Abstract
The need for models of dengue disease has reached a pinnacle as the transmission of this mosquito-borne virus has increased dramatically. Little is known about the mechanisms that lead to dengue fever and its more severe form, dengue hemorrhagic fever; this is owing to the fact that only humans show signs of disease. In the past 5 years, research has better identified the initial target cells of infection, and this has led to the development of models of infection in primary human cell cultures. Mouse–human chimeras, containing these target cells, have also led to progress in developing animal models. These advances should soon end the stalemate in testing antivirals and vaccine preparations that had necessarily been done in incomplete or irrelevant models.
Introduction
Dengue viruses (serotypes 1–4) are flaviviruses that cause dengue fever (DF), the most prevalent arthropod-borne viral disease of humans. Approximately 50 million cases of DF occur per year and 2.5 billion persons are at risk of infection [1]. DF is an acute, self-limited febrile illness, with myalgia, rash and some mild hemorrhages, but infection might also result in a more severe presentation, dengue hemorrhagic fever (DHF), with thrombocytopenia and capillary leakage, and might proceed to a life-threatening hypovolemic shock, dengue shock syndrome (DSS). Approximately 250,000 cases of DHF/DSS occur per year, but this number has increased substantially owing to the global growth of populations, urbanization, the spread of the main mosquito vector (Aedes aegypti) and the spread of distinct virus variants [1]. Despite the worldwide morbidity caused by dengue virus infection, neither the pathogenesis nor the transmission of these viruses is well understood. These studies have been hampered by the lack of in vitro and in vivo models of disease; only recently have some potential models been described. Therefore, what is currently known has been derived from long-term observation and indirect associations of human infections with virus transmission, host immune status, clinical presentation and vector densities. The role of sequential dengue infections (up to four infections possible, once by each serotype) or other immunological factors in increasing dengue virus pathogenicity in the same host has not been proven directly. This is an important consideration in the development of multivalent dengue vaccine preparations, which could prove dangerous to vaccinees if they induce subneutralizing levels of antibodies and enhance disease severity instead [2].
In vitro models
The pathology of tissue specimens from DF and DHF patients suggests that lymph nodes, spleen, liver and bone marrow are involved or affected by dengue virus replication, whereas studies of live patients showed that a variety of cell types such as dendritic cells (DCs), monocytes, macrophages, lymphocytes and endothelial cells become infected at varying rates during the course of disease. Many primary cell cultures, including epithelial, endothelial and fibroblasts have been shown to support viral replication in vitro, albeit at low titers. Therefore, a range of permanent human cell lines are now used for propagation of virus strains, diagnostic assays, antiviral studies and virus receptor and replication studies. However, most of these studies have used laboratory-adapted dengue virus strains because they can infect a broad range of cell lines, grow to high titers and form plaques, which makes titrating the virus easier. Results obtained with high-passage dengue virus strains might differ from those obtained with low-passage isolates because dominant mutations that confer phenotypes that might not be physiologically relevant are acquired in vitro (i.e. plaquing phenotype or variation in glycosylation of viral envelope). The use of low-passage virus strains in these studies is therefore desirable, but can be difficult, especially with isolates directly from clinical specimens. If the viruses do not grow well or form plaques, the best measuring tests are quantitative RT-PCR of viral RNA or flow cytometry of infected cells [3]. Three studies have demonstrated that dengue virus isolates vary in their capacity to infect the same cell type in vitro, depending on virus genotype and passage history [4–6]. The first targets of dengue virus replication, after mosquito bite, were postulated to be monocytes or macrophages and numerous studies focused on this cell lineage. However, dengue virus has been shown to infect monocytes relatively poorly in comparison to DCs, including Langerhans cells and monocyte-derived DCs [7]. Primary DC cultures can be derived from blood monocytes in vitro, by bead purification and cytokine stimulation, and this system can be used to compare dengue virus replication. The in vitro replication rate of isolates has been used to extrapolate to replication rates in vivo (viral load in human blood), and therefore serves as a virulence marker. However, it is still unclear if virus levels in human blood directly correlate with severity of disease, although most prospective studies tend to show a correlation. Because vascular leakage is a hallmark of DHF, clinical features and histopathological studies have provided clues that derangement of the normal regulatory function of endothelial cells, rather than endothelial cell death, underlies the hemorrhagic diathesis of dengue. Therefore, numerous studies have focused on measuring cytokine levels in patients, and correlating these with cells infected in vitro, and the effects of virus replication and cytokines on endothelial cell permeability. Unfortunately, these studies have many technical difficulties in measuring permeability in human endothelial cell monolayers and the viruses used have had very high cell passage [8]. Primarily because of this lack of in vitro models of dengue disease, the development and characterization of dengue antivirals has been slow, with few studies using primary human cells or low passage virus strains to measure inhibition of replication in more relevant systems.
In vivo models
Numerous attempts have been made to develop animal models of DF/DHF, especially during the beginning of the 20th century. Several studies, reviewed by Simmons et al. [9] in 1931, showed that a broad range of animals, namely chickens, pigeons, other fowl, goats, dogs, pigs, lizards, guinea pigs, rabbits, rats, mice and hamsters show no signs of dengue disease, even after inoculation of high virus doses, by different routes.
Pathogenesis in primates
The genetic proximity of nonhuman primates (NHP) to humans makes it plausible that they show a similar pathogenesis of disease. The detection of dengue antibodies in sera of NHP in sylvatic or rural settings of Asia and Africa suggests that they are involved in virus transmission; it is not known whether this infection represents ‘spillover’ infection from human cycles, via mosquito, or whether a dengue virus cycle occurs naturally in these animals [10,11].
The first documented inoculation of NHP with dengue virus was done by Lavinder et al. [12] in 1914, when they injected nine rhesus macaques (Macaca mulatta) subcutaneously or intravenously with blood from dengue patients; neither fever nor other symptoms of disease appeared. Two other studies showed that if dengue virus is serially passaged from human volunteers to rhesus monkeys and back to humans by subcutaneous inoculation of serum from the viremic stage of disease, humans show all classic signs of DF but no signs were observed in the monkeys [13,14]. Dinger et al. let infected mosquitoes feed on rhesus and cynomolgus macaques (Macaca fascicularis); none of the animals showed signs of dengue. When infected monkeys were used to feed mosquitoes, at different time points, and these mosquitoes were then allowed to feed on human volunteers, none of the volunteers got sick [15]. Despite these disappointing results, several studies using different virus serotypes and doses were done during the last century, using a variety of species of the Cercopithecinae subfamily, including rhesus macaques, cynomolgus macaques, Japanese macaques (Macaca fuscata), green monkeys (Cercopithecus aethiops), patas monkeys (Erythrocebus patas), yellow baboons (Papio cynocephalus) and mangabeys (Cercocebus spp.). These studies showed that these animals are susceptible, in terms of forming a low (<100 PFU/ml) to undetectable viremia and a consistent antibody response, but none of these species showed overt signs of disease [13,16]. Several studies using a total of 45 night monkeys (Aotus spp.), 27 squirrel monkeys (Saimiri sciureus), 15 cotton-top marmosets (Saguinus oedipus), 11 white face monkeys (Cebus capucinus), four black spider monkeys (Ateles fusciceps), four Saimiri monkeys (Saimiri örstedii), three marmosets (Marikini geoffroyi), two howler monkeys (Alouatta palliata) and two red spider monkeys (Ateles geoffroyi) demonstrated that all three families of New World monkeys are susceptible to infection (viremia and antibody response), but none manifested clinically apparent or subclinically detectable disease after inoculation by peripheral routes [17–20]. Three studies have been conducted using a total of 16 chimpanzees (Pan troglodytes) and 39 white-handed gibbons (Hylobates lar) [21–23]. No clinical signs that could be definitively attributed to the virus were noted in any of these apes; furthermore, limited availability and ethical concerns make it impractical to use these endangered animals in research.
Pathogenesis in mice
The lack of a non-human, dengue-susceptible species has driven the development of a broad range of murine models. Although various murine models have been reported (Table 1), none recreate the full spectrum of dengue disease (DF and DHF/DSS). Two different approaches have been taken to develop a murine model: to induce human-like disease in immunocompetent mice and to mimic human disease in knockout mice or immunodeficient mice that are engrafted with human cells.
Table 1.
Characteristics of murine models of dengue virus infection
| Parameter | Model | |||||
|---|---|---|---|---|---|---|
| Immunocompetent mice | Immunodeficient/knockout mice | |||||
| BALB/c | A/J | AG129 | SCID-PBL | SCID cell lines | NOD/SCID-hu | |
| Availability of human cells | n/aa | n/a | n/a | ++ | +++ | + |
| Availability of mice | +++b | +++ | +++ | + | + | + |
| Relevance to dengue infection | + | ++ | ++ | ++ | + | +++ |
| Evaluation of innate system | + | + | +++ | + | + | +++ |
| Evaluation of vaccine/antiviral | + | + | ++ | ++ | ++ | +++c |
| References | [26] | [26,27] | [28] | [30] | [31–34] | [43] |
| Supplier | A | A | A | Ad | Ad | Bd |
Suppliers: A: Jackson laboratory, http://www.jax.org/and B: Taconic, http://www.taconic.com/.
n/a, not applicable.
+, poor; ++, moderate and +++, good.
If full engraftment with T cells is achieved.
Transplants have to be done separately.
Immunocompetent mouse models
Although many studies suggest that laboratory mouse strains are permissive to dengue virus infection and replication, it has been shown repeatedly that there are no overt signs of disease and wild type viruses replicate to such low titers in mouse tissues that they are barely detectable. To overcome this barrier, researchers have used either very high doses of virus (8 log10 PFU), mouse-adapted dengue virus strains or an intracranial route of inoculation. However, high virus doses and several modes of infection, such as intracranial, intraperitoneal or intravenous are physiologically irrelevant to what is seen in nature, during which each mosquito might inject between 4.0 and 5.4 log10 PFU into the human dermis. In addition, early studies by Sabin [24,25] revealed that virulent, wild type dengue viruses produced no disease upon inoculation of human volunteers after seven or more intracranial passages in mice, and are therefore significantly attenuated. Thus, it is assumed that the mouse-adaptation of dengue virus strains results in a loss of human pathogenic properties and that the use of these strains might not reflect natural events.
The most promising model in immunocompetent mice is the A/J mouse strain; it presents with some thrombocytopenia, as seen in humans, and is more sensitive to dengue infection than BALB/c or B6 mice [26]. Intravenous injection of A/J mice with non-mouse-adapted dengue serotype 2 virus induced paraplegia in a subset (55%) of mice on day 8–14 p.i. and a transient thrombocytopenia on days 10–13 p.i.; increased hematocrit and decreased white blood cell counts were noted with onset of paralysis (not a DF/DHF sign in humans). Antiplatelet antibody was also generated, and increased numbers of splenic natural killer (NK) cells and B cells could be demonstrated after infection [26,27]. However, these mice had to be inoculated with a large quantity of virus (8 log10 PFU per mouse) intravenously, to induce signs; viremia was low and transient, and could only be detected on day 2 p.i. with a sensitive RT-PCR.
Immunocompromised mouse models
The knockout AG129 mouse strain lacks interferon (IFN)-α/β and IFN-γ receptor genes; this model was proposed because IFN-deficient mice show an increased susceptibility to virus infection and there is evidence that α, β and γ interferons might be involved in human dengue pathogenesis. Intraperitoneally administered, mouse-adapted, serotype 2 virus (6 log10 PFU) was uniformly lethal in these mice; hind-leg paralysis and blindness occurred at day 7 p.i. and they died on day 12 [28], but only when a mouse-adapted virus strain was used. Virus levels in blood peaked to 1000 PFU/ml on day 3 and dropped sharply thereafter; low levels persisted until day 9. Further studies have demonstrated that IFN-α/β and IFN-γ receptors have crucial nonoverlapping functions in resolving primary dengue infection in this model, and it might be useful for studying the immunopathogenesis of dengue [27].
Severe combined immunodeficient (SCID) mice transplanted with human continuous cell lines, primary cells or organized tissues have been used to study the replication of other viruses, which lack a suitable small animal model [29]. SCID mice, which lack humoral and cellular immunity owing to the absence of mature T or B cells, can support dengue-susceptible human cell line xenografts. SCID mice reconstituted with human peripheral blood lymphocytes (hu-PBL-SCID) have been widely used for studies of HIV pathogenesis and were first evaluated as a dengue model in 1995 [30]. SCID mice were injected intraperitoneally with human PBL, and after successful engraftment, were infected intraperitoneally with 4.9–5.9 log10 PFU of dengue serotype 1 virus. Unfortunately, none of the animals showed any kind of clinical signs and only 5 of 82 animals showed virus replication in tissues, with low viremias (<5 PFU/ml). It was suggested that the main reason for the low infection rate was a scanty number of appropriate human target cells in the reconstituted mice; low virus titers can also be attributed to the fact that the infection rate of lymphocytes is low compared to monocytes and DCs, as PBL contain mostly lymphocytes. Subsequently, SCID mice were engrafted intraperitoneally with K562 cells, an erythroleukemia cell line [31]. After injection of dengue serotype 2 virus into the peritoneal tumor masses, these mice showed paralysis and died 2 weeks p.i.; virus was detected in the tumor masses, peripheral blood and brain. In another model, HepG2 cells from a human hepatocarcinoma cell line were transplanted into SCID mice [32]. When these mice were infected intraperitoneally with serotype 2 virus, high virus titers were detected in serum, liver and brain when paralysis appeared. A similar approach was used in SCID mice transplanted with Huh-7 cells, a hepatoma cell line [33]. After injection of serotype 4 virus into the peritoneal tumor masses, mice developed viremia of up to 8 log10 PFU/ml on days 6–7 p.i. and virus was detected in brain, liver and tumor homogenates. All of the models using SCID mice transplanted with human tumor cells allow the evaluation of virus replication/attenuation in vivo and are therefore useful for vaccine studies [34]. However, it remains questionable whether tumor cells mimic the different susceptible tissues in the human body and if differentiation-related factors play a role in virus replication [35,36].
The NOD/SCID mouse
In contrast to the severe deficiency in their adaptive immune system, SCID mice have a normal to enhanced innate immune system (especially natural killer cells) that hinders the engraftment of hematopoietic tissues; the engraftment levels in these mice are extremely low and variable [37]. Therefore, the SCID mutation was back-crossed onto a non-obese diabetic (NOD) strain background; the NOD strain has defects in NK cell function, antigen-presenting cell development and function, and genetically lacks C5, resulting in a deficiency in hemolytic complement [38]. The resulting NOD/SCID strain combines the defects in innate and adaptive immune system and can therefore be reconstituted at high levels with a broad variety of human cells and tissues, such as bone marrow, PBL, skin, erythrocytes and umbilical cord blood [39]. Since the cellular targets of dengue virus were postulated to be monocytes or macrophages, numerous studies have focused on viral replication in cells of this lineage [40]. However, dengue viruses were subsequently shown to infect and replicate comparatively poorly in primary monocytes, and recent reports have indicated that immature DCs, such as Langerhans cells in the skin, are the major replication site after mosquito bite [7]. A mouse model reconstituted with high levels of human DCs was therefore desirable. Recent reports have demonstrated that NOD/SCID mice reconstituted with human CD34+ hematopoietic progenitor cells (Fig. 1) have high levels of human CD45+ (a panleukocyte marker) cells in a broad variety of tissues such as bone marrow, spleen, skin, lungs, liver and heart, and sometimes in gut, kidney and pancreas [41,42]. High levels of DC reconstitution and development were obtained in multiple organs but mainly in spleen, bone marrow and blood (Fig. 2). It was also shown that these mice develop a full repertoire of human DCs, up to 24 weeks after transplant, indicating ongoing and sustained production of DCs after initial engraftment. Moreover, systemic production of human cytokines was induced after mice were injected with different pathogens, indicating DCs were functional [41]. We have inoculated reconstituted NOD/SCID mice subcutaneously with a low dose (approximately 4.7 log10 PFU) of a low passage dengue serotype 2 strain, thus mimicking mosquito transmission. These mice developed clinical signs of DF such as fever, thrombocytopenia and erythema as seen in humans [43]. Viremia levels peaked on day 2–6 p.i. and were undetectable by day 18. In addition, viral positive strand RNA was detected by quantitative RT-PCR in spleen, skin and liver; negative strand RNA, as a surrogate of viral replication, was found in spleen and skin. Furthermore, body weight decreased dramatically (up to 20%) in some of the mice, probably due to fever and lethargy. Other findings include inflammatory cells and dengue antigen in liver and spleen, liver enzyme elevation and increased fibrinolysis. The lack of murine immune cells in this model allows us to study solely the immunological functions of the human engrafted cells, factors or tissues, without the confounding effects of the murine system. It is therefore a valuable model to study pathogenesis and test antidengue products.
Figure 1.
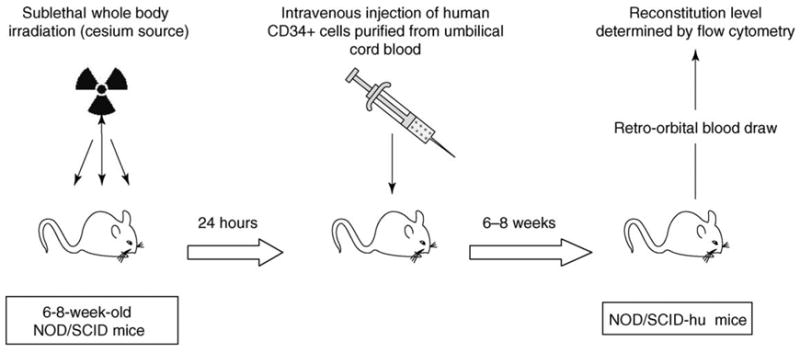
Reconstitution of NOD/SCID mice with human hematopoietic progenitor cells. Irradiated NOD/SCID mice are injected with CD34+ cells purified from human umbilical cord blood. Six to 8 weeks later reconstitution levels of human CD45+ cells (panleukocyte marker) and CD11c+/CD123+ cells (DC marker) are determined by flow cytometry. Details are provided in Refs [41,42].
Figure 2.
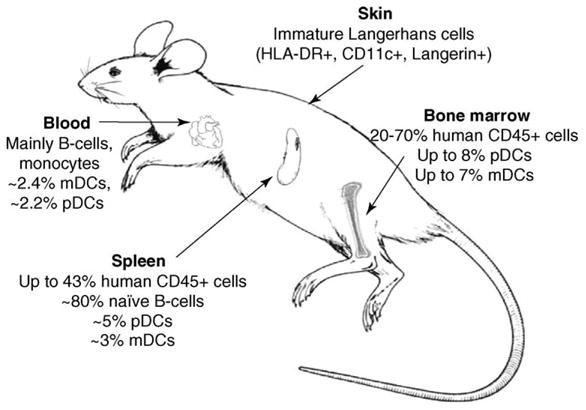
Levels of human cells in reconstituted NOD/SCID mice. NOD/SCID mice reconstituted with CD34+ cells develop functional subsets of human dendritic cells in a variety of tissues. They have high percentages of human plasmacytoid dendritic cells (pDCs) and myeloid dendritic cells (mDCs) in bone marrow, spleen and blood. Immature Langerhans cells can be detected in the skin. Details are provided in Refs [41,42].
In silico models
The evolutionary and epidemiological relationships of dengue viruses have been studied with computer algorithms that make estimations based on nucleotide or amino acid sequences. These models have shown that specific dengue virus genetic variants or genotypes have established transmission cycles on other continents, are responsible for outbreaks of more severe disease (DHF/DSS) and have helped track the displacement of one genotype by another, more virulent genotype. However, some of these algorithms have also been used to extrapolate on the occurrence of intramolecular recombination in dengue RNA genomes [44], the ancient origin of epidemic strains [45] and the movement of genotypes within a country or from one country to another [46], without the support of molecular studies of viruses or sufficient field sampling and epidemiologic studies of the transmission cycles. Therefore, it is unclear whether these phenomena actually occur in nature. These reservations also apply to the study of the effects of antibody-dependent enhancement of dengue virus infection of monocytes in the host (demonstrated only in vitro), especially when estimating its long-range effects on virus epidemiology and population dynamics [47]. Other computer models of dengue virus infection have included clinical applications, such as distinguishing dengue from other fevers, based on logistic regression analysis of clinical and laboratory features [48], and measuring pathophysiological parameters of severe dengue (DHF/DSS) and risk in DF patients, using neural network programs to predict outcome, and to determine patient management or treatment [49]. It remains to be seen whether these models can explain or predict underlying pathological processes that are still mysteries; this further highlights the need for animal models of disease.
Conclusions
Understanding dengue virus pathogenesis has been extremely difficult because only humans seem to develop this disease; what little is known to date has been derived from years of painstaking, indirect, observational studies. Only recently have potential in vitro markers of severity of disease been reported, but these tests use primary human cells and low passage virus isolates that are difficult to grow and measure because of implicit variability. Also, because no other animal besides humans shows signs of disease, the latest developments in animal models have tended toward making human–mouse chimeras that show signs of dengue fever. So far these chimeras have only the innate arm of the human immune system functioning but new mouse strains and transplant techniques have the promise of creating a mouse with a functioning human adaptive immune response. It is this area of research that has the most potential for explosive advances, as these chimeras could very soon replace the inadequate in vivo models available now. Thus, we predict that soon we might overcome the barriers to dengue research that have plagued the field for the past 60 years.
Acknowledgments
We thank D. Jones for library assistance. Funding was from the National Institutes of Health (grant AI50123).
Footnotes
Section Editors:
Enitan Carrol – Malawi–Liverpool–Wellcome Trust Clinical Research, Blantyre and Liverpool, Malawi and UK Andrew Riordan – Birmingham Heartlands Hospital, Birmingham, UK
References
- 1.World Health Organization. Dengue/dengue hemorrhagic fever. Fact Sheet no. 117. 2002 http://www.who.int/mediacentre/factsheets/fs117/en/
- 2.Halstead SB, et al. The future of dengue vaccines. Lancet. 2002;360:1243–1245. doi: 10.1016/S0140-6736(02)11276-1. [DOI] [PubMed] [Google Scholar]
- 3.Lambeth CR, et al. Flow cytometry-based assay for titrating dengue virus. J Clin Microbiol. 2005;43:3267–3272. doi: 10.1128/JCM.43.7.3267-3272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cologna R, et al. Selection for virulent dengue viruses occurs in humans and mosquitoes. J Virol. 2005;79:853–859. doi: 10.1128/JVI.79.2.853-859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cologna R, et al. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J Virol. 2003;77:3929–3938. doi: 10.1128/JVI.77.7.3929-3938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamond MS, et al. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000;74:4957–4966. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marovich M, et al. Human dendritic cells as targets of dengue virus infection. J Investig Dermatol Symp Proc. 2001;6:219–224. doi: 10.1046/j.0022-202x.2001.00037.x. [DOI] [PubMed] [Google Scholar]
- 8.Dewi BE, et al. In vitro assessment of human endothelial cell permeability: effects of inflammatory cytokines and dengue virus infection. J Virol Methods. 2004;121:171–180. doi: 10.1016/j.jviromet.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Simmons JS, et al. Experimental studies on dengue. Phillippine J Sci. 1931;44:189–248. [Google Scholar]
- 10.Rudnick A, et al. Possible jungle dengue – recent studies and hypotheses. Jpn J Med Sci Biol. 1967;20:69–74. [PubMed] [Google Scholar]
- 11.Kilbourn AM, et al. Health evaluation of free-ranging and semi-captive orangutans (Pongo pygmaeus pygmaeus) in Sabah, Malaysia. J Wildl Dis. 2003;39:73–83. doi: 10.7589/0090-3558-39.1.73. [DOI] [PubMed] [Google Scholar]
- 12.Lavinder CH, et al. The etiology of dengue. J Infect Dis. 1914;15:341–346. [Google Scholar]
- 13.Blanc G, et al. Recherches experimentales sur la sensibilite des singes inferieurs au virus de la Dengue. Acad Dermatol Sci. 1929;188:468–470. [Google Scholar]
- 14.Findlay GM. The relation between dengue and rift valley fever. Trans R Soc Trop Med Hyg. 1932;26:157–160. [Google Scholar]
- 15.Dinger JE, et al. Dengue und Gelbfieber. Arch Schiffs Trop Hyg. 1931;35:497–526. [Google Scholar]
- 16.Halstead SB, et al. Studies on the pathogenesis of dengue infection in monkeys. I Clinical laboratory responses to primary infection. J Infect Dis. 1973;128:7–14. doi: 10.1093/infdis/128.1.7. [DOI] [PubMed] [Google Scholar]
- 17.Rosen L. Experimental infection of New World monkeys with dengue and yellow fever viruses. Am J Trop Med Hyg. 1958;7:406–410. doi: 10.4269/ajtmh.1958.7.406. [DOI] [PubMed] [Google Scholar]
- 18.Scherer WF, et al. Cross-protection studies and search for subclinical disease in new world monkeys infected sequentially with different immunologic types of dengue viruses. Am J Epidemiol. 1972;95:67–79. doi: 10.1093/oxfordjournals.aje.a121372. [DOI] [PubMed] [Google Scholar]
- 19.Schiavetta AM, et al. Variable susceptibility of the owl monkey (Aotus nancymae) to four serotypes of dengue virus. Contemp Top Lab Anim Sci. 2003;42:12–20. [PubMed] [Google Scholar]
- 20.Kochel TJ, et al. Cross-serotype neutralization of dengue virus in Aotus nancymae monkeys. J Infect Dis. 2005;191:1000–1004. doi: 10.1086/427511. [DOI] [PubMed] [Google Scholar]
- 21.Paul JR, et al. Experimental attempts to transmit phlebotomus and dengue fever to chimpanzees. Proc Soc Exp Biol. 1948;68:193–198. doi: 10.3181/00379727-68-16431. [DOI] [PubMed] [Google Scholar]
- 22.Scherer WF, et al. Experimental infection of chimpanzees with dengue viruses. Am J Trop Med Hyg. 1978;27:590–599. doi: 10.4269/ajtmh.1978.27.590. [DOI] [PubMed] [Google Scholar]
- 23.Whitehead RH, et al. Sequential dengue virus infections in the white-handed gibbon (Hylobates lar) Am J Trop Med Hyg. 1970;19:94–102. doi: 10.4269/ajtmh.1970.19.94. [DOI] [PubMed] [Google Scholar]
- 24.Sabin AB. Recent advances in our knowledge of dengue and sandfly fever. Am J Trop Med Hyg. 1955;4:198–207. doi: 10.4269/ajtmh.1955.4.198. [DOI] [PubMed] [Google Scholar]
- 25.Sabin AB, et al. Production of immunity to dengue with virus modified by propagation in mice. Science. 1945;101:640–642. doi: 10.1126/science.101.2634.640. [DOI] [PubMed] [Google Scholar]
- 26.Huang KJ, et al. Manifestation of thrombocytopenia in dengue-2-virus-infected mice. J Gen Virol. 2000;81 (Pt 9):2177–2182. doi: 10.1099/0022-1317-81-9-2177. [DOI] [PubMed] [Google Scholar]
- 27.Shresta S, et al. Early activation of natural killer and B cells in response to primary dengue virus infection in A/J mice. Virology. 2004;319:262–273. doi: 10.1016/j.virol.2003.09.048. [DOI] [PubMed] [Google Scholar]
- 28.Johnson AJ, et al. New mouse model for dengue virus vaccine testing. J Virol. 1999;73:783–786. doi: 10.1128/jvi.73.1.783-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosier DE. Human xenograft models for virus infection. Virology. 2000;271:215–219. doi: 10.1006/viro.2000.0336. [DOI] [PubMed] [Google Scholar]
- 30.Wu SJ, et al. Evaluation of the severe combined immunodeficient (SCID) mouse as an animal model for dengue viral infection. Am J Trop Med Hyg. 1995;52:468–476. doi: 10.4269/ajtmh.1995.52.468. [DOI] [PubMed] [Google Scholar]
- 31.Lin YL, et al. Study of dengue virus infection in SCID mice engrafted with human K562 cells. J Virol. 1998;72:9729–9737. doi: 10.1128/jvi.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.An J, et al. Development of a novel mouse model for dengue virus infection. Virology. 1999;263:70–77. doi: 10.1006/viro.1999.9887. [DOI] [PubMed] [Google Scholar]
- 33.Blaney JE, Jr, et al. Genetic basis of attenuation of dengue virus type 4 small plaque mutants with restricted replication in suckling mice and in SCID mice transplanted with human liver cells. Virology. 2002;300:125–139. doi: 10.1006/viro.2002.1528. [DOI] [PubMed] [Google Scholar]
- 34.Blaney JE, Jr, et al. Recombinant, live-attenuated tetravalent dengue virus vaccine formulations induce a balanced, broad, and protective neutralizing antibody response against each of the four serotypes in rhesus monkeys. J Virol. 2005;79:5516–5528. doi: 10.1128/JVI.79.9.5516-5528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin YL, et al. Infection of five human liver cell lines by dengue-2 virus. J Med Virol. 2000;60:425–431. doi: 10.1002/(sici)1096-9071(200004)60:4<425::aid-jmv10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 36.Phoolcharoen W, et al. Internalization of the dengue virus is cell cycle modulated in HepG2, but not Vero cells. J Med Virol. 2004;74:434–441. doi: 10.1002/jmv.20195. [DOI] [PubMed] [Google Scholar]
- 37.Greiner DL, et al. SCID mouse models of human stem cell engraftment. Stem Cells. 1998;16:166–177. doi: 10.1002/stem.160166. [DOI] [PubMed] [Google Scholar]
- 38.Serreze DV, et al. NOD marrow stem cells adoptively transfer diabetes to resistant (NOD × NON)F1 mice. Diabetes. 1988;37:252–255. doi: 10.2337/diab.37.2.252. [DOI] [PubMed] [Google Scholar]
- 39.Meyerrose TE, et al. Immune-deficient mouse models for analysis of human stem cells. Biotechniques. 2003;35:1262–1272. doi: 10.2144/03356ss06. [DOI] [PubMed] [Google Scholar]
- 40.Bunyaratvej A, et al. Dengue viruses induce cell proliferation and morphological changes of endothelial cells. Southeast Asian J Trop Med Public Health. 1997;28 (Suppl 3):32–37. [PubMed] [Google Scholar]
- 41.Cravens PD, et al. Development and activation of human dendritic cells in vivo in a xenograft model of human hematopoiesis. Stem Cells. 2005;23:264–278. doi: 10.1634/stemcells.2004-0116. [DOI] [PubMed] [Google Scholar]
- 42.Palucka AK, et al. Human dendritic cell subsets in NOD/SCID mice engrafted with CD34+ hematopoietic progenitors. Blood. 2003;102:3302–3310. doi: 10.1182/blood-2003-02-0384. [DOI] [PubMed] [Google Scholar]
- 43.Bente DA, et al. Dengue fever in humanized NOD/SCID mice. J Virol. 2005;79:13797–13799. doi: 10.1128/JVI.79.21.13797-13799.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Worobey M, et al. Widespread intra-serotype recombination in natural populations of dengue virus. Proc Natl Acad Sci U S A. 1999;96:7352–7357. doi: 10.1073/pnas.96.13.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang E, et al. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol. 2000;74:3227–3234. doi: 10.1128/jvi.74.7.3227-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carrington CV, et al. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J Virol. 2005;79:14680–14687. doi: 10.1128/JVI.79.23.14680-14687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cummings DA, et al. Dynamic effects of antibody-dependent enhancement on the fitness of viruses. Proc Natl Acad Sci U S A. 2005;102:15259–15264. doi: 10.1073/pnas.0507320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chadwick D, et al. Distinguishing dengue fever from other infections on the basis of simple clinical and laboratory features: application of logistic regression analysis. J Clin Virol. 2006;35:147–153. doi: 10.1016/j.jcv.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Ibrahim F, et al. A novel dengue fever (DF) and dengue haemorrhagic fever (DHF) analysis using artificial neural network (ANN) Comput Methods Programs Biomed. 2005;79:273–281. doi: 10.1016/j.cmpb.2005.04.002. [DOI] [PubMed] [Google Scholar]


