Abstract
Biofilm communities cultivated in rotating annular bioreactors using water from the South Saskatchewan River were assessed for the effects of seasonal variations and nutrient (C, N, and P) additions. Confocal laser microscopy revealed that while control biofilms were consistently dominated by bacterial biomass, the addition of nutrients shifted biofilms of summer and fall water samples to phototrophic-dominated communities. In nutrient-amended biofilms, similar patterns of nitrification, denitrification, and hexadecane mineralization rates were observed for winter and spring biofilms; fall biofilms had the highest rates of nitrification and hexadecane mineralization, and summer biofilms had the highest rates of denitrification. Very low rates of all measured activities were detected in control biofilms (without nutrient addition) regardless of season. Nutrient addition caused large increases in hexadecane mineralization and denitrification rates but only modest increases, if any, in nitrification rates, depending upon the season. Generally, both alkB and nirK were more readily PCR amplified from nutrient-amended biofilms. Both genes were amplified from all samples except for nirK from the fall control biofilm. It appears that bacterial production in the South Saskatchewan River water is limited by the availability of nutrients and that biofilm activities and composition vary with nutrient availability and time of year.
In lotic ecosystems, such as rivers and streams, microorganisms can be found in two states, as free-living planktonic cells or attached to various surfaces, where these sessile cells form biofilms (12). In the past, most investigations of the activities and compositions of microbial communities in lotic ecosystems have focused on planktonic rather than biofilm microorganisms. This was due to methodological (10) and conceptual (13) constraints. Advanced microscopic (fluorescent in situ hybridization and confocal laser scanning microscopy) (36) and molecular biological (1) techniques have revolutionized the study of complex microbial communities, such as biofilms. It is now recognized that biofilms form on most surfaces exposed to water (55) and thus are fundamental components of aquatic ecosystems. Not only are biofilm microorganisms at the base of aquatic food webs (26), but it is likely that bacteria residing within biofilms play a key role in the degradation of organic pollutants and in biogeochemical cycles (13).
Among the bacterial activities that have significant implications for ecosystem health and stability, the nitrogen cycle is recognized as being of fundamental importance (54). Nitrification and denitrification are complementary and essential steps in the global nitrogen cycle that are dependent on bacteria. Nitrification is the oxidation of ammonium (NH4+) to nitrite (NO2−) and then to nitrate (NO3−). Oxidation of NH4+ or NO2− provides the only source of energy for nitrifying bacteria (46) and thus is essential to their survival. Nitrification is performed by a limited number of bacterial species (27) and can be severely affected by environmental stress. Denitrification is the consecutive reduction of nitrate (NO3−), to nitrite (NO2−), nitric oxide (NO), nitrous oxide (N2O), and finally, dinitrogen gas (N2). Denitrification has a significant role in the oxidation of organic compounds in anaerobic habitats, as this process yields more energy than fermentation processes (10).
The organic matter utilized can include organic pollutants, such as those arising from industrial activities. Hexadecane (C16H34) is a model compound representing the aliphatic hydrocarbons found in crude oil (4) and is a major component of diesel fuel (39). Petroleum hydrocarbons such as hexadecane may find their way into rivers as a result of refining (24), leakage from storage tanks and distribution systems (37), or accidental spills (52). As a consequence, the ability to catabolize hexadecane is widespread among bacteria (11).
The activities of biofilm bacteria may be affected by natural variations and anthropogenic sources. In lotic ecosystems, environmental conditions, such as temperature and nutrients, vary considerably with the season (26). Industrial, municipal, and agricultural activities introduce nutrients, such as organic carbon, inorganic nitrogen, and phosphate, into rivers (14). This can lead to eutrophication and alteration of aquatic species distribution and diversity (53). Despite the importance of these ecosystems, little is known about lotic biofilm communities or the effects of changing natural and anthropogenic conditions on them.
The objectives of the present study were to assess the impacts of seasonal variations and nutrient inputs on the physiological activities and genetic compositions of replicated river biofilms cultivated in rotating annular reactors (mesocosms). The activities and compositions of biofilms were evaluated by using a combination of physiological and molecular biology techniques, testing for the effects of changing conditions upon hexadecane mineralization, nitrification, and denitrification.
MATERIALS AND METHODS
Biofilm cultivation.
Raw water samples were taken from the South Saskatchewan River (Saskatoon, Saskatchewan, Canada) at different seasons throughout a year, as previously described (40), and were used as inocula for the experiments. General water analyses were carried out according to procedures described previously (16). Table 1 summarizes the chemistry of the feed waters for the bioreactors. Biofilms were grown in rotating annular bioreactors on polycarbonate strips, as described in detail previously (34). In brief, the reactors were constructed using a rotating inner solid polycarbonate cylinder machined to allow the insertion of 12 removable polycarbonate slides; the outer cylinder was a commercially available 1-liter glass jar with a threaded mouth. Mixing was established in the reactor through the rotation of the inner cylinder and the circulation of water through the reactor from top to bottom. The experimental setup included a 10-liter reservoir for the raw water, a peristaltic pump for recirculating the water, and the rotating annular reactor with a motor and a control for adjusting the speed of the rotating inner cylinder. The reactor was operated at 132 rpm (surface velocity, 1.6 km h−1). The reactor volume was 500 ml, and flow through the reactors was 1 liter day−1. The water in the reservoir was replaced with fresh river water every 7 days.
TABLE 1.
Results of analysis of South Saskatchewan River waters
| Season | Dates of growth | Tempa (°C) | pH | Turbidity (NTUb) | DOCc (mg liter−1) | TKNd (mg liter−1) | NH4+ (mg liter −1) | Inorganic Ne (mg liter−1) | PO43− (mg liter−1) | HPCf (CFU ml−1) | Chl ag (mg liter−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spring | 5 Apr-28 May 2001 | 11 ± 5 (3-16) | 8.45 | 4.36 | 3.2 | 0.25 | 0.08 | 0.03 | <0.05 | 905 | 3.7 |
| Summer | 19 May-7 Jul 1999 | 17 ± 3 (12-22) | 8.24 | 4.9 | 3.7 | 0.50 | 0.10 | 0.54 | <0.05 | 245 | 2.1 |
| Fall | 10 Nov 1999-3 Jan 2000 | 0.7 ± 1.1 (4-0) | 8.56 | 3.4 | 3.6 | 0.86 | 0.02 | 0.12 | 0.06 | 970 | 1.7 |
| Winter | 11 Jan-4 Mar 2001 | 0.3 ± 0.1 (0.2-0.6) | 8.37 | 0.6 | 3.1 | 0.27 | 0.05 | 0.09 | <0.05 | 84 | 0.8 |
Temp, in situ temperature (±1 standard deviation; values in parentheses are minima and maxima measured).
NTU, nephelometric turbidity units.
DOC, dissolved organic carbon.
TKN, total Kjeldahl nitrogen.
Inorganic N, sum of NO2− and NO3−.
HPC, heterotrophic plate count.
Chl a, chlorophyll a.
The biofilms were grown for 8 weeks on the dates shown in Table 1. For each season, three replicate bioreactors were operated with river water alone (no nutrient addition), and three replicate bioreactors were operated with the addition of the following nutrients (final concentrations): glucose at 67 μM carbon, ammonium chloride at 80 μM nitrogen, and potassium phosphate monobasic at 5 μM phosphorus (38). The experiments carried out in this study are most relevant to mixed phototrophic-heterotrophic biofilms on rocks, plants, and river bed surfaces in rivers but do not address those events occurring within the sediments.
Microcosm preparation.
After the 8-week period of growth in the bioreactors, fresh biofilm samples on polycarbonate strips were used to assess the impact of nutrient status and seasonal variations on hexadecane mineralization, nitrification, and denitrification in microcosms. To prepare the microcosms, the polycarbonate strips were aseptically cut (2 cm2), and each piece with its associated biofilm was transferred to a 27-ml crimpable glass vial with 5 ml of the same river water supplemented with the same nutrient concentrations used in the bioreactors. All microcosms were incubated statically, in the dark, at room temperature under the conditions described below. The polycarbonate strips containing biofilm were also frozen immediately at −80°C for subsequent molecular analyses.
Hexadecane mineralization.
To monitor hexadecane mineralization, nutrient-amended and unamended microcosms received a 1-ml glass tube containing 0.5 ml of 1 N KOH (ACS grade; BDH, Toronto, Ontario, Canada) as a 14CO2 trap. Each microcosm bottle received a mixture of radioactive [1-14C]hexadecane (specific activity, 2.2 mCi mmol−1; Sigma-Aldrich, Mississauga, Ontario, Canada) and nonradioactive hexadecane (Anachemia, Montreal, Quebec, Canada) at a final concentration of 100 mg of hexadecane liter−1 and 50,000 dpm. Incubation was aerobic. The production of 14CO2 was measured as follows: periodically, the KOH was aspirated by syringe, the tubes were rinsed with 0.5 ml of KOH, and then 0.5 ml of fresh 1 N KOH was added. The two KOH aliquots were combined, added to 10 ml of scintillation cocktail (ACS; Amersham, Oakville, Ontario, Canada), and counted by liquid scintillation spectrometry (Tri-Carb 2100TR; Packard Instruments, Downers Grove, Ill.) (22). Mineralization results are expressed as the cumulative percentage of 14CO2 produced from the [1-14C]hexadecane initially added.
Nitrification.
Following the preparation of nutrient-amended and unamended microcosms, NH4Cl (BDH) was added to microcosms without KOH traps at a final concentration of 5 mM. The microcosms were capped with 20-mm-diameter butyl stoppers (Fisher, Fair Lawn, N.J.) and remained air filled for aerobic incubation. Acetylene (C2H2; Prodair, Montreal, Quebec, Canada) was added to the headspaces of half of the microcosms at a final partial pressure of 2 kPa (48). C2H2 prevents oxidation of NH4+ via inhibition of ammonium monooxygenase (41).
Periodically, 0.75 ml of the liquid phase was withdrawn and added to 0.75 ml of autoclaved deionized water. The mixture was agitated and centrifuged for 10 min at 4°C and 16,060 × g. After centrifugation, 1 ml of sample was transferred into a high-pressure liquid chromatography (HPLC) vial, which was sealed with a Teflon-lined cap. These samples were used to measure the consumption of NH4+ during nitrification. A volume of 0.4 ml of each of the above-mentioned centrifuged samples was mixed with 0.6 ml of autoclaved deionized water (1/5 dilution) and used to measure the production of NO3− during nitrification (49). The samples were analyzed by HPLC.
Denitrification.
Microcosms prepared as described above, without a KOH trap and with or without nutrient amendment, received 5 mM NaNO3 (BDH). The microcosms were capped with 20-mm-diameter rubber stoppers (Bellco Glass Inc., Vineland, N.J.), and their atmospheres were made anaerobic by evacuating (5 min each) and refilling them to 1 atmosphere three times with N2 (prepurified [99.998%]; Praxair, Mississauga, Ontario, Canada) in order to eliminate dissolved gases in the water. C2H2 was added to the headspaces of half of the microcosms at a final partial pressure of 2 kPa (48). C2H2 prevents the reduction of N2O to N2 via inhibition of N2O reductase (56). Denitrification was measured by determination of N2O production in the presence of C2H2. Periodically, gas samples (0.2 ml) were withdrawn from the headspaces of the microcosms and analyzed for N2O on an 8610C gas chromatograph (SRI, Torrance, Calif.) with a thermal conductivity detector and an electron capture detector in parallel, as previously described (47). The N2O concentration (parts per million per volume [ppmv], milliliters of NO2 per milliliter of headspace] was calculated based on a standard curve (47).
HPLC.
Ions were analyzed by HPLC (model SP8800; Spectra-Physics) with ion detection using a conductivity detector (model 431; Waters-Millipore). For cations (NH4+), 20 μl of a 1/2 dilution of the liquid phase of the microcosms was injected into the HPLC system through a Hamilton (Reno, Nev.) column (PRP-X200; 250 by 41 mm; stainless steel) packed with 10-μm-diameter spherical poly(styrene-divinylbenzene)sulfonate at 40°C with 30% methanol in 4 mM nitric acid at 2 ml minute−1 as the mobile phase. For anions (NO3−), 100 μl of a 1/5 dilution of the liquid phase was injected into the HPLC system through a Hamilton column (PRP-X100; 250 by 41 mm; stainless steel) packed with 10-μm-diameter spherical poly(styrenedivinylbenzene)trimethylammonium) at 40°C with 2.5% methanol in 4 mM para-hydroxybenzoic acid (adjusted to pH 8.5 with NaOH) as the mobile phase (A. Corriveau, personal communication).
Total community DNA extraction.
For each bioreactor, a frozen polycarbonate strip was aseptically cut (2 cm2) and transferred into a 50-ml polypropylene tube (Falcon; Becton Dickinson, Franklin Lanes, N.J.). Cells from the frozen biofilm samples were lysed by enzymatic treatment as performed by Fortin et al. (20), except that the volumes of reagents used were five times smaller, agitation during lysozyme treatment was at 200 rpm, and proteinase K was added to a final concentration of 200 μg ml−1. DNA extraction was performed by phenol-chloroform treatment as described by Ausubel et al. (5), except that incubation after cetyltrimethylammonium bromide addition was for 30 min. The DNA was precipitated overnight at −20°C with an equal volume of cold 100% isopropanol and then centrifuged for 30 min at 4°C (17,400 × g). The pellets were washed three times with 2 ml of 70% cold ethanol, centrifuged for 5 min at 4°C (17,400 × g), and air dried. The DNA was resuspended in 300 μl of 10 mM Tris-0.1 mM EDTA (pH 8) with gentle agitation at room temperature for 1 h, followed by 1 h on ice. The DNA was precipitated overnight at −20°C with 1/10 volume of 3 M sodium acetate (pH 5.2) and 2.5 volumes of cold 100% ethanol. The precipitated DNA was recovered by centrifugation for 15 min at 4°C (16,060 × g), washed three times with 500 μl of 70% ethanol at room temperature, air dried, and dissolved in 30 μl of Tris-EDTA.
PCR amplification.
To assess the metabolic potential of the biofilm, extracts of the following target genes were analyzed by PCR amplification: the alkB gene, coding for alkane hydroxylase, a monooxygenase that is the first enzyme of the hexadecane mineralization pathway (32), and the nirK gene, coding for copper-containing nitrite reductase, which reduces nitrite to nitric oxide during denitrification (10).
The PCR amplification procedure was modified from previously published procedures (8, 28). An aliquot of extract (0.5 to 2 μl of undiluted or diluted [1/10 or 1/100] DNA) was added to a final volume of 50 μl of reaction mixture (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) containing 10 mM Tris-HCl (pH 9); 50 mM KCl; 1.5 mM MgCl2; 200 μM (each) dATP, dTTP, dGTP, and dCTP; and 1 μM each primer (forward and reverse). Table 2 describes the consensus primers used for amplification of the selected bacterial genes. The tubes were placed into a DNA thermal cycler (Perkin-Elmer Cetus, Montreal, Quebec, Canada) and heated to 96°C for 5 min. The temperature was then brought down to 80°C, and 2.5 U of Taq DNA polymerase (Amersham Pharmacia Biotech Inc.) in 10× PCR buffer (10 mM Tris-HCl [pH 9.0], 500 mM KCl, 15 mM MgCl2) was added. The PCR conditions were 30 cycles of 1 min at 94°C (denaturing), 1 min at 60°C for alkB or 65°C for nirK (annealing), and 1 min at 72°C (extension), followed by a final extension of 3 min at 72°C. DNA was electrophoresed in 1.4% agarose (Wisent, St-Bruno, Quebec, Canada) gels with 1× TAE buffer (50) using 2.5 μg of GeneRuler 100-bp DNA ladder (MBI Fermentas, Vilnius, Lithunia) as a molecular weight marker. The gels were stained with ethidium bromide and photographed.
TABLE 2.
Oligonucleotide primers used for PCR amplification of bacterial genes.
| Target gene | Primer | Position (length [nt]) | Consensus sequencea | Reference organism | PCR fragment size (bp) | Target bacteria (target enzyme) |
|---|---|---|---|---|---|---|
| alkB | Forward | 495-521 (27) | 5′-CIG IIC ACG AII TIG GIC ACA AGA AGG-3′ | Rhodococcus sp. strain Q15 | 549 | Hexadecane degraders (alkane hydroxylase) |
| Reverse | 1018-1044 (27) | 5′-IGC ITG ITG ATC III GTG ICG CTG IAG-3′ | ||||
| nirK | Forward | 560-589 (30) | 5′-GGG CAT GAA CGG CGC GCT CAT GGT GCT GCC-3′ | P. aureofaciens ATCC 13985 | 376 | Nitrite oxidizers (Cu-nitrite reductase) |
| Reverse | 906-935 (30) | 5′-CGG GTT GGC GAA CTT GCC GGT GGT CCA GAC-3′ |
I, inosine.
Confocal microscopy and image analyses.
The algal and bacterial biomass in river biofilms was determined by direct imaging via confocal laser microscopy in conjunction with digital image analyses as described previously (33). In brief, polycarbonate strips were cut into 1-cm2 subsamples, and autofluorescence of the algal cells was used to image the photosynthetic biomass while application of the nucleic acid stain Syto 9 (Molecular Probes Inc. Eugene, Oreg.) was used to detect and image bacterial cells in the river biofilms. The image stacks were then analyzed using semiautomated macros developed for NIH Image version 1.62 (a public domain program developed at the National Institutes of Health and available at http://rsb.info.nih.gov/nih-image/).
RESULTS
Hexadecane mineralization.
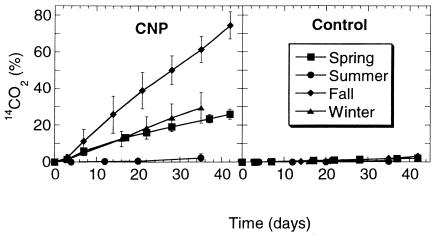
In the presence of nutrients (CNP), biofilm from river water collected in the fall showed the highest hexadecane mineralization rate, and the summer biofilm showed the lowest activity (Fig. 1). Intermediate between these two extremes were the winter and the spring biofilms. Mineralization began immediately after the addition of substrate, with the exception of summer biofilm, and no plateau was reached after 35 to 42 days of incubation. The combination of nutrients (CNP) enhanced hexadecane mineralization, since only very weak activity was observed in the absence of nutrients (control biofilms). In autoclaved microcosms, the cumulative background of 14C radioactivity found in the KOH traps at the end of the incubation period was always less than 0.4% of that initially added.
FIG. 1.
Effects of seasonal variations and nutrients on mineralization of [14C]hexadecane (100 mg liter−1; 50,000 dpm) by bioreactor biofilms. Each datum point is the average of six replicate microcosms, and the bars represent 1 standard deviation.
PCR amplification of alkB DNA was better with extracts from biofilms grown in the presence of nutrients than in their absence, implying that the hexadecane-degrading populations were more abundant when nutrients were present (Fig. 2). Globally, this correlates well with Fig. 1, in which mineralization was greater with CNP than control biofilms. With CNP biofilms, the best amplification for the alkB gene was with the summer biofilm, closely followed by the winter biofilm. Fall and, particularly, spring biofilms had lower amplification yields. With control biofilms (in the absence of nutrients), the best amplification was obtained with the winter biofilm, and the other seasonal biofilms had similar amplification yields. With the summer and fall biofilms in the presence and absence of nutrients, there was clearly PCR-inhibitory material present in the extracts, since there was no amplification using undiluted extracts.
FIG. 2.
PCR amplification of alkB from spring, summer, fall, and winter biofilms with (CNP) and without (Con) nutrient amendments. Total-community DNA extracts were undiluted (0) or diluted 10−1 (−1) or 10−2 (−2) prior to PCR amplification using the alkB consensus primers. Lanes M, molecular size markers; lane +, positive control (50 ng of Pseudomonas oleovorans DNA template); lane −, negative control (no DNA). The arrows indicate the positions of the 549-bp alkB gene amplicon, and the asterisks indicate the positions of the 500-bp fragments of the molecular size markers.
Nitrification.
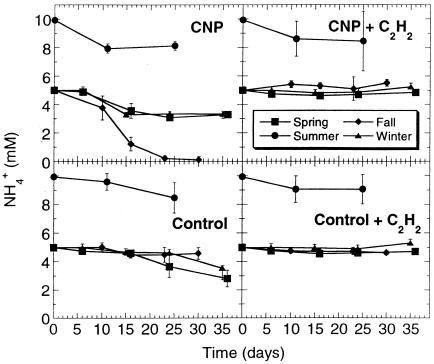
Figure 3 shows that NH4+ oxidation was initiated earlier with biofilms grown in the presence of nutrients but reached similar levels, whatever the nutrient status, at the end of the incubation period. The only exception was with the fall biofilms, where all NH4+ was oxidized by CNP biofilms but no significant activity was observed in the absence of nutrients. No decrease in NH4+ concentration was detected in the presence of C2H2, whether nutrients were present or not. In contrast with hexadecane mineralization, the presence of combined nutrients was not required by the seasonal biofilms to oxidize NH4+, except with the fall biofilm.
FIG. 3.
Effects of seasonal variations and nutrients on NH4+ oxidation during nitrification by bioreactor biofilms. Each datum point is the average of six replicate microcosms, and the bars represent 1 standard deviation.
In the presence of CNP, biofilm from river water collected in the fall showed the highest NH4+ oxidation rate, and the summer biofilm showed the lowest activity. Intermediate between these two extremes were the winter and the spring biofilms. This pattern is the same as the one obtained for hexadecane mineralization. Typically, a lag phase of ∼1 week was required before NH4+ oxidation could be detected, and a plateau was reached between 10 and 16 days of incubation. In the absence of nutrients (control biofilms), a different seasonal pattern was observed. Biofilm from river water collected in the spring showed the highest NH4+ oxidation rate, closely followed by the winter and then the summer biofilms. No activity was detected with the fall biofilm. A lag phase of 11 to 25 days occurred, but no plateau was reached at the end of the incubation period.
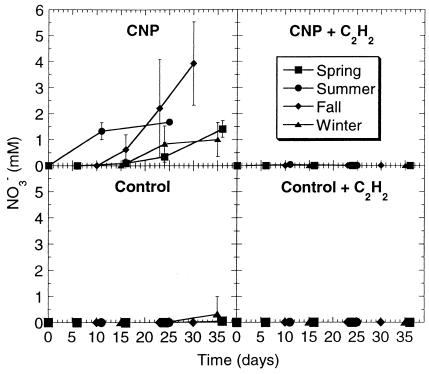
The presence of nutrients clearly enhanced NO3− production, since no activity was observed with biofilms grown in the absence of CNP (Fig. 4). This was also the case with hexadecane degraders, but not with NH4+ oxidizers. In the presence of CNP, the portrait of seasonal NO3− production was less clear than the one obtained with the previously described activities. Biofilms developing from river water collected in the summer initiated NO3− production earliest (no apparent lag phase), whereas fall biofilm clearly showed the highest activity after a lag phase of 10 days. After a 16-day lag phase, spring and winter biofilms had weak activities, although similar to each other, and winter biofilms reached a plateau after 24 days. No NO3− production was detected in the presence of C2H2, whether nutrients were present or not.
FIG. 4.
Effects of seasonal variations and nutrients on NO3− production during nitrification by bioreactor biofilms. Each datum point is the average of six replicate microcosms, and the bars represent 1 standard deviation.
Denitrification.
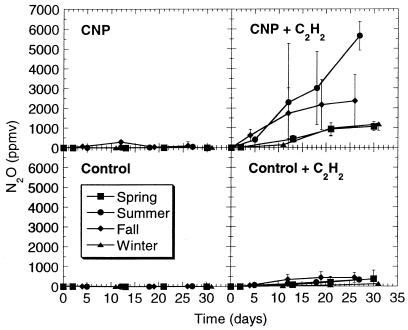
The combination of nutrients dramatically increased N2O production by summer biofilm, markedly enhanced the activity of fall biofilm, and stimulated spring and winter biofilms (Fig. 5). In the presence of CNP (with C2H2), summer and fall biofilms had similar activities until day 12. After that, fall biofilm reached a plateau, whereas the activity of summer biofilm increased until the end of the incubation period. No apparent lag phase was observed with fall biofilm. As was the case with all the activities assessed in the present study, winter and spring biofilms had similar activities when CNP were present. In the case of denitrification, these biofilms had the lowest activity and reached a plateau at the end of the incubation period. In the absence of nutrients (with C2H2), the biofilms had weak denitrification rates that were similar to each other. In the absence of C2H2, only a small amount of N2O accumulated, whether nutrients were present or not.
FIG. 5.
Effects of seasonal variations and nutrients on N2O production during denitrification by bioreactor biofilms. Each datum point is the average of six replicate microcosms, and the bars represent 1 standard deviation.
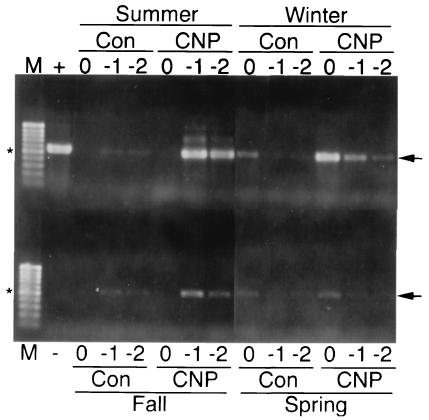
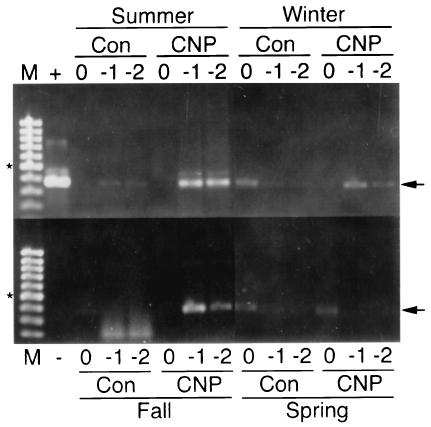
With the exception of spring biofilm, PCR amplification of nirK DNA was better with extracts from CNP than with control biofilms (Fig. 6), implying that copper-containing nitrite reductase (nirK) bacteria were more abundant when nutrients were present. This correlates well with Fig. 5, in which denitrification was greater with CNP than with control biofilms. In the presence of CNP, the best amplification for the nirK gene was with the summer biofilm, followed by the fall biofilm, whereas winter and, particularly, spring biofilms had lower amplification yields. Biofilms grown in the absence of nutrients had poor and similar amplification yields, and the poorest amplification was obtained with DNA extracts from fall biofilms. In both cases (CNP and control biofilms), a strong relationship between denitrification activity and nirK band intensity was observed, although the significance of this is not known. With the summer and fall biofilms in the presence and absence of nutrients, there was clearly PCR-inhibitory material present in the extracts, since there was no amplification using undiluted extract. This was also the case with alkB PCR. Inhibition of the nirK PCR with CNP winter biofilm was also observed.
FIG. 6.
PCR amplification of nirK from spring, summer, fall, and winter biofilms with (CNP) and without (Con) nutrient amendments. Total-community DNA extracts were undiluted (0) or diluted 10−1 (−1) or 10−2 (−2) prior to PCR amplification using the nirK consensus primers. Lanes M, molecular size markers; lane +, positive control (50 ng of Pseudomonas aureofaciens ATCC 13985 DNA template); lane −, negative control (no DNA). The arrows indicate the positions of the 376-bp nirK gene amplicon, and the asterisks indicate the positions of the 500-bp fragments of the molecular size markers.
Algal and bacterial biomass.
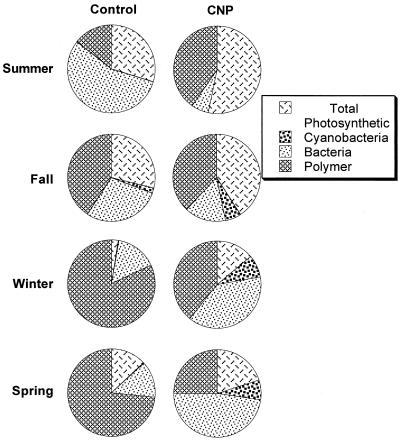
Digital image analyses indicated that the algal/bacterial ratios in control biofilms were as follows: spring, 1:1; summer, 1:2; fall, 1:1; and winter, 1:5 (Fig. 7). Bacteria dominated in winter and, to a lesser extent, in summer biofilms, whereas spring and fall biofilms had equal proportions of algal and bacterial biomass. In biofilms with nutrient additions (CNP), the corresponding ratios were spring, 1:2; summer, 8:1; fall, 3:1; and winter, 1:2. The addition of nutrients shifted summer and, to a lesser extent, fall biofilms to a phototrophic-dominated community, reduced the bacterial ratio in winter biofilm, and doubled the bacterial ratio in spring biofilm.
FIG. 7.
Digital image analysis of the algal-bacterial population compositions of seasonal biofilms grown with (CNP) and without (Control) nutrients. Total photosynthetic populations were observed by autofluorescence, and Syto9 was used to stain and image bacterial cells.
DISCUSSION
Seasonal variations.
The microcosm assay and PCR amplification results demonstrate that the biofilms that developed from river water collected at different times of the year had different microbial activities and community compositions. This implies that the microbial populations within biofilms would vary considerably over the course of a year and, as a consequence, response to stress would vary with season as well.
Our results reveal that in the presence of nutrients, the seasonal order was the same (fall > winter = spring) for all the activities studied and for algal/bacterial ratios and that summer biofilm did not follow any specific pattern. Even though river biofilms are dominated by bacteria (21, 40), our results are consistent with the tight linkages between the heterotrophic and phototrophic components of biofilm communities. For example, bacteria have been shown to utilize the extracellular substances released by algae in biofilms (23). Furthermore, variation in the heterotrophic activities of biofilms in streams has been most closely linked to variation in chlorophyll a levels (7). In the present study, ratios of algae to bacteria in the seasonal biofilms were used to indicate fundamental changes in the microbial community, which may be linked to detectable changes in heterotrophic activity, such as hexadecane mineralization and denitrification.
Biofilms from river water collected in the winter and spring showed very similar activities, especially when nutrients were present. Such similarity can be explained by the fact that no increase in the river flow resulting from ice and snow thaw was observed during the spring experiment. Accordingly, an examination of the analyses of the river water indicates relatively similar physical and chemical conditions (Table 1) and algal/bacterial ratios (Fig. 7) between winter and spring, showing again the linkage between heterotrophs and autotrophs. Since biofilms were grown at a constant temperature in the bioreactors, river water temperature differences between winter and spring did not likely influence bacterial activity and composition.
Since only a few studies regarding the impact of seasonal variations on in situ or replicated river biofilms have been published, it is difficult to generalize about typical or characteristic seasonal microbial flora and environmental conditions that may be found in a river over the course of a year. For example, in the present study, denitrification by biofilms was lowest in the spring, but a peak in denitrification activity by sediments from the Swale-Ouse River (England) was observed in spring (43). It is generally accepted that, in aquatic ecosystems, the main parameters influencing bacterial production are temperature (18) and the availability of nutrients (31). Some authors have defined a “warm” period by higher water temperature and nutrient concentrations, resulting in favorable conditions for microbial activity. The same authors defined a “cold” period by lower water temperature and nutrient availability, conditions which are less favorable for microbial activity (26). Accordingly, bacterioplanktonic production in an Alabama stream (35), periphytic biomass in Kennett River (United Kingdom) (19), and the occurrence of epilithic sodium dodecyl sulfate-degrading bacteria in Ely River (United Kingdom) (2) were highest in the summer and/or fall. In contrast, planktonic bacterial abundance showed no clear seasonal pattern in the Danube River (Austria) (25). In general, variations in biofilm productivity and activity could not be correlated with any physical or chemical factor. In the present study, nutrient concentrations did not follow any pattern throughout the year. This lack of variation is attributable to the presence of a large reservoir and a hydropower generation site 75 km upstream of the sampling area. As a consequence, the chemistry of the release waters from this reservoir are essentially constant, and severe drought in the drainage of the South Saskatchewan River has also greatly reduced inputs from sources other than mountain snow melt.
In the present study, we carried out experiments by removing water from the riverine habitat to use as a source of inoculum and by growing biofilms under constant conditions of light, temperature, and flow regime regardless of the season of experimentation. Strikingly, unique communities developed in each season, indicating changes in the nature of the microorganisms available to form periphyton or epilithon biofilms with time. A number of studies (30, 36, 42) have demonstrated the high bacterial diversity and dynamic nature of river biofilms.
Nutrients.
The microcosm assays, PCR amplification results, and microscopic analyses of community structure showed that the addition of nutrients (CNP) to the bioreactor water had a great stimulatory effect on biofilm activities and an impact on its composition. This suggests that a major factor limiting bacterial productivity in the South Saskatchewan River water is the presence of utilizable nutrients.
The response of biofilm bacteria to nutrient status may be explained in terms of r and K strategists (3, 15). Considering the nutrient aspect, hexadecane degraders and denitrifiers behaved as r strategists: in the presence of supplied nutrients, their population sizes and activities were much higher than when nutrients were absent. This may be due to the ability of these heterotrophic r strategists to catabolize readily available organic matter, such as added glucose, or to exploit carbon reserves resulting from increases in algal biomass and activity. CNP clearly stimulated denitrification, as very little N2O accumulated in the absence of CNP, and enhanced nirK PCR amplificon yields. In a study of the impact of organic compounds commonly found in agricultural pollutants on sediments of the Upper Bann River (Ireland), it was shown that glucose was efficient in stimulating denitrification, glycine had a limited stimulatory effect, and formate and acetate partially inhibited denitrification (29). In the present study, only a small amount of N2O accumulated in the absence of C2H2, whether nutrients were present or not. This suggests that either the process of complete denitrification was active (reducing NO3− to N2O and then to N2) or that NO3− was assimilated.
In contrast, nitrifying bacteria are K strategists, as they have very low growth rates in comparison to r strategists, with doubling times usually between 11 and 50 h (46). Because oxidation of NH4+ or NO2− provides the only source of energy for nitrifying bacteria (46) and thus is essential to their survival, these reactions may have been used as a source of energy for nonbiosynthetic processes (transport of solutes and macromolecule synthesis) rather than biomass production (44). This would explain why NH4+ oxidation was active in both the presence and absence of CNP. No decrease in NH4+ concentration was detected in the presence of C2H2, whether nutrients were present or not. This suggests that NH4+ assimilation was negligible compared to NH4+ oxidation and that nitrification was performed by chemolithotrophic bacteria rather than heterotrophic bacteria. Moreover, the accumulation of NO2− in the absence of C2H2, whether nutrients were present or not (data not shown), shows that nitrification was occurring. Nitrite oxidizers are known to be more sensitive to adverse conditions than NH4+ oxidizers (9), which would explain why no NO3− was produced in the absence of CNP. This suggests that conditions were not favorable for NO2− oxidizers unless nutrients were present.
Evidence suggests that rivers may be N or P limited when not impacted by human activity (51). Considering a typical molar C/N/P ratio of ∼50:10:1 in bacteria (17) and assuming a 50% assimilation efficiency for C (38), C was in large excess in the South Saskatchewan River water compared to N and P (Table 1). Bacterial activity and positive PCR amplification were restored by the addition of CNP to the water, resulting in a C/N/P ratio closer to the typical molar ratio and reducing the N and P limitation. N (45) and P (38) limitations for biofilm bacteria were also observed in other river waters. In the South Saskatchewan River, the addition of nutrients had a significant effect on the algae-to-bacteria ratio, substantially increasing the proportion of algae in the river biofilms, as well as increasing the measured activity levels. These observations are consistent with the close coupling of bacterial and algal growth in river biofilms and are in keeping with other studies (6, 7).
Conclusions.
This study revealed that the seasonal pattern in bacterial activity and in the occurrence of nirK in the South Saskatchewan River biofilms was as follows: fall > winter = spring. The results also showed that nutrients (CNP) were limiting for algal and bacterial activity and production. The community response to nutrients varied with season, and community composition was an important variable in response to environmental stress. Work is in progress to study biofilm variations over 2 years and to characterize the impact of separate nutrients on South Saskatchewan River biofilm activity and composition.
Acknowledgments
We gratefully acknowledge Suzanne Labelle, Anca Mihoc, Claude Masson, Danielle Ouellette, Sylvie Sanschagrin, and Gesine Wisse for excellent technical assistance; Nathalie Fortin for providing expertise in DNA extraction; George Swerhone for biofilm cultivation; and Alain Corriveau for performing HPLC analyses.
This research was supported by Health Canada through the Toxic Substances Research Initiative, by the National Water Research Institute, by Environment Canada, and by FCAR-MRST (M.R.C.).
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. J., M. J. Day, N. J. Russell, and G. F. White. 1988. Temporal and geographical distributions of epilithic sodium dodecyl sulfate-degrading bacteria in a polluted South Wales river. Appl. Environ. Microbiol. 54:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews, J. H., and R. F. Hall. 1986. r and K selection and microbial ecology. Adv. Microb. Ecol. 9:99-147. [Google Scholar]
- 4.Atlas, R. M., and R. Bartha. 1993. Microbial Ecology, 3rd ed. The Benjamin/Cummings Publishing Company, Inc., Redwood City, Calif.
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. Moore, J. G. Seidman, J. A. Smith, and E. K. Struhl. 1990. Short protocols in molecular biology, vol. 1. Massachussetts General Hospital, Harvard Medical School, Cambridge, Mass.
- 6.Battin, T. J. 2000. Hydrodynamics is a major determinant of streambed biofilms: from the sediment to the reach scale. Limnol. Oceanogr. 45:1308-1319. [Google Scholar]
- 7.Battin, T. J., A. Wille, B. Sattler, and R. Psenner. 2001. Phylogenetic and functional heterogeneity of sediment biofilms along environmental gradients in a glacial stream. Appl. Environ. Microbiol. 67:799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berthelet, M., and C. W. Greer. 1996. Detection of catabolic genes in soil using the polymerase chain reaction, p. 635-644. In Moo-Young, M., W. A. Anderson, and A. M. Chakrabarty (ed.), Environmental bio/technology: principles and applications. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 9.Bollag, J. M., and E. J. Kurek. 1980. Nitrite and nitrous oxide accumulation during nitrification in the presence of pesticide derivatives. Appl. Environ. Microbiol. 39:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bothe, H., G. Jost, M. Schloter, B. B. Ward, and K. P. Witzel. 2000. Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol. Rev. 24:673-690. [DOI] [PubMed] [Google Scholar]
- 11.Britton, L. N. 1984. Microbial degradation of aliphatic hydrocrabons, p. 89-129. In D. T. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, Inc., New York, N.Y.
- 12.Costerton, J. W., K.-J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 13.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutil, C., R. Chabot, S. Boivin, Y. Comeau, and G. Gagné. 2002. Le traitement du lisier de porc à la ferme avec la solution Biofertile. Vecteur Environ. 35:28-31. [Google Scholar]
- 15.Ellis, B. D., P. Butterfield, W. L. Jones, G. A. McFeters, and A. K. Camper. 2000. Effects of carbon sources, carbon concentration, and chlorination on growth related parameters of heterotrophic biofilm bacteria. Microb. Ecol. 38:330-347. [DOI] [PubMed] [Google Scholar]
- 16.Environment Canada. 1992. Analytical methods manuals. Inland Waters Directorate, Water Quality Branch, Environment Canada, Ottawa, Ontario.
- 17.Fagerbakke, K. M., M. Heldal, and S. Norland. 1996. Content of carbon, nitrogen, oxygen, sulfur and phosphorus in native aquatic and cultured bacteria. Aquat. Microb. Ecol. 10:15-27. [Google Scholar]
- 18.Felip, M., M. L. Pace, and J. J. Cole. 1996. Regulation of planktonic bacterial growth rates: the effect of temperatures and resources. Microb. Ecol. 31:15-28. [DOI] [PubMed] [Google Scholar]
- 19.Flynn, N. J., D. L. Snook, A. J. Wade, and H. P. Jarvie. 2002. Macrophyte and periphyton dynamics in a UK Cretaceous chalk stream: the River Kennet, a tributary of the Thames. Sci. Total Environ. 282-283:143-157. [DOI] [PubMed]
- 20.Fortin, N., R. R. Fulthorpe, G. D. Allen, and C. W. Greer. 1998. Molecular analysis of bacterial isolates and total community DNA from kraft pulp mill effluent treatment systems. Can. J. Microbiol. 44:537-546. [PubMed] [Google Scholar]
- 21.Geesey, G. G., R. Mutch, J. W. Costerton, and R. B. Green. 1978. Sessile bacteria: an important component of the microbial population in small mountain streams. Limnol. Oceanogr. 23:1214-1223. [Google Scholar]
- 22.Greer, C. W., L. Masson, Y. Comeau, R. Brousseau, and R. Samson. 1993. Application of molecular biology techniques for isolating and monitoring pollutant-degrading bacteria. Water Pollut. Res. J. Can. 28:275-287. [Google Scholar]
- 23.Haak, S. K., and G. A. McFeters. 1982. Nutritional relationships among microorganisms in an epilithic biofilm community. Microb. Ecol. 8:115-126. [DOI] [PubMed] [Google Scholar]
- 24.Heitkamp, M. A., and B. T. Johnson. 1984. Impact of an oil field effluent on microbial activities in a Wyoming river. Can. J. Microbiol. 30:786-792. [DOI] [PubMed] [Google Scholar]
- 25.Hoch, B., B. Berger, G. Kavka, and G. J. Herndl. 1995. Remineralization of organic matter and degradation of the organic fraction of suspended solids in the River Danube. Aquat. Microb. Ecol. 9:279-288. [Google Scholar]
- 26.Iriberri, J., B. Ayo, M. Unanue, I. Barcina, and L. Egea. 1993. Channeling of bacterioplanktonic production toward phagotrophic flagellates and ciliates under different conditions in a river. Microb. Ecol. 26:111-124. [DOI] [PubMed] [Google Scholar]
- 27.Jetten, M. S. M., S. Logemann, G. Muyzer, L. A. Robertson, S. de Vries, M. C. M. van Loosdrecht, and J. G. Kuenen. 1997. Novel principles in the microbial conversion of nitrogen compounds. Antonie Leeuwenhoek 71:75-93. [DOI] [PubMed] [Google Scholar]
- 28.Juck, D., T. C. Charles, L. G. Whyte, and C. W. Greer. 2000. Polyphasic microbial community analysis of petroleum hydrocarbon-contaminated soils from two northern Canadian communities. FEMS Microbiol. Ecol. 33:241-249. [DOI] [PubMed] [Google Scholar]
- 29.Kelso, B. H. L., R. V. Smith, and R. J. Laughlin. 1999. Effects of carbon substrates on nitrite accumulation in freshwater sediments. Appl. Environ. Microbiol. 65:61-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenzaka, T., N. Yamaguchi, K. Tani, and M. Nasu. 1998. rRNA-targeted fluorescent in situ hybridization analysis of bacterial community structure in river water. Microbiology 144:2085-2093. [DOI] [PubMed] [Google Scholar]
- 31.Kirchman, D. L. 1994. The uptake of inorganic nutrients by heterotrophic bacteria. Microb. Ecol. 28:255-271. [DOI] [PubMed] [Google Scholar]
- 32.Kok, M., R. Oldenhuis, M. P. G. van der Linden, P. Raatjees, J. Kingma, P. H. van Lelyveld, and B. Witholt. 1989. The Pseudomonas oleovorans alkane hydroxylase gene. J. Biol. Chem. 264:5435-5441. [PubMed] [Google Scholar]
- 33.Lawrence, J. R., T. R. Neu, and G. D. W. Swerhone. 1998. Application of multiple parameter imaging for the quantification of algal, bacterial and exopolymer components of microbial biofilms. J. Microbiol. Methods 32:253-261. [Google Scholar]
- 34.Lawrence, J. R., G. D. W. Swerhone, and T. R. Neu. 2000. A simple rotating annular reactor for replicated biofilm studies. J. Microbiol. Methods 42:214-224. [DOI] [PubMed] [Google Scholar]
- 35.Mann, C. J., and R. G. Wetzel. 1995. Dissolved organic carbon and its utilization in a riverine wetland ecosystem. Biogeochemistry 31:99-120. [Google Scholar]
- 36.Manz, W., K. Wendt-Potthoff, T. R. Neu, U. Szewzyk, and J. R. Lawrence. 1999. Phylogenetic composition, spatial structure, and dynamics of lotic bacterial biofilms investigated by fluorescent in situ hybridization and confocal laser scanning microscopy. Microb. Ecol. 37:225-237. [DOI] [PubMed] [Google Scholar]
- 37.Massol-Deya, A. A., J. Whallon, R. F. Hickey, and J. M. Tiedje. 1995. Channel structures in aerobic biofilms of fixed-film reactors treating contaminated groundwater. Appl. Environ. Microbiol. 61:769-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohamed, M. N., J. R. Lawrence, and R. D. Robarts. 1998. Phosphorus limitation of heterotrophic biofilms from the Fraser river, British Columbia, and the effect of pulp mill effluent. Microb. Ecol. 36:121-130. [DOI] [PubMed] [Google Scholar]
- 39.Morrison, R. T., and R. N. Boyd. 1983. Organic chemistry, 4th ed. Allyn and Bacon, Toronto, Canada.
- 40.Neu, T. R., and J. R. Lawrence. 1997. Development and structure of microbial stream biofilms as studied by confocal laser scanning microscopy. FEMS Microbiol. Ecol. 24:11-25.
- 41.Oremland, R. S., and D. G. Capone. 1988. Use of “specific” inhibitors in biogeochemistry and microbial ecology. Adv. Microb. Ecol. 10:285-383. [Google Scholar]
- 42.O'Sullivan, L. A., A. J. Weightman, and J. C. Fry. 2002. New degenerate Cytophaga-Flexibacter-Bacteroides-specific 16S ribosomal DNA-targeted oligonucleotide probes reveal high bacterial diversity in River Taff epilithon. Appl. Environ. Microbiol. 68:201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pattinson, N. P., R. Garcia-Ruiz, and B. A. Whitton. 1998. Spatial and seasonal variation in denitrification in the Swale-Ouse system, a river continuum. Sci. Total Environ. 210-211:289-305.
- 44.Pelczar, M. J., E. C. S. Chan, and N. R. Krieg. 1986. Microbiology, 5th ed. McGraw-Hill, Inc., New York, N.Y.
- 45.Podemski, C. L., and J. M. Culp. 1996. Nutrient and contaminant effects of bleached kraft mill effluent on benthic algae and insects of the Athabasca River, p. 571-580. In M. K. Servos, J. H. Carey, and G. J. Van Der Kraak (ed.), Environmental fate and effects of pulp and paper mill effluents. St. Lucie Press, Delray Beach, Fla.
- 46.Prosser, J. I. 1989. Autotrophic nitrification in bacteria. Adv. Microb. Physiol. 30:125-181. [DOI] [PubMed] [Google Scholar]
- 47.Roy, R., and C. W. Greer. 2000. Hexadecane mineralization and denitrification in two diesel fuel-contaminated soils. FEMS Microbiol. Ecol. 32:17-23. [DOI] [PubMed] [Google Scholar]
- 48.Roy, R., and R. Knowles. 1995. Differential inhibition by allylsulfide of nitrification and methane oxidation in freshwater sediment. Appl. Environ. Microbiol. 61:4278-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roy, R., and R. Knowles. 1994. Effects of methane metabolism on nitrification and nitrous oxide production in polluted freshwater sediment. Appl. Environ. Microbiol. 60:3307-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual., 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Scrimgeour, G. J., and P. A. Chambers. 2000. Cumulative effects of pulp mill and municipal effluents on epilithic biomass and nutrient limitation in a large northern river ecosystem. Can. J. Fish. Aquat. Sci. 57:1342-1354. [Google Scholar]
- 52.Sotsky, J. B., C. W. Greer, and R. M. Atlas. 1994. Frequency of genes in aromatic and aliphatic hydrocarbon biodegradation pathways within bacterial populations from Alaskan sediments. Can. J. Microbiol. 40:981-985. [DOI] [PubMed] [Google Scholar]
- 53.Trudgill, S. T., D. E. Walling, and B. W. Webb. 1999. Water quality, processes, and policy. John Wiley and Sons, Chichester, United Kingdom.
- 54.Ward, B. B. 1996. Nitrification and denitrification: probing the nitrogen cycle in aquatic environments. Microb. Ecol. 32:247-261. [DOI] [PubMed] [Google Scholar]
- 55.Wilderer, P. A., and W. G. Charaklis. 1989. Structure and function of biofilms, p. 5-17. In W. G. Charaklis and P. A. Wilderer (ed.), Structure and function of biofilms. John Wiley and Sons, New York, N.Y.
- 56.Yoshinari, T. D., and R. Knowles. 1976. Acetylene inhibition of nitrous oxide reduction by denitrifying bacteria. Biochem. Biophys. Res. Commun. 69:705-710 [DOI] [PubMed]