellets (P) were separated by SDS-PAGE, and the indicated proteins were detected by Western blot analysis. Components of TORC1 and/or TORC2 are indicated by brackets.
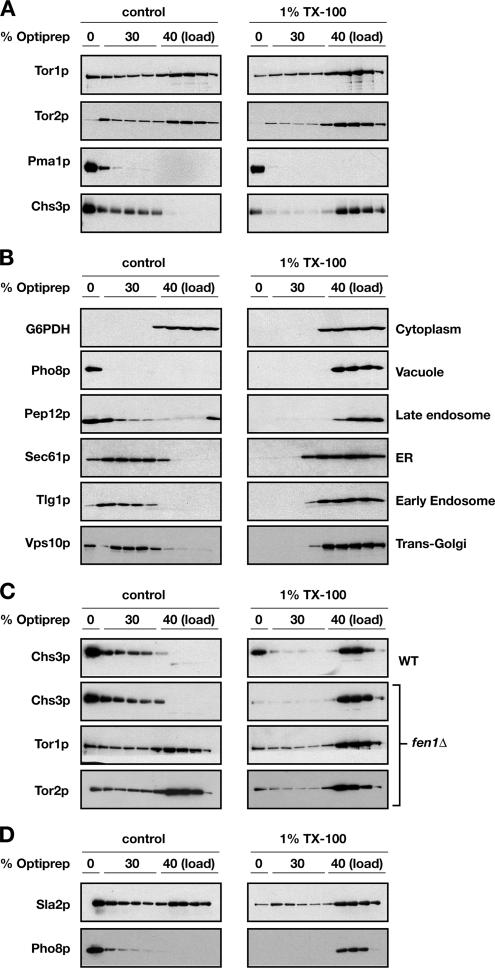
Insolubility of membrane proteins in the presence of TX-100 is indicative of possible association with membrane microdomains that have been variously termed lipid rafts, detergent-insoluble glycolipid-enriched complexes (DIGs), or, more generally, detergent-resistant membranes (DRMs) (Bagnat et al., 2000; Simons and Vaz, 2004). One hallmark behavior of DRMs is their characteristic low buoyant density after detergent extraction and centrifugation in iodixanol (e.g., OptiPrep) step gradients (Bagnat et al., 2000). Thus, to test whether Tor1p and/or Tor2p are associated with these operationally defined DRMs, we adjusted both untreated as well as TX-100–treated cell extracts to 40% OptiPrep and resolved them on 0–40% OptiPrep discontinuous step gradients (see Materials and Methods). After centrifugation and fractionation, SDS-PAGE and Western blot analysis were used to determine the location of Tor1p and Tor2p as well as a number of different specific marker proteins (Figure 2, A and B).
Figure 2.
A portion of Tor1p and Tor2p associate with detergent-resistant membranes that are distinct from plasma membrane rafts. (A) Cell extracts of W303a-derived strains were treated with Triton X-100 (right) or with an equal volume of TNEX buffer (left) and subjected to 0–40% OptiPrep density gradient. Fractions were collected from the top of the gradient, and proteins were separated by SDS-PAGE. Localization of detergent-resistant fraction of Tor1p and Tor2p proteins on the gradient is clearly distinct from that of plasma membrane rafts represented by Pma1p and Chs3p markers. (B) Cell extractions and gradients were performed as described in A. Localization of the membranes associated with different organelle markers was dramatically affected by TX-100 treatment. Cytoplasmic marker Zwf1p (G6PDH) was included as a nonmembrane-associated control. (C) Cell extracts were prepared from fen1Δ strain (PLY644) as well as from the respective wild-type parent and subjected to OptiPrep gradient as described in A. Deletion of FEN1 gene, involved in sphingolipid biosynthesis, only partially affects the floatation profile of Tor1p and Tor2p, whereas the floatation of Chs3p is abolished when the extract is treated with TX-100. (D) Sla2p displays a gradient profile similar to that of the TOR proteins. Alkaline phosphatase Pho8p detected in the same experiment is shown as an example of a protein that does not associate with TX-100–resistant membranes. We note that a variable amount of Tor1p and Tor2p occurs in the 0% OptiPrep fractions, which we attribute to the difficulty in fractionation of the 0%30% interface.
As expected, the plasma membrane proton ATPase Pma1p, a representative marker for DRMs (Bagnat et al., 2000), was localized at the top of gradients, corresponding to 0% OptiPrep fractions, in both untreated as well as in TX-100–treated extracts (Figure 2A). A similar behavior was also observed for another plasma membrane marker protein Chs3p (Ziman et al., 1996) (Figure 2A). By contrast, marker proteins for several distinct membrane compartments were observed to float in untreated extracts but not in TX-100–treated extracts, where in the latter case they largely cofractionated with the cytoplasmic marker protein Zwf1p (G6PDH) (Figure 2B). Interestingly, both Tor1p and Tor2p displayed a unique behavior, in comparison with each of the marker proteins described above, in that a portion of both proteins was spread throughout the 30% OptiPrep fractions and in a form that was largely resistant to TX-100 extraction (Figure 2A). Similar profiles were also observed for both Kog1p and Avo3p, components of TORC1 and TORC2, respectively (data not shown). These results demonstrate that a portion of both TORC1 and TORC2 are localized to a TX-100–resistant membrane environment that is distinct from DRMs that contained Pma1p and Chs3p. Accordingly, we differentiate between plasma membrane-DRMs (PM-DRMs) and TOR-DRMs to describe Pma1p/Chs3p-containing fractions versus Tor1p/Tor2p-containing fractions, respectively.
Sphingolipids are important for the formation and integrity of yeast DRMs (Simons and Vaz, 2004). Accordingly, we asked whether the TX-100-resistant behavior of Tor1p and Tor2p was altered in extracts prepared from a strain deleted for FEN1, which encodes fatty acid elongase required for normal levels of sphingolipids that contain very long chain fatty acids (Oh et al., 1997; Kohlwein et al., 2001; Obeid et al., 2002). We were particularly interested in comparing the behavior of the Tor proteins to Chs3p, which displayed an overlapping profile on OptiPrep gradients in the absence of TX-100 treatment (Figure 2A). We observed that the TX-100–resistant behavior of Chs3p was abolished in extracts prepared from a fen1Δ mutant (Figure 2C). By contrast, a significant amount of both Tor1p and Tor2p continued to float on OptiPrep gradients in detergent-treated extracts prepared from this mutant (Figure 2C), confirming the unique behavior of TOR-DRMs.
Proteomic Analysis of PM- and TOR-DRMs
To further characterize TOR-DRMs, we took advantage of its unique floatation profile after TX-100 treatment to isolate and identify cofractionating proteins in the upper 30% OptiPrep fractions using tandem MS/MS spectrometry (see Materials and Methods). For comparison, we also analyzed in parallel the protein composition of PM-DRMs from 0% OptiPrep fractions. Remarkably, the majority of proteins identified by mass spectrometry uniquely within PM-DRM fractions corresponded to known plasma membrane proteins, including a number of proteins that have previously been shown to reside in DRMs, such as Pma1p, Nce101p/Nce2p, Gas1p (Bagnat et al., 2000), and Sur7p (Malinska et al., 2004) as well as glycosylphosphatidylinositol-anchored proteins Ecm33p and Utr2p (Table 2).
Table 2.
Proteins recovered from PM-DRM fractionsa
| Name | Description/gene product | No. of unique peptides | Sequence coverage (%) | Assigned subcellular localizationb |
|---|---|---|---|---|
| Rho1 | GTP-binding protein of the rho subfamily of Ras-like proteins | 5 | 23.4 | Bud neck/bud tip/peroxiome/mitochondria |
| Gas1 | β-1.3-Glucanosyltransferase | 15 | 16.5 | Plasma membrane/mitochondria |
| Ecm33 | GPI-anchored protein | 6 | 15.0 | Plasma membrane/mitochondria |
| Pma1 | Plasma membrane H+-ATPase | 22 | 13.5 | Plasma membrane |
| Gas5 | Putative 1,3-β-glucanosyltransferase | 5 | 10.1 | Cell wall |
| Sso2 | Syntaxin homologues (post-Golgi target membrane-associated soluble N-ethylmaleimide-sensitive factor-attachment protein receptors) | 3 | 9.2 | ER/cytoplasm |
| Hxt2 | High-affinity glucose transporter | 6 | 9.1 | Plasma membrane |
| Ctr1 | High-affinity copper transporter | 3 | 8.6 | Plasma membrane |
| Ste2 | Receptor for α-factor pheromone | 2 | 8.1 | Plasma membrane |
| Hxt3 | Low-affinity glucose transporter | 5 | 7.8 | Plasma membrane |
| Fre1 | Ferric reductase and cupric reductase | 6 | 7.4 | Plasma membrane |
| Sur7 | Multicopy suppressor of rvs167 mutation | 3 | 7.0 | Mitochondria/cell cortex |
| Nce102 | Component of the DIGs | 2 | 6.9 | ER/mitochondria/cytoplasm |
| Gas3 | Putative 1,3-β-glucanosyltransferase | 3 | 6.7 | Cell wall |
| Mrh1 | Membrane protein Related to Hsp30p | 2 | 5.9 | Plasma membrane/mitochondria |
| Hxt1 | Low-affinity glucose transporter | 4 | 4.4 | Plasma membrane |
| Utr2 | Putative glycosidase, GPI-anchored protein | 2 | 4.3 | Cell wall/septin ring |
| Ptr2 | Peptide transporter | 2 | 4.2 | Plasma membrane |
| Hxt4 | High-affinity glucose transporter | 4 | 4.1 | Plasma membrane |
| Chs1 | Chitin synthase I | 3 | 3.5 | Plasma membrane |
| Slg1 | Protein involved in cell wall integrity and stress response | 2 | 3.4 | Actin cap |
| Gyp5 | GTPase-activating protein for yeast Rab family members | 2 | 3.1 | Plasma membrane/bud neck/bud tip/Golgi |
| Tpo3 | Polyamine transport protein | 2 | 3.1 | Plasma membrane/vacuole |
| 3Hxt7 | High-affinity glucose transporter | 3 | 2.8 | Plasma membrane/mitochondria |
| Hxt6c | High-affinity glucose transporter | 3 | 2.8 | Plasma membrane/mitochondria |
| Hxt5 | Hexose transporter with moderate affinity for glucose | 3 | 2.7 | Plasma membrane |
| Dnf2 | P-type ATPase, a potential aminophospholipid translocase | 3 | 2.4 | Plasma membrane |
| Ena1d | P-type ATPase sodium pump | 2 | 1.9 | Plasma membrane |
| Ena2d | P-type ATPase sodium pump | 2 | 1.9 | Plasma membrane/mitochondria |
| Ena5d | Protein with similarity to P-type ATPase sodium pumps | 2 | 1.9 | Plasma membrane |
| Gsc2 | Catalytic subunit of 1,3-β-glucan synthase | 2 | 1.3 | Actin cap |
| Yor1 | Transporter of the ATP-binding cassette (ABC) family | 2 | 1.2 | Plasma membrane |
| Fks1 | Catalytic subunit of 1,3-β-d-glucan synthase | 2 | 1.1 | Actin cap/actin cortical patch |
| Snq2 | ABC transporter | 3 | 1.1 | Plasma membrane/mitochondria |
aProteins listed represent the most abundant nonribosomal peptide species identified by mass spectrometry of trypsin-digested proteins obtained from PM-DRMs. Parameters listed are number of unique peptides and percentage of coverage of the primary sequence of the protein in terms of unique peptides.
bInformation obtained from SGD (www.yeastgenome.org).
cUnable to distinguish between these proteins.
dUnable to distinguish between these proteins.
By contrast, proteins identified within TOR-DRMs corresponded to a broad distribution of functional activities as well as organelle locations (Supplemental Table 1); however, we observed that a significant number of these proteins have been implicated in processes related to actin polarization and/or endocytosis (Table 3). To confirm these proteomic results, we constructed a strain that expressed a Myc epitope-tagged version of SLA2, encoding a representative member from the list, and we examined its behavior by Western blot analysis after fractionation of untreated and TX-100–treated extracts on OptiPrep gradients (Figure 2D). Indeed, this protein displayed a profile remarkably similar to Tor1p and Tor2p in that a significant portion localized to the upper 30% OptiPrep fractions in both extracts (compare Figure 2A with D). A similar result was obtained when we examined a Myc-epitope tagged version of RVS161, which encodes another protein identified in TOR-DRMs (Table 3; data not shown). We conclude from these results that we have identified within TOR-DRMs a set of proteins that share common biochemical properties with TORC1 and TORC2 as defined by this assay.
Table 3.
Proteins involved in actin polarization and/or dynamics recovered from TOR-DRMsa
| Name | Description/gene product | No. of unique peptides | Sequence coverage (%) | Essential gene?b |
|---|---|---|---|---|
| Cct8 | Subunit of the cytosolic chaperonin Cct ring complex | 4 | 16.5 | Yes |
| Slt2 | Serine/threonine mitogen-activated protein kinase, suppressor of lyt2 | 3 | 14.5 | No |
| Rvs161 | BAR adaptor protein | 2 | 11.7 | No |
| Tpd3 | Protein phosphatase 2A regulatory subunit A | 2 | 10.1 | No |
| Srv2 | Adenylyl cyclase-associated protein | 3 | 10.1 | No |
| Sac6 | Fimbrin, actin bundling protein | 3 | 9.7 | No |
| Ypk1 | Serine/threonine protein kinase | 2 | 5.4 | No |
| She4 | Protein binding to myosin motor domains | 2 | 4.4 | No |
| Myo2 | Class V myosin | 3 | 4.0 | Yes |
| Sla2 | Adaptor protein that links actin to clathrin and endocytosis | 2 | 3.4 | No |
| Cdc24 | Guanine nucleotide exchange factor for Cdc42p | 2 | 3.3 | Yes |
| Sro7 | Suppressor of rho3 | 2 | 3.3 | No |
| Bem2 | Rho GTPase activating protein | 2 | 1.6 | No |
aProteins listed are a subset of nonribosomal peptide species identified by mass spectrometry of trypsin-digested proteins obtained from TOR-DRMs (see Supplemental Table 1 for a complete list). Parameters listed are number of unique peptides and percentage of coverage of the protein's primary sequence in terms of unique peptides.
bInformation obtained from SGD (www.yeastgenome.org).
Genetic Interactions between TORC1 and TORC2 and Actin/Endocytosis-related Genes
We took a genetic approach to test for functional associations between components listed in Table 3 and TOR by assaying for synthetic sick/lethal (SSL) interactions. Specifically, we examined the growth phenotypes of strains deleted individually for each of the nonessential genes listed in Table 3 (a total of 10 genes) in combination with mutations of representative components of TORC1 and TORC2. For TORC1, we examined deletions of two nonessential genes, TOR1 and TCO89 (Loewith et al., 2002; Reinke et al., 2004), whereas for TORC2 we examined deletions of two nonessential genes, AVO2 and BIT61 (Loewith et al., 2002; Reinke et al., 2004) as well as a novel temperature-sensitive allele of the essential gene AVO3 (see Materials and Methods). In each case, pairwise combinations of heterozygous diploid strains were constructed, followed by sporulation and tetrad dissection. We then assessed the degree of SSL interaction for each gene pair by genotyping the resulting spores and by inspection of individual spore sizes that formed at different temperatures.
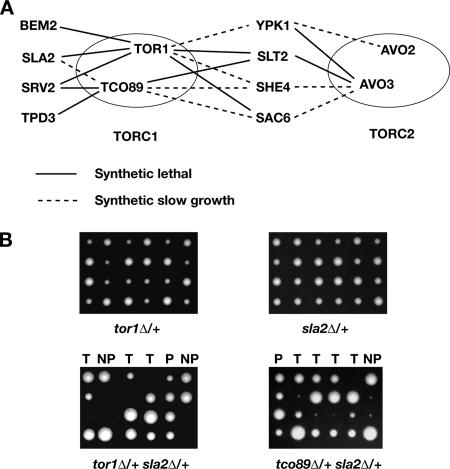
From among the 60 heterozygous diploids strains examined, 10 synthetic lethal and eight synthetic slow-growth interactions were detected (summarized in Figure 3A). An example of both of these types of interactions is shown in Figure 3B, where simultaneous deletion of SLA2 and TOR1 results in synthetic lethality, whereas deletion of SLA2 and TCO89 results in a synthetic slow-growth phenotype. Some form of synthetic interaction was observed between one or more TORC1 or TORC2 components, and eight of the 10 genes tested from Table 3 (no detectable synthetic interactions were observed for RVS161 or SRO7). Four genes displayed synthetic interactions with both TORC1 as well as TORC2 components, whereas four genes displayed synthetic interactions exclusively with TORC1 components (Figure 3A). We note that the total number of interactions observed was significantly higher than predicted estimates of the total density of SSL interactions in the yeast genome (∼1:200; Tong et al., 2004), suggesting our proteomics approach led to an enrichment of gene products that interact functionally with TORC1 and TORC2.
Figure 3.
Multiple genetic interactions between both TORC1 and TORC2 components and genes involved in actin cytoskeleton organization or/and endocytosis. (A) Genetic interactions between several components of Tor complexes and nonessential genes of proteins recovered from Tor-containing fraction by mass spectrometry for which products are involved in cytoskeleton organization or/and endocytosis. Continuous lines represent synthetic lethality, and dashed lines represent synthetic slow-growth interactions. (B) SLA2 is one of the genes that show synthetic interactions specifically with the components of TORC1. Examples of tetrad dissections of heterozygous diploids of single mutants: tor1Δ, sla2Δ as well as those double mutants: tor1Δ sla2Δ and tco89Δ sla2Δ. Note that tor1Δ is synthetically lethal with sla2Δ, whereas tco89Δ displays a synthetic slow growth defect with sla2Δ. T, tetratype; P, parental ditype; NP, nonparental ditype.
TORC1 Influences Actin Polarity in Response to Nutrient Limitation
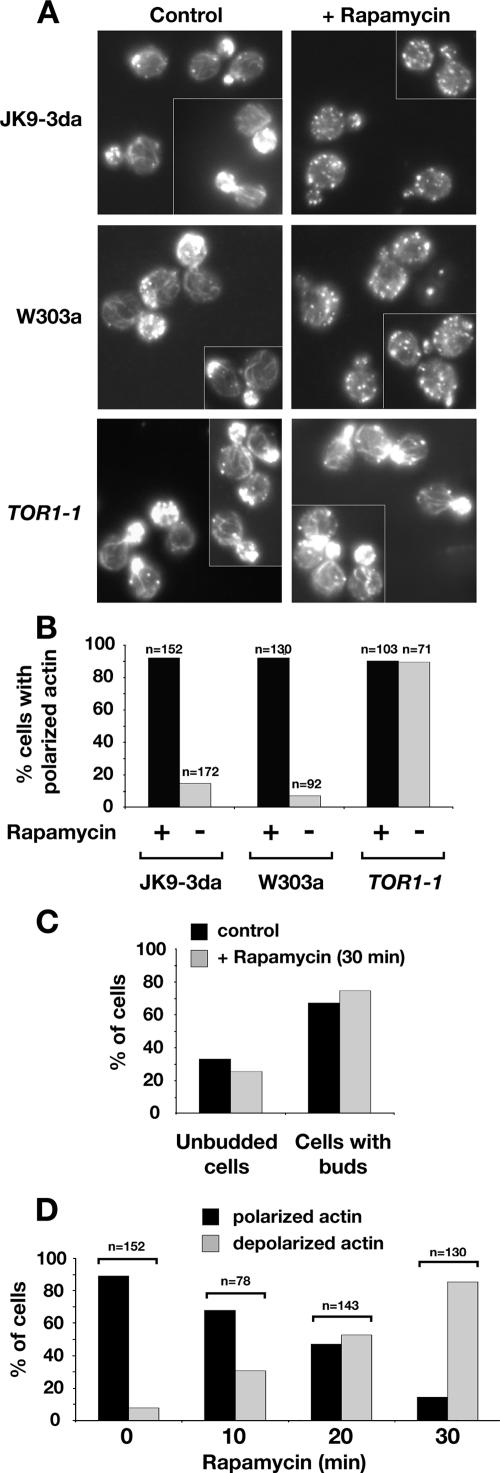
Our observation that TORC1 components display a significant number of SSL interactions with genes involved in actin polarization is consistent with a previous report that rapamycin treatment results in significant depolarization of the actin cytoskeleton (Torres et al., 2002). We confirmed this finding for two different wild-type strains, W303a and JK9-3da, where rapamycin treatment of cells for 30 min resulted in nearly complete depolarization of actin (Figure 4, A and B). Importantly, the budding index of cells did not change appreciably after drug treatment during this experiment, demonstrating that depolarization was not a secondary consequence of a rapamycin-induced cell cycle arrest, which remained a formal criticism of the previous study cited above (Figure 4C). Moreover, we observed significant depolarization after a period of rapamycin treatment as brief as 10 min (Figure 4D). By contrast, rapamycin treatment of cells expressing a dominant rapamycin resistant TOR1-1 allele had no significant effect on actin polarization, confirming that rapamycin-induced depolarization is due to inhibition of TORC1 (Figure 4, A and B).
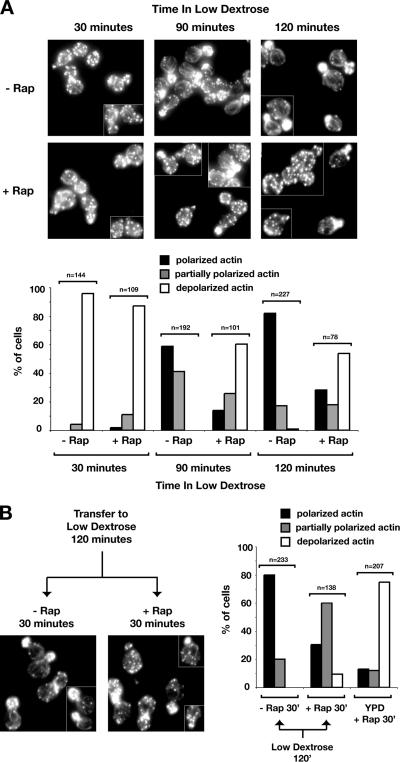
Figure 4.
Actin cytoskeleton organization is rapidly affected by specific inhibition of TORC1 by rapamycin and its reorganization may be part of the starvation response. (A) Distribution of polymerized actin upon rapamycin treatment in wild-type strains of W303a and Jk9-3da backgrounds and in a strain that carries rapamycin insensitive TOR1-1 allele (JH1-11c). Cells were grown in YPD and treated either with rapamycin or dimethyl sulfoxide (DMSO) for 30 min. Cells were fixed, stained with rhodamine-phalloidin, and visualized by fluorescence microscopy as described in Materials and Methods. All fluorescence images are Z-series stacks of eight to 12 0.2-μm steps projected to a single plane. (B) Percentage of small- and midsize-budded cells that display polarized distribution of actin upon rapamycin treatment in the experiment shown in A. (C) Depolarization of the actin cytoskeleton is not a secondary effect of a cell cycle arrest, because the percentage of unbudded cells does not change within 30 min of rapamycin treatment. Wild-type Jk9-3da cells were grown to early log phase, treated with rapamycin, and processed as described in A. (D) Time course of depolarization of the actin cytoskeleton upon rapamycin treatment. Wild-type Jk9-3da cells were treated with rapamycin for the amount of time indicated and processed as described in A.
A recent study demonstrated that starvation for glucose results in rapid depolarization of the actin cytoskeleton, followed by slower repolarization, suggesting that transient remodeling of the cytoskeleton represents an important part of the cellular response to a sudden change in carbon availability (Uesono et al., 2004). In this study, the kinetics of depolarization after glucose starvation was observed to be considerably faster than that caused by rapamycin treatment, leading to the conclusion that TOR was unlikely to mediate actin depolarization caused by glucose starvation (Uesono et al., 2004). However, a possible requirement for TOR during repolarization of the actin cytoskeleton was not addressed in this study. Given our present findings, we decided to test directly whether rapamycin treatment affected the kinetics of actin repolarization after glucose starvation. The results of this experiment are shown in Figure 5A.
Figure 5.
Remodeling of actin cytoskeleton in response to glucose starvation. (A) Rapamycin treatment significantly diminishes repolarization of the actin cytoskeleton that occurs after a shift of cells from high to low dextrose-containing media. Wild-type Jk9-3da cells were grown in rich YPD medium and pretreated with rapamycin or DMSO for 30 min before the shift to YPD containing 0.05% glucose for the time indicated. (B) Rapamycin treatment does not significantly perturb the actin cytoskeleton once it is remodeled after transfer to low glucose conditions. Cells were shifted from high (2%) to low (0.05%) glucose medium for 120 min in the absence of rapamycin, followed by either rapamycin or DMSO treatment for an additional 30 min.
First, we observed that the actin cytoskeleton became completely depolarized after 30 min of incubation of cells in media that lacked glucose, in agreement with previous findings (Figure 5A) (Uesono et al., 2004). Significant repolarization was observed after 90 min of incubation in glucose-free media and nearly complete repolarization was observed at 120 min (Figure 5A). We note that this rate of repolarization was considerably faster than that reported previously, where it took approximate 5 h to significantly restore actin polarization (Uesono et al., 2004). We have determined that this difference is attributable to differences in strain background, where we used JK9–3da for this experiment, whereas the previous study used W303a (data not shown). Importantly, we found that addition of rapamycin to cells before a shift to glucose-free media strongly impeded the kinetics as well as extent of actin repolarization, demonstrating that a functional TORC1 is indeed required for actin remodeling in response to glucose starvation (Figure 5A).
Given these results, we next wanted to ask whether there was a continued requirement for TOR once the actin cytoskeleton became repolarized after incubation in glucose-free media. We therefore starved cells for glucose for 120 min, followed by incubation for an additional 30 min either in the absence or presence of rapamycin (Figure 5B). We observed that the effect of rapamycin on actin depolarization was significantly attenuated, in comparison with an identical time of drug treatment of cells grown in YPD (Figure 5B, right). Thus, we conclude from these results that once actin becomes repolarized after glucose starvation, a requirement for TOR is much less critical for maintenance of polarity.
TORC1 Influences a Late Step of Endocytosis
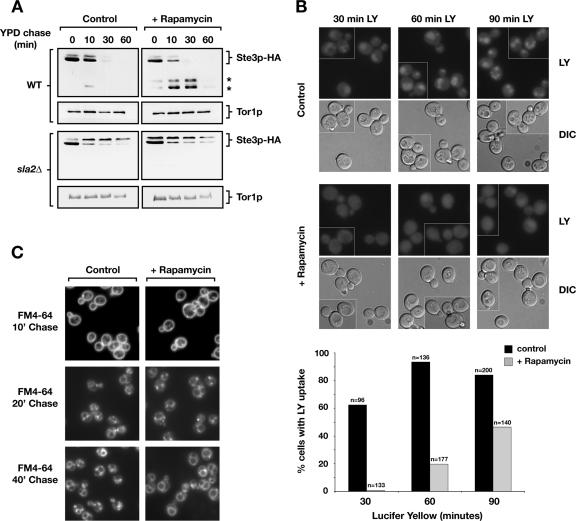
Many of the genes that displayed SSL interactions with TORC1 components (Figure 3) have also been identified as being involved in receptor-mediated endocytosis (Kubler and Riezman, 1993; Wendland et al., 1996; Penalver et al., 1997; Wesp et al., 1997; deHart et al., 2003). We therefore examined the effect of rapamycin on the internalization of HA-epitope–tagged, GAL-regulated Ste3p, by using a galactose shut-off assay in conjunction with Western blot analysis to monitor protein turnover after internalization (Walther et al., 2006). No significant differences were observed in the rates of internalization within rapamycin-treated versus untreated cells, as judged by the disappearance of full-length Ste3p-HA after transfer of cells from galactose- to glucose-containing media (Figure 6A). This result is consistent with previous observations that rapamycin treatment does not impair α factor internalization (deHart et al., 2003). However, we did observe a reproducible accumulation of faster mobility species of Ste3p-HA in rapamycin-treated cells (Figure 6A, top right, denoted by asterisks). These species most likely correspond to C-terminal fragments of Ste3p that have been observed previously to accumulate in mutants impaired for delivery of Ste3p to the vacuolar lumen (Chen and Davis, 2002; Shaw et al., 2003). As expected, appearance of these faster mobility intermediates required internalization of Ste3p-HA, because they did not occur in rapamycin-treated sla2Δ cells, where uptake of Ste3p is greatly diminished (Figure 6A).
Figure 6.
Rapamycin delays fluid phase endocytic trafficking of lucifer yellow to the vacuole as well as degradation but not internalization of Ste3p-HA. (A) Wild-type and sla2Δ cells carrying plasmid pGAL-STE3-HA were grown overnight in medium containing 2% galactose to induce STE3-HA expression, and they were treated with rapamycin or DMSO for 30 min. After a shift to YPD, aliquots were removed at indicated times, and proteins were extracted and analyzed by Western blotting to detect Ste3p-HA. The star symbol indicates presumed degradation intermediates of Ste3p-HA. (B) LY trafficking to the vacuole is delayed upon rapamycin treatment of wild-type strain (LHY291). Cells were grown in YPD to OD600 = 0.3–0.5, and they were either treated with rapamycin or DMSO. Cells were processed as described in Materials and Methods, and they were incubated with LY at 30°C for the indicated minutes. Quantification for this experiment is shown below. (C) Bulk membrane flow rate is not affected by rapamycin treatment as assayed by staining with the lipophilic dye FM4-64. Cells were grown overnight in YPD, treated with rapamycin or DMSO for 30 min, and further processed as described previously (Vida and Emr, 1995).
To investigate these results further, we examined the effect of rapamycin treatment on the uptake of LY, a marker for fluid phase endocytosis that transits throughout the endocytic pathway and that accumulates within the vacuole (Dulic et al., 1991). Here, we observed a significant delay in LY accumulation after rapamycin treatment, where only limited uptake was observed when cells were incubated with the dye for 60 min, a time corresponding to maximum uptake in untreated cells (Figure 5, B and C). A larger percentage of rapamycin-treated cells eventually displayed LY accumulation after an extended period of incubation with dye (90 min); however, the level of staining was always much more diffuse compared with untreated cells (Figure 6B). Together with our results with Ste3p-HA, we conclude that inhibition of TORC1 with rapamycin results in a significant delay in endocytosis but at a step that is likely to occur after internalization. By contrast to these results, rapamycin treatment resulted in no significant delay in the rate of accumulation of the lipophilic membrane marker FM4-64 at the vacuole, indicating that inhibition of TORC1 perturbs a limited scope of events related to endocytic trafficking to the vacuole (Figure 6C).
A genetic Network for TORC1 and Actin/Endocytosis Genes
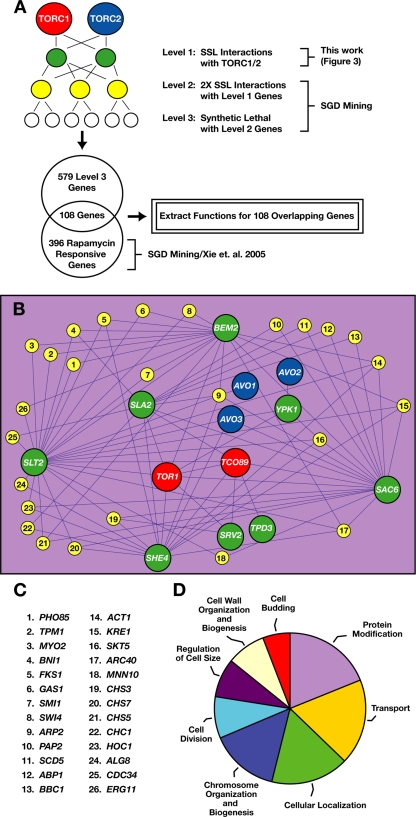
Given our results described above, we sought to gain additional insight into the scope of cellular processes potentially influenced by both TORC1 and actin patch/endocytosis-related components. We explored a number of different approaches for meta-analysis, and we surveyed several interaction data sets available through the Saccharomyces Genome Database (SGD), and ultimately we devised the following regime to construct a network of genetic interactions (Figure 7). First, we defined our SSL interactions, described above, as LEVEL 1 of this network (Figure 7A) (Supplemental Table 2). Next, we identified through SGD 26 genes that display SSL interactions with at least two LEVEL 1 components and we defined these collectively as LEVEL 2 (Figure 7, B and C) (Supplemental Table 2). Not surprisingly, this list was enriched for genes implicated in actin organization (Figure 7C). We next identified through SGD 579 genes that display strictly synthetic lethal interactions with one or more LEVEL 2 genes, which we defined as LEVEL 3 (Supplemental Table 2). To narrow our focus to genes related specifically to TORC1, we filtered these LEVEL 3 genes through a list of 396 recently identified gene deletions that display altered (primarily increased) sensitivities to rapamycin (Xie et al., 2005) (Figure 7A). This latter step resulted in the identification of 108 genes that represent several distinct functional categories, as defined by the Gene Ontology index in SGD (Figure 7D and Supplemental Table 2).
Figure 7.
Network of genetic interactions involving TORC1 components and genes implicated in cytoskeleton organization. (A) Diagram of different levels of genetic interactions involving TORC1 used to extract functions and/or pathways that are interdependent with actin–cytoskeleton components. LEVEL 3 genes were filtered through the list of rapamycin-responsive genes (Xie et al., 2005) where individual gene deletions display rapamycin-hypersensitive or resistant phenotypes. Filtering was performed using a Perl script developed in our laboratory (available upon request). Gene functions (standardized GO terms) were extracted using the Gene Ontology tool (GO_term finder) available through the SGD (http://db.yeastgenome.org/cgi-bin/GO/goTermFinder). (B) Network of genetic interactions was built using Cytoscape 2.1 software (http://www.cytoscape.org) by using manual alterations. Nodes and connecting edges denote genes and their genetic interactions, respectively. Red and blue nodes specify components of TORC1and TORC2, respectively. Green nodes (LEVEL 1 genes) represent genes encoding proteins recovered from TOR-DRMs that displayed SSL interactions with TORC1 and/or TORC2 genes. Yellow nodes (LEVEL 2 genes) depict genes that display two or more SSL interactions with LEVEL 1 genes. (C) List of LEVEL 2 genes depicted in B. (D) Diagram of pathways extracted from our network of genetic interactions as described in A. The transport function is shown in detail in Figure 8A. GO terms were clustered based on hierarchical tree of GO biological process annotations.
To examine the specificity of this network, we repeated the step of collecting synthetic lethal interactions via SGD beyond LEVEL 3 for three additional iterations (LEVELS 4–6) before filtering the results through the list of rapamycin-responsive genes identified by Xie et al. (2005). In particular, we wanted to determine whether our approach would ultimately link all 396 of these genes within the network, or, alternatively, whether this network would continue to identify only a subset of rapamycin responsive genes. Indeed, we observed that the network reached a plateau after the fifth LEVEL, and it included ∼250 rapamycin-responsive genes (Supplemental Table 2), which represented less than two thirds of the total number of genes identified by Xie et al. (2005). Additional functional categories derived from LEVELS 4 and 5 included processes linked to DNA metabolism as well as vacuole organization and biogenesis, autophagy, and cellular stress responses (Supplemental Table 2). Importantly, many processes commonly associated with TORC1 signaling, including carbon regulation as well as amino acid metabolism were not identified by this process, despite the fact that genes involved in these processes are included within the data set by Xie et al. (2005). Thus, these results suggest that the TORC1-actin/endocytosis network we have described involves a specific subset of rapamycin-responsive genes.
Testing the Network: Novel Genetic Interactions between TORC1 and Vesicular Trafficking Genes
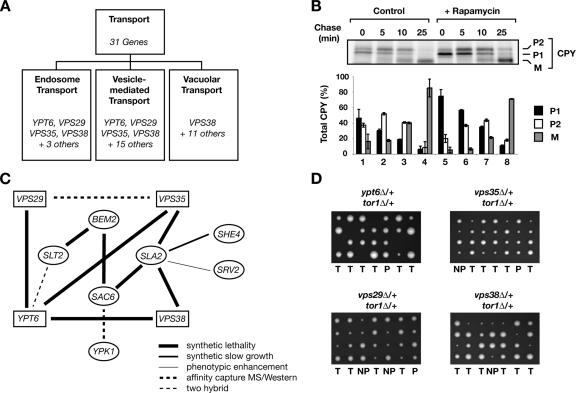
A number of the processes listed in Figure 7D, including regulation of cell size, cell wall organization, and protein modification, have been shown previously to be regulated by TORC1. By contrast, several genes identified by this analysis cluster within the general category of “transport” and include genes that have not been linked previously to TORC1, except by virtue that their deletion results in hypersensitivity to rapamycin (Figure 7D) (Xie et al., 2005). Thus, to investigate this category further, we partitioned all genes within transport into distinct subcategories using the Gene Ontology index in SGD (Figure 8A). This approach identified three subcategories related to endosomal and vesicular transport, two of which possessed four genes in common: YPT6, VPS29, VPS35, and VPS38 (Figure 8A). Each of these genes has been implicated in the trafficking of a number of specific substrates, including CPY, which passes through the early secretory pathway and is targeted to the vacuole via the late endocytic pathway (Vashist et al., 2001). We therefore examined the effect of rapamycin treatment on the trafficking of CPY, which can be monitored using pulse-chase analysis as a series of post-translational glycosylation and proteolytic cleavage events as it transitions from two distinct precursor (P1 and P2) forms to a fully processed, mature (M) form. Indeed, we observed a mild but reproducible delay in CPY trafficking, particularly at the earliest step of conversion of P1 to P2 forms, in cells that had been treated with rapamycin, in comparison with untreated cells (Figure 8B). We note that this level of defect in CPY maturation is remarkably similar to that observed for cells deleted for the CHC1 gene that encodes clathrin heavy chain (Payne et al., 1988) as well as a number of mild mutant alleles of genes within the YPT gene family (Singer-Kruger et al., 1994).
Figure 8.
Genetic interactions network suggests a link between TORC1 and vesicular trafficking. (A) Detailed representation of the Transport function that is part of the diagram shown in Figure 7D. YPT6, VPS29, VPS35, and VPS38 genes fall into multiple subcategories related to transport. (B) CPY trafficking is delayed upon rapamycin treatment. Wild-type W303a cells were grown in SCD medium lacking methionine and cysteine, treated with DMSO or rapamycin for 30 min, and then metabolically labeled with [35S]methionine/cysteine for 10 min and chased in the presence of the excess of cold methionine/cysteine at 30°C for the indicated minutes. Proteins were immunoprecipitated using α-CPY antibodies, resolved on SDS-PAGE, and analyzed using a PhosphorImager. Quantification of data for three independent experiments is shown, where black bars denote ER precursor (P1), white bars denote Golgi-modified forms (P2), and gray bars denote mature (M) forms of CPY. Error bars denote standard deviations. (B) Network of genetic and physical interactions (derived from SGD) that link highly represented genes in the transport category (squares) with LEVEL 1 genes (ovals). (C) TOR1 genetically interacts with highly represented genes in the transport category. tor1Δ is synthetically lethal with deletion of YPT6, and displays synthetic slow growth interactions with VPS29, VPS35, and VPS38 genes as shown by tetrad dissection of respective heterozygous diploids. The plate combining tor1 and ypt6 deletions was incubated at 32°C, whereas plates combining tor1 and vps deletions were incubated at 30°C.
In addition to synthetic lethal interactions with LEVEL 2 genes (required for their inclusion in the network), each of the four genes, YPT6, VPS29, VPS35, and VPS38, are also tightly linked to several LEVEL 1 genes, both by SSL interactions as well as by other genetic and biochemical interactions (Figure 8C). Thus, to directly test for their potential relationship to TORC1, we assayed for SSL interactions after sporulation of diploid strains, each deleted for one of these four nonessential genes in combination with deletion of TOR1. Indeed, we observed a synthetic lethal interaction between tor1Δ and ypt6Δ as well as significant synthetic slow-growth interactions with each of the three vps gene deletions (Figure 8D). Together, these results demonstrate a remarkably tight series of functional interactions involving TORC1 and actin/endocytosis genes throughout this network.
Cofractionation of TORC1 with Distinct Organelle Markers
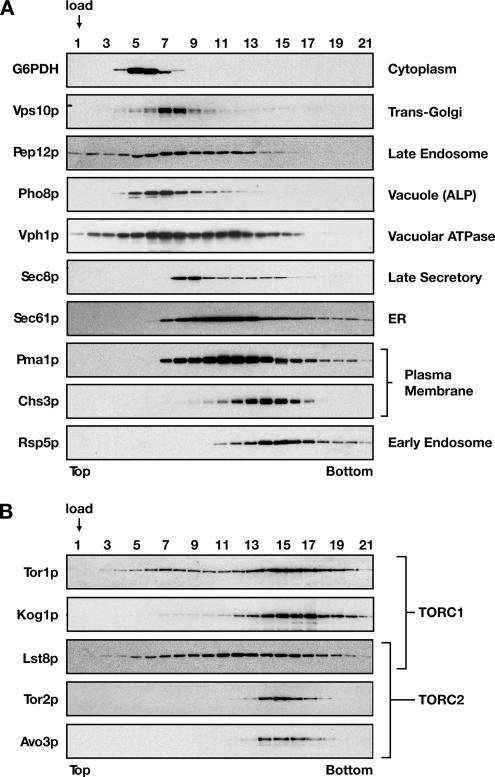
Given the spectrum of trafficking events that are predicted to be influenced by TORC1, both by our network analysis as well as results from other laboratories, we sought to revisit the question as to which intracellular membrane compartment(s) is associated with this complex. Results of previous studies are consistent with TORC1 components being localized to membranes related to the secretory and/or endosomal pathways (Kunz et al., 2000; Chen and Kaiser, 2003; Wedaman et al., 2003). However, as most biochemical fractionation approaches have used sucrose gradient floatation assays, it has been difficult to distinguish between these different compartments, due to the similar behavior of representative secretory and endosomal protein markers used for these assays. Accordingly, we explored a variety of gradient methods to maximize separation of different organelle markers from spheroplasted cell extracts, and we found that sorbitol overlay gradients (Cleves et al., 1991; McGee et al., 1994; Zinser and Daum, 1995) (see Materials and Methods) provided an effective means to distinguish several organelles, in particular markers for the trans-Golgi (Vps10p) versus the early endosome (Rsp5p) (Figure 9A).
Figure 9.
Cofractionation of TORC1 and TORC2 components on sorbitol overlay gradients. Cell extracts were prepared from W303a-derived strains and subjected to sorbitol overlay gradients, as described in Materials and Methods. After centrifugation, gradients were fractionated and analyzed by SDS-PAGE followed by Western blot analysis, probing either for representative marker proteins for different secretory and endocytic membrane compartments (A) or representative components of TORC1 and/or TORC2, as indicated (B).
Under these conditions, components of both TORC1 and TORC2 displayed significant overlap with the early endosome marker Rsp5p (Figure 9, A and B). In particular, we observed that both Avo3p, a component that is unique to TORC2, as well as Tor2p, which is preferentially associated with TORC2 (Loewith et al., 2002; Reinke et al., 2004), displayed tight sedimentation profiles that most closely aligned with peak fractions for Rsp5p (Figure 9, A and B). By contrast, a significant portion of the TORC1 components Tor1p, Lst8p, and, to a lesser extent, Kog1p, showed a more diffuse sedimentation profile that was overlapping with several other organelle markers, including the trans-Golgi, endoplasmic reticulum (ER), and vacuole (Figure 9B). A control experiment using a Percoll gradient floatation assay confirmed that a significant portion of each TORC1 and TORC2 components examined behaved as a membrane-associated protein (Supplemental Figure 1). Moreover, subsequent analysis of two distinct Tor1p-containing fractions (7 and 15), taken from the sorbitol overlay gradient and used in an OptiPrep underlay assay, confirmed that a significant portion of Tor1p from each sorbitol overlay gradient fraction was membrane associated (our unpublished data). Together, these results are consistent with the conclusion that both TORC1 and TORC2 are likely to associate with an endosome-like compartment and that TORC1 associates with other compartments as well. This conclusion is consistent with our previous immunoelectron microscopy results where Tor2p displayed a more concise, punctate localization pattern adjacent to the plasma membrane, whereas Tor1p displayed a more dispersed localization pattern (Wedaman et al., 2003). Moreover, these results are consistent with previous findings that TORC1 components are also localized to the vacuole (Huh et al., 2003; Reinke et al., 2004; Araki et al., 2005).
DISCUSSION
We have combined biochemical, genetic, and bioinformatics approaches to learn more about the scope of cellular activities regulated by TORC1 in yeast. In particular, we have described a network of genetic interactions that links TORC1 together with components involved in cortical actin patch dynamics and endocytosis; moreover, we have provided evidence that rapamycin treatment perturbs a number of trafficking events that are distinct from internalization at the plasma membrane. Together with recent findings that TORC1 also influences actin polarity as well as cell wall integrity signaling (Torres et al., 2002; Reinke et al., 2004), two processes previously ascribed strictly to TORC2, our results underscore the emerging connection with respect to cellular processes regulated by both TORC1 and TORC2 in yeast. This conclusion is consistent with recent findings for mammalian TOR signaling, where mTORC1 as well as mTORC2 have now been implicated in the regulation of polarized cell growth and/or cell movement as well as processes related to endocytosis (Berven et al., 2004; Jacinto et al., 2004; Sarbassov et al., 2004; Hennig et al., 2006). Moreover, these results are consistent with the unexpected degree of cross talk that has been demonstrated recently for these complexes in mammalian cells, where, for example, the AGC kinase family member PKB/Akt has been shown to act both downstream of mTORC2 as well as upstream of mTORC1 (Guertin and Sabatini, 2005; Sarbassov et al., 2005).
Based on our biochemical results, we distinguish detergent-resistant membranes that contain TOR from classically defined PM-DRMs that contain Pma1p as well as several other plasma membrane proteins, based on their differences in behavior on OptiPrep gradients as well as their response to perturbation of the sphingolipid content within cells. Cofractionation of proteins within TOR-DRMs could result from physical associations with either TORC1 or TORC2. For example, within TOR-DRMs, we identified Tpd3p, a structural component of the Pph21/22 phosphatase complex that is regulated by Tap42p and that acts downstream of TORC1 (Crespo and Hall, 2002). In this regard, Tap42p has recently been shown to interact directly with TORC1 within a detergent-resistant membrane fraction that is likely to correspond to TOR-DRMs, we have described here (Yan et al., 2006). Alternatively, cofractionation could be indicative of a shared biochemical environment that exists in the absence of direct physical interactions. In this regard, we have been unable to observe any significant association between Tor1p and Sla2p, as monitored by coimmunoprecipitation experiments (our unpublished data). Given the significant number of SSL interactions detected between TORC1 and components identified within TOR-DRMs, however, we suggest there are nevertheless significant functional links between proteins that cofractionate within TOR-DRMs.
Results of our genetic analyses, in combination with database mining, affirm the view that TORC1 affects a wide spectrum of cellular functions (Figure 7). This conclusion is further supported by our cell fractionation data that TORC1 may interact with a number of intracellular membrane compartments in addition to the plasma membrane (Figure 9). This latter conclusion is consistent with a recent report that yeast TORC1 is regulated by Pmr1p, a Golgi-localized Ca++/Mn++ ATPase (Devasahayam et al., 2006), as well as recent findings that mTOR interacts with both the ER as well as Golgi, in part due to the presence of specific sequences within N-terminal HEAT repeats (Liu and Zheng, 2007). Moreover, the utility of meta-analysis to extend our genetic interactions to construct an interaction network (Figure 7D) was demonstrated by the discovery of novel SSL interactions between TOR1 and components involved in intracellular trafficking (Figure 8). The interaction network we have described is based on a combination of SSL interactions as well as hypersensitivity of mutant strains to rapamycin (Xie et al., 2005). Thus, genes identified through this analysis may either represent components that function within the same pathway as TORC1, or, alternatively, within pathways that act in parallel to TORC1.
Previous studies have demonstrated that inhibition of TORC1 with rapamycin alters the intracellular trafficking of distinct nutrient-regulated amino acid permeases, including the tryptophan permease Tat2p and the general amino acid permease Gap1p, by altering their sorting at the trans-Golgi to the plasma membrane versus the vacuole (Schmidt et al., 1998; Chen and Kaiser, 2003). As these alterations mimic the physiological consequences of changes in amino acid and/or nitrogen source availability for these permeases, these results suggest that TORC1 is involved in modulating protein trafficking in response to nutritional signals (Wullschleger et al., 2006). Similarly, our findings reported here that rapamycin treatment significantly delays repolarization of the actin cytoskeleton after glucose starvation suggests TORC1 signaling may also be involved in cytoskeletal rearrangements that occur in response to changes in nutrient availability. According to this view, it is possible that other effects of rapamycin described here, including actin depolarization and delayed accumulation of lucifer yellow within the vacuole, are also in part consequences of complex cellular reorganization that occurs in response to nutrient deprivation in yeast. This notion is consistent with involvement of the actin cytoskeletal network in distinct membrane and protein trafficking events, many of which are important for cellular responses to starvation, including remodeling of the cell wall, as well as vacuole-related events during specific forms of autophagy (Hamasaki et al., 2005; Levin, 2005; Reggiori et al., 2005; He and Klionsky, 2007). Moreover, this view could account for the wide range of cellular processes that are coupled to both TORC1 and actin, as identified by our meta-analysis (Figure 7). Interestingly, TORC1 and components that regulate both the actin cytoskeleton as well as endocytosis are also linked in terms of the cell integrity response pathway, whereby plasma membrane and/or cell wall stability is modulated in response to thermal or osmotic stress (Torres et al., 2002; Reinke et al., 2004; Levin, 2005). These findings are consistent with proposals that there may be a common “regulatory architecture” whereby cells interpret and respond to nutritional as well as other environmental stresses (Levin, 2005). Our findings reported here underscore the important relationship between TORC1 and actin within this architecture.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Schekman for α-Pma1p and α-Chs3p antibodies, P. Walter for α-Sec61p antibodies and for plasmid pGAL-STE3-HA, and H. Pelham for plasmid pRS416-TLG1-MYC. We thank M. Madonna for establishing conditions for use of sorbitol overlay gradients and for generating preliminary results by using this assay. We thank K. Kaplan and the Kaplan laboratory for assistance with microscopy. This work was sponsored by National Science Foundation grant MCB-1031221 and American Cancer Society Research Scholar grant RSG-04-075-01-TBE (to T.P.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-03-0274) on May 16, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Araki T., Uesono Y., Oguchi T., Toh E. A. LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet. Syst. 2005;80:325–343. doi: 10.1266/ggs.80.325. [DOI] [PubMed] [Google Scholar]
- Audhya A., Loewith R., Parsons A. B., Gao L., Tabuchi M., Zhou H., Boone C., Hall M. N., Emr S. D. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 2004;23:3747–3757. doi: 10.1038/sj.emboj.7600384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M., Keranen S., Shevchenko A., Shevchenko A., Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berven L. A., Willard F. S., Crouch M. F. Role of the p70(S6K) pathway in regulating the actin cytoskeleton and cell migration. Exp. Cell Res. 2004;296:183–195. doi: 10.1016/j.yexcr.2003.12.032. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Cardenas M., Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C., Altman R., Schieltz D., Yates J., Kellogg D. R. The septins are required for the mitosis-specific activation of the Gin4 kinase. J. Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. J., Kaiser C. A. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 2003;161:1–15. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. C., Powers T. Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr. Genet. 2006:1–13. doi: 10.1007/s00294-005-0055-9. [DOI] [PubMed] [Google Scholar]
- Chen L., Davis N. G. Ubiquitin-independent entry into the yeast recycling pathway. Traffic. 2002;3:110–123. doi: 10.1034/j.1600-0854.2002.030204.x. [DOI] [PubMed] [Google Scholar]
- Cleves A. E., McGee T. P., Whitters E. A., Champion K. M., Aitken J. R., Dowhan W., Goebl M., Bankaitis V. A. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo J. L., Hall M. N. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2002;66:579–591. doi: 10.1128/MMBR.66.4.579-591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHart A.K.A., Schnell J. D., Allen D. A., Tsai J.-Y., Hicke L. Receptor internalization in yeast requires the Tor2-Rho1 signaling pathway. Mol. Biol. Cell. 2003;14:4676–4684. doi: 10.1091/mbc.E03-05-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devasahayam G., Ritz D., Helliwell S. B., Burke D. J., Sturgill T. W. Pmr1, a Golgi Ca2+/Mn2+-ATPase, is a regulator of the target of rapamycin (TOR) signaling pathway in yeast. Proc. Natl. Acad. Sci. USA. 2006;103:17840–17845. doi: 10.1073/pnas.0604303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como C. J., Arndt K. T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- Dulic V., Egerton M., Elguindi I., Raths S., Singer B., Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- Düvel K., Santhanam A., Garrett S., Schneper L., Broach J. R. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell. 2003;11:1467–1478. doi: 10.1016/s1097-2765(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Guertin D. A., Sabatini D. M. An expanding role for mTOR in cancer. Trends Mol. Med. 2005;11:353–361. doi: 10.1016/j.molmed.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hamasaki M., Noda T., Baba M., Ohsumi Y. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 2005;6:56–65. doi: 10.1111/j.1600-0854.2004.00245.x. [DOI] [PubMed] [Google Scholar]
- Harsay E., Bretscher A. Parallel secretory pathways to the cell surface in yeast. J. Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E., Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J. Cell Biol. 2002;156:271–285. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Klionsky D. J. Atg9 trafficking in autophagy-related pathways. Autophagy. 2007;3:271–274. doi: 10.4161/auto.3912. [DOI] [PubMed] [Google Scholar]
- Helliwell S. B., Howald I., Barbet N., Hall M. N. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics. 1998;148:99–112. doi: 10.1093/genetics/148.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell S. B., Wagner P., Kunz J., Deuter-Reinhard M., Henriquez R., Hall M. N. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell. 1994;5:105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig K. M., Colombani J., Neufeld T. P. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J. Cell Biol. 2006;173:963–974. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:681–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jacinto E., Guo B., Arndt K. T., Schmelzle T., Hall M. N. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- Jacinto E., Loewith R., Schmidt A., Lin S., Ruegg M. A., Hall A., Hall M. N. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Broach J. R. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J. R., Schneper L., Broach J. R., Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Fujioka Y., Suzuki N. N., Inagaki F., Wullschleger S., Loewith R., Hall M. N., Ohsumi Y. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein S. D., Eder S., Oh C. S., Martin C. E., Gable K., Bacikova D., Dunn T. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:109–125. doi: 10.1128/MCB.21.1.109-125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubler E., Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J., Henriquez R., Schneider U., Deuter-Reinhard M., Movva N. R., Hall M. N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- Kunz J., Schneider U., Howald I., Schmidt A., Hall M. N. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 2000;275:37011–37020. doi: 10.1074/jbc.M007296200. [DOI] [PubMed] [Google Scholar]
- Levin D. E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Tsang C. K., Watkins M., Bertram P. G., Zheng X. F. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Liu X., Zheng X. F. Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol. Biol. Cell. 2007;18:1073–1082. doi: 10.1091/mbc.E06-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Malinska K., Malinsky J., Opekarova M., Tanner W. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 2004;117:6031–6041. doi: 10.1242/jcs.01493. [DOI] [PubMed] [Google Scholar]
- Marion R. M., Regev A., Segal E., Barash Y., Koller D., Friedman N., O'Shea E. K. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee T. P., Skinner H. B., Whitters E. A., Henry S. A., Bankaitis V. A. A phosphatidylinositol transfer protein controls the phosphatidylcholine content of yeast Golgi membranes. J. Cell Biol. 1994;124:273–287. doi: 10.1083/jcb.124.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulet J. M., Martin D. E., Loewith R., Hall M. N. Mutual antagonism of target of rapamycin and calcineurin signaling. J. Biol. Chem. 2006;281:33000–33007. doi: 10.1074/jbc.M604244200. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Adolf G., Lydall D., Seddon A. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SW15 nuclear entry. Cell. 1990;62:631–647. doi: 10.1016/0092-8674(90)90110-z. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Wong E. D., Meeusen S., Wagner J. A. Studying the behavior of mitochondria. Methods Enzymol. 2002;351:381–393. doi: 10.1016/s0076-6879(02)51859-0. [DOI] [PubMed] [Google Scholar]
- Obeid L. M., Okamoto Y., Mao C. Yeast sphingolipids: metabolism and biology. Biochim. Biophys. Acta. 2002;1585:163–171. doi: 10.1016/s1388-1981(02)00337-2. [DOI] [PubMed] [Google Scholar]
- Oh C. S., Toke D. A., Mandala S., Martin C. E. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Baker D., van Tuinen E., Schekman R. Protein transport to the vacuole and receptor-mediated endocytosis by clathrin heavy chain-deficient yeast. J. Cell Biol. 1988;106:1453–1461. doi: 10.1083/jcb.106.5.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalver E., Ojeda L., Moreno E., Lagunas R. Role of the cytoskeleton in endocytosis of the yeast maltose transporter. Yeast. 1997;13:541–549. doi: 10.1002/(SICI)1097-0061(199705)13:6<541::AID-YEA112>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Pringle J. R., Preston R. A., Adams A. E., Stearns T., Drubin D. G., Haarer B. K., Jones E. W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. J. Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Shintani T., Klionsky D. J. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A., Anderson S., McCaffery J. M., Yates J., 3rd, Aronova S., Chu S., Fairclough S., Iverson C., Wedaman K. P., Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- Rohde J. R., Cardenas M. E. Nutrient signaling through TOR kinases controls gene expression and cellular differentiation in fungi. Curr. Top. Microbiol. Immunol. 2004;279:53–72. doi: 10.1007/978-3-642-18930-2_4. [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Schmelzle T., Beck T., Martin D. E., Hall M. N. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 2004;24:338–351. doi: 10.1128/MCB.24.1.338-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T., Helliwell S. B., Hall M. N. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell. Biol. 2002;22:1329–1339. doi: 10.1128/mcb.22.5.1329-1339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Beck T., Koller A., Kunz J., Hall M. N. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Bickle M., Beck T., Hall M. N. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Kunz J., Hall M. N. TOR2 is required for organization of the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. D., Hama H., Sohrabi F., DeWald D. B., Wendland B. PtdIns(3,5)P2 is required for delivery of endocytic cargo into the multivesicular body. Traffic. 2003;4:479–490. doi: 10.1034/j.1600-0854.2003.t01-1-00106.x. [DOI] [PubMed] [Google Scholar]
- Simons K., Vaz W. L. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- Singer-Kruger B., Stenmark H., Dusterhoft A., Philippsen P., Yoo J. S., Gallwitz D., Zerial M. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J. Cell Biol. 1994;125:283–298. doi: 10.1083/jcb.125.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenova S. D., Dunn R., Adler A. S., Hicke L. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. J. Biol. Chem. 2004;279:16017–16025. doi: 10.1074/jbc.M313479200. [DOI] [PubMed] [Google Scholar]
- Tabuchi M., Audhya A., Parsons A. B., Boone C., Emr S. D. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 2006;26:5861–5875. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Torres J., Di Como C. J., Herrero E., Angeles de la Torre-Ruiz M. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 2002;277:43495–43502. doi: 10.1074/jbc.M205408200. [DOI] [PubMed] [Google Scholar]
- Uesono Y., Ashe M. P., Toh E. A. Simultaneous yet independent regulation of actin cytoskeletal organization and translation initiation by glucose in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:1544–1556. doi: 10.1091/mbc.E03-12-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Kim W., Belden W. J., Spear E. D., Barlowe C., Ng D. T. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T. C., Brickner J. H., Aguilar P. S., Bernales S., Pantoja C., Walter P. Eisosomes mark static sites of endocytosis. Nature. 2006;439:998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- Wang H., Jiang Y. The Tap42-protein phosphatase type 2A catalytic subunit complex is required for cell cycle-dependent distribution of actin in yeast. Mol. Cell. Biol. 2003;23:3116–3125. doi: 10.1128/MCB.23.9.3116-3125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedaman K. P., Reinke A., Anderson S., Yates J. I., McCaffery J. M., Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisae. Mol. Biol. Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B., McCaffery J. M., Xiao Q., Emr S. D. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J. Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesp A., Hicke L., Palecek J., Lombardi R., Aust T., Munn A. L., Riezman H. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell. 1997;8:2291–2306. doi: 10.1091/mbc.8.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xie M. W., Jin F., Hwang H., Hwang S., Anand V., Duncan M. C., Huang J. Insights into TOR function and rapamycin response: chemical genomic profiling by using a high-density cell array method. Proc. Natl. Acad. Sci. USA. 2005;102:7215–7220. doi: 10.1073/pnas.0500297102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Shen X., Jiang Y. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 2006;25:3546–3555. doi: 10.1038/sj.emboj.7601239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., Chuang J. S., Schekman R. W. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell. 1996;7:1909–1919. doi: 10.1091/mbc.7.12.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E., Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]
- Zurita-Martinez S. A., Cardenas M. E. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell. 2005;4:63–71. doi: 10.1128/EC.4.1.63-71.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.