Abstract
The present study describes the 19F nuclear magnetic resonance analysis of the conversion of 3-halocatechols to lactones by purified chlorocatechol 1,2-dioxygenase (ClcA2), chloromuconate cycloisomerase (ClcB2), and chloromuconolactone dehalogenase (ClcF) from Rhodococcus opacus 1cp grown on 2-chlorophenol. The 3-halocatechol substrates were produced from the corresponding 2-halophenols by either phenol hydroxylase from Trichosporon cutaneum or 2-hydroxybiphenyl 3-mono-oxygenase from Pseudomonas azelaica. Several fluoromuconates resulting from intradiol ring cleavage by ClcA2 were identified. ClcB2 converted 2-fluoromuconate to 5-fluoromuconolactone and 2-chloro-4-fluoromuconate to 2-chloro-4-fluoromuconolactone. Especially the cycloisomerization of 2-fluoromuconate is a new observation. ClcF catalyzed the dehalogenation of 5-fluoromuconolactone to cis-dienelactone. The ClcB2 and ClcF-mediated reactions are in line with the recent finding of a second cluster of chlorocatechol catabolic genes in R. opacus 1cp which provides a new route for the microbial dehalogenation of 3-chlorocatechol.
The aerobic microbial degradation of haloaromatics can proceed through formation of intermediate halocatechols that are mostly converted by the so-called modified ortho cleavage pathway. The general features of this pathway are intradiol ring cleavage of the halocatechols by chlorocatechol dioxygenase (ClcA; EC 1.13.11.1) to give halomuconates, isomerization of the halomuconates by chloromuconate cycloisomerase (ClcB; EC 5.5.1.7) to give (halo)dienelactones, and further conversion by dienelactone hydrolase (ClcD; EC 3.1.1.45) and maleylacetate reductase (EC 1.3.1.32) to give 3-oxoadipate (1, 7, 8, 27, 31, 35).
For several Proteobacteria it was shown that 2-chloro-cis,cis-muconate, the ring cleavage product of 3-chlorocatechol, is converted by ClcB to 2-chloro- or 5-chloromuconolactone (17, 35, 43, 44) and that the latter compound is dehalogenated by the same enzyme to trans-dienelactone (trans-4-carboxymethylenebut-2-en-4-olide) (44). All (chloro)muconate lactonizing enzymes studied so far are not active with 2-fluoromuconate (38, 42), and in Pseudomonas B13, 2-fluoromuconate was identified as a dead-end metabolite from 2- and 3-fluorobenzoate catabolism (36).
Among the Actinobacteria, Rhodococcus opacus 1cp grown on 4-chlorophenol was shown to convert 4-chlorocatechol via 3-chloromuconate and cis-dienelactone to maleylacetate (16, 18, 19, 38). The ClcB and ClcD involved in this pathway are more specific than their proteobacterial counterparts, with ClcB being unable to dehalogenate 2-chloro-cis,cis-muconate (38). Recently, a variant of R. opacus 1cp that uses 2-chlorophenol for growth was isolated (20, 21). This strain was shown to contain, besides ClcA2, ClcB2, and ClcD2 activities, 5-chloromuconolactone dehalogenase (ClcF) activity also (21, 22). ClcB2 and ClcF are both needed in this R. opacus 1cp variant to carry out the proteobacterial ClcB activity, i.e., the conversion of 2-chloromuconate to cis-dienelactone. The enzymes of this new modified ortho cleavage pathway of 3-chlorocatechol are only distantly related to known chlorocatechol enzymes, whereas ClcF is distantly related to muconolactone isomerase (CatC; EC 5.3.3.4) (22).
In this study we have addressed the substrate specificity of ClcB2 and ClcF from 2-chlorophenol-grown R. opacus 1cp in further detail. To that end, several fluoromuconates were enzymatically produced from halophenols and 19F nuclear magnetic resonance (NMR) was used for the identification of reaction products. The 19F NMR technique combines high selectivity with high sensitivity and is especially useful for the in situ detection of unstable intermediates (2, 4, 5, 6, 24). It is shown for the first time that 2-fluoromuconate is converted by ClcB2 to 5-fluoromuconolactone and that the latter compound is dehalogenated by ClcF to cis-dienelactone. The results are discussed in relation to the conversion of halomuconates and halomuconolactones by isofunctional enzymes.
MATERIALS AND METHODS
Chemicals.
2-Chloro-5-fluorophenol, 2-chloro-4-fluorophenol, 4-chloro-2-fluorophenol, and 4-fluorophenol were obtained from Fluorochem (Old Glossop, Derbyshire, United Kingdom). Catechol, 3-fluorocatechol, 2-chlorophenol, 2,4-dichlorophenol and 2-hydroxybiphenyl were purchased from Aldrich (Steinheim, Germany). Dithiothreitol, NADH, and NADPH were from Boehringer (Mannheim, Germany). Ascorbic acid was obtained from Merck. Chromatography materials were from Amersham Pharmacia Biotech AB (Uppsala, Sweden). 2-Chloromuconate and cis-dienelactone were kindly provided by M. Schlömann.
Microorganism and cultivation conditions.
R. opacus 1cp can utilize 2,4-dichlorophenol and 4-chlorophenol as growth substrates (12). After a long adaptation, the variant utilizing 2-chlorophenol, which specifically induces ClcA2, ClcB2, ClcD2, and ClcF (21, 22), was obtained (20). Biomass was obtained as described earlier (21).
Enzyme activity.
The activity of enzymes was determined spectrophotometrically at 25°C. The activity of ClcA2 was monitored at 260 nm by using a ɛ260 of 16.8 mM−1 cm−1 for the formation of muconate from catechol and a ɛ260 of 17.1 mM−1 cm−1 for the formation of 2-chloromuconate from 3-chlorocatechol (7, 32). The activity of ClcB2 was determined by monitoring the decrease in absorbance of 2-chloromuconate at 260 nm (ɛ260 = 17.1 mM−1 cm−1) (35). The activity of ClcF was determined by monitoring the increase in absorption at 280 nm due to the conversion of 5-chloromuconolactone to cis-dienelactone (ɛ260 = 17.0 mM−1 cm−1) (33).
Enzyme purification.
Phenol hydroxylase from Trichosporon cutaneum CBS2466 was purified, essentially as described by Sejlitz and Neujahr (37). The final preparation had a specific activity of 5.0 μmol of NADPH oxidized min−1 mg−1 and was free of any detectable catechol dioxygenase activity. 2-Hydroxybiphenyl 3-mono-oxygenase from Pseudomonas azelaica HBP1 was purified from recombinant Escherichia coli JM101 harboring the hbpA gene (39). The final preparation had a specific activity of 5.8 μmol of NADH oxidized min−1 mg−1 (40).
ClcA2, ClcB2, ClcD2, and ClcF were purified as described earlier (21). The specific activity of ClcA2 was 16.1 U mg−1 with catechol and 11.5 U mg−1 with 3-chlorocatechol. Purified ClcA2 migrated as a single band in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with an apparent molecular mass of 29.5 kDa (21). Analytical gel filtration on Superdex S-200 revealed one peak with an apparent mass of 66 ± 4 kDa, indicating that native ClcA2 is a dimer composed of two identical subunits. This conclusion is supported by the preliminary analysis of ClcA2 crystals (10).
The specific activity of ClcB2 with 2-chloromuconate was 18 U mg−1. Purified ClcB2 migrated as a single band in SDS-PAGE with an apparent molecular mass of 40 kDa (21). Analytical gel filtration on Superdex S-200 revealed one peak with an apparent mass of 340 ± 10 kDa (mean ± standard deviation). This and the high sequence similarity to known bacterial (chloro)muconate cycloisomerases (22) suggest that native ClcB2 is a homo-octamer (14).
The specific activity of ClcF with 5-chloromuconolactone was 82.7 U mg−1. Purified ClcF migrated as a single band in SDS-PAGE with an apparent molecular mass of 12 kDa (21). Analytical gel filtration on Superdex S-200 revealed one peak with an apparent mass of 66 ± 5 kDa. Similar values were reported for the related muconolactone isomerase from Ralstonia eutropha JMP134, which is assumed to be a homodecamer (28). From this and the high sequence similarity with other muconolactone isomerases (22), it is proposed that native ClcF may have a similar quaternary structure (15).
19F NMR studies.
Incubations for 19F NMR analysis were performed in 100 mM Tris-HCl (pH 7.2). This pH was selected because phenol hydroxylase (23, 37), 2-hydroxybiphenyl mono-oxygenase (13, 39), and ClcA2 (21) have a pH optimum around pH 7.6 whereas ClcB2 has a pH optimum around pH 6.2 (21). Reactions were started with 0.70 mM of the relevant fluorophenol and 1 mM NAD(P)H or with 0.75 mM 3-fluorocatechol. Generally, enzymes were added to give the following (active-site) concentrations: 1.6 μM 2-hydroxybiphenyl mono-oxygenase, 4.7 μM phenol hydroxylase, 1.4 μM ClcA2, 0.5 μM ClcB2, and 1.3 μM ClcF. After each enzyme addition, the reaction mixtures were incubated for 30 min at 30°C unless indicated otherwise. Immediately thereafter, the incubation mixtures were frozen in liquid nitrogen and stored at −20°C until analysis.
19F NMR analysis was performed on a Bruker DPX 400 NMR spectrometer as described previously (2, 3, 11, 24). Chemical shifts are reported relative to CFCl3. The resonance of the internal standard 4-fluorobenzoate was set at −114.2 ppm with respect to CFCl3. 19F NMR chemical shifts of the various fluorine-containing compounds were identified on the basis of authentic reference compounds, as described previously (3, 24) or as described in the present study. The detection limit of an overnight 19F NMR measurement is 1 μM.
High-pressure liquid chromatography (HPLC) analysis.
Reactions of phenol hydroxylase and 2-hydroxybiphenyl mono-oxygenase with 4-fluorophenol and 2-chloro-4-fluorophenol were analyzed on a Waters M600-PDA system equipped with a Waters 996 photodiode-array detector. Reaction products were separated with a 4.6- by 150-mm Alltima C18 reverse-phase column (Alltech) running in 40% methanol containing 0.65% acetic acid at a flow rate of 1.0 ml/min. Reaction mixtures contained 500 μM substrate, 1 mM NAD(P)H, and 2 U of enzyme in 0.5 ml of air-saturated 50 mM phosphate buffer (pH 7.2), containing 1 mM dithiothreitol and 1 mM ascorbate. Reactions were stopped by the addition of 50 μl of 50% (vol/vol) trichloroacetic acid, and precipitated protein was removed by centrifugation. Typical retention times were as follows: 4-fluorophenol, 9.17 min; 2-chloro-4-fluorophenol, 22.52 min; 4-fluorocatechol, 5.46 min; 3-chloro-5-fluorocatechol, 14.95 min.
For HPLC product analysis of the sequential conversion of 3-fluorocatechol by ClcA2, ClcB2, and ClcF, samples were taken every 20 min and directly injected onto a 4.6- by 150-mm Alltima C18 column (Alltech). The flow rate was 1 ml/min with 23% (vol/vol) methanol in water containing 0.1% trifluoroacetic acid as the mobile phase. 3-Fluorocatechol and 5-fluoromuconolactone were detected at 210 nm, and 2-fluoromuconate and cis-dienelactone were detected at 260 nm. Typical retention times were as follows: 3-fluorocatechol, 9.1 min; 2-fluoromuconate, 8.3 min; 5-fluoromuconolactone, 2.1 min; cis-dienelactone, 7.3 min. The formation of 2-fluoromuconate and 5-fluoromuconolactone was confirmed by 1H-coupled 19F-NMR analysis.
RESULTS
Enzymatic production of fluorocatechols.
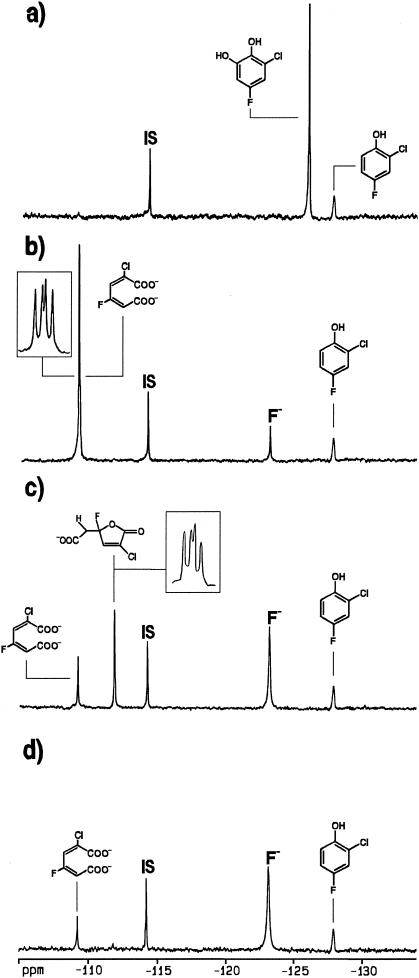
Several fluorocatechols were produced by ortho-hydroxylation of fluorophenols by using either phenol hydroxylase from T. cutaneum or 2-hydroxybiphenyl mono-oxygenase from P. azelaica. Figure 1a presents, as an example, the 19F NMR spectrum recorded after incubation of 2-chloro-4-fluorophenol (−128.0 ppm) with 2-hydroxybiphenyl mono-oxygenase. Based on the fact that incorporation of a hydroxyl moiety in a fluorophenol ortho, meta, or para with respect to the fluorine substituent shifts the 19F NMR chemical shift value by −23.1 ± 0.3 ppm, +1.3 ± 0.4 ppm, and −11.2 ± 0.7 ppm, respectively (24), the catechol product with its resonance at −126.7 ppm was assigned to 3-chloro-5-fluorocatechol. The possibility that 2-chloro-4-fluorophenol is converted by 2-hydroxybiphenyl mono-oxygenase via oxidative dehalogenation to 4-fluorocatechol (−126.7 ppm [24]) was excluded by HPLC analysis (see Materials and Methods). Besides the production of 3-chloro-5-fluorocatechol, 2-hydroxybiphenyl mono-oxygenase was used for the preparation of 3-fluorocatechol (−140.5 ppm) from 2-fluorophenol (−141.9 ppm) and 3-chloro-6-fluorocatechol (−142.6 ppm) from 2-chloro-5-fluorophenol (−119.3 ppm). Besides the production of 4-fluorocatechol from 4-fluorophenol, phenol hydroxylase was used for the preparation of 5-chloro-3-fluorocatechol (−139.5 ppm) from 4-chloro-2-fluorophenol (−138.8 ppm).
FIG. 1.
19F NMR analysis of the incubation of 2-chloro-4-fluorophenol with the following: 1.6 μM 2-hydroxybiphenyl mono-oxygenase (a), 1.4 μM ClcA2 (b), 0.5 μM ClcB2 (c), and 1.3 μM ClcF (d).
Ring cleavage of halocatechols by ClcA2.
3-Fluorocatechol, 3-chloro-6-fluorocatechol, 3-chloro-5-fluorocatechol, and also 5-chloro-3-fluorocatechol were converted by ClcA2 to the corresponding fluoromuconates. Assuming similar extinction coefficients for the muconate products at 260 nm (7, 32), 3-fluorocatechol was the best substrate, showing a relative rate of about 50% of the rate observed with 3-chlorocatechol. 3-Chloro-6-fluorocatechol, 3-chloro-5-fluorocatechol, and 5-chloro-3-fluorocatechol were more slowly converted, showing relative rates of about 25, 10, and 5% compared to the rate with 3-chlorocatechol.
Figure 1b presents the 19F NMR spectral data of the conversion of 3-chloro-5-fluorocatechol. The splitting pattern of the muconate product (−109.2 ppm) reveals 3J(4F,3H) and 3J(4F,5H) coupling constants of 30.7 and 21.5 Hz, respectively, reflecting 19F NMR characteristics similar to those of 3-fluoromuconate and different from those of 2-fluoromuconate (Table 1) (2). This, together with previous assignments (2), identifies the product of the ClcA2-mediated conversion of 3-chloro-5-fluorocatechol as 2-chloro-4-fluoromuconate. The 19F NMR characteristics of the other newly identified 2-halomuconate products are summarized in Table 1.
TABLE 1.
19F NMR chemical shift values and identification of fluoromuconates and fluoromuconolactones
| Compound | Chemical shift (ppm) | Coupling constants (Hz) | Conversion by ClcB2 |
|---|---|---|---|
| 4-Chloro-2-fluoromuconate | −111.1 | 3J (2F,3H) (20.9) | No |
| 2-Chloro-4-fluoromuconate | −109.2 | 3J (4F,3H) (30.7) | Yes |
| 3J (4F,5H) (21.5) | |||
| 2-Chloro-5-fluoromuconate | −112.0 | 3J (2F,3H) (20.1) | Yesa |
| 2-Fluoromuconateb | −112.0 | 3J (2F,3H) (20.7) | Yes |
| 2-Chloro-4-fluoro-muconolactone | −111.7 | 3J (4F,5H) (19.9) | Product from 2-chloro-4-fluoromuconate |
| 3J (4F,5H) (13.8) | |||
| 5-Fluoromuconolactone | −201.0 | 2J (5F,5H) (47.4) | Product from 2-fluoromuconate |
| 3J (5F,4H) (25.2) |
Conversion was far less than for 2-chloro-4-fluoromuconate or 2-fluoromuconate, hampering identification of the corresponding muconolactone.
19F NMR data from reference 2.
Conversion of 2-halomuconates by ClcB2.
The conversion of 2-halomuconates by ClcB2 was studied at pH 7.2, one pH unit above the optimum pH for ClcB2 (21), but closer to pH 7.6, the optimum pH for phenol hydroxylase (37) and 2-hydroxybiphenyl mono-oxygenase (39). 19F NMR analysis of blank reactions without added ClcB2 revealed that the 2-halomuconates are rather stable at pH 7.2, showing less than 10% signal loss after 10 h of incubation.
ClcB2 of R. opacus 1cp was previously reported to possess a narrow substrate specificity (21). 2-Chloro-cis,cis-muconate is a good substrate for ClcB2, but the enzyme is not active with cis,cis-muconate, 3-chloromuconate, 3-methylmuconate, and 2,4-dichloromuconate. By using the newly identified fluoromuconates (Table 1), the substrate specificity of ClcB2 was further defined. Incubations of 2-chloro-4-fluoromuconate and 4-chloro-2-fluoromuconate with ClcB2 revealed significant conversion of the first (Fig. 1c) but not of the latter compound (Table 1). Thus, replacement of the chlorine substituent in 2,4-dichloromuconate at C4 but not at C2 by a fluorine converts the dihalomuconate into a substrate for ClcB2.
Theoretically, ClcB2 could catalyze the isomerization of 2-chloro-4-fluoro-cis,cis-muconate to the cis,trans- or trans,cis-muconate isomer instead of converting it to the muconolactone (34). However, based on the J values observed this can be excluded. Especially the loss of the large 3J(4F,3H) coupling of around 30 Hz, also present in other 3- or 4-halomuconates (2), and the occurrence of a small 3J(4F,5H) coupling of 13.8 Hz in the product formed eliminate isomerization to other muconates as an alternative for the formation of the muconolactone.
Originally, an unstable nature of muconolactones at physiological pH was postulated (35, 36). However, more recent investigations revealed that at least some muconolactones are rather stable under these conditions. At pH 7.5, 5-chloromuconolactone had a half-life of 99 h (41), whereas 4-fluoromuconolactone showed a half-life of 43 h at pH 7.2 (32). Furthermore, a number of dimethylmuconolactones and diastereomeric 5-chloro-3-methyl-muconolactones were obtained and characterized at pH 7.4 to 7.5 (25, 26).
A comparison of the 19F NMR data in Fig. 1c to those presented in Fig. 1b reveals a 70% decrease in the area of the 19F NMR signal of 2-chloro-4-fluoromuconate and a concomitant appearance of a signal at −111.7 ppm with an intensity representing about 20% of the 2-chloro-4-fluoromuconate disappearance. In addition, a significant increase in the intensity of the fluoride anion peak at −123.0 ppm is observed to an extent representing about 80% of the 2-chloro-4-fluoromuconate disappearance. The 1H-coupled 19F NMR splitting pattern of the resonance at −111.7 ppm reveals 3J(4F,5H) coupling constants of 19.9 and 13.8 Hz. These values are similar to that previously observed for 4-fluoromuconolactone (3). Taking into account that (i) due to the essential axial position of the fluorine substituent with respect to the lactone ring the vicinal C3-H, C4-F couplings of 4-fluoromuconolactones are generally not detected (3, 32) and that (ii) the anisochrony in the diastereotypic methylene protons results in unequal 3JF-H coupling values (3, 32), the product formed from 2-chloro-4-fluoromuconate by ClcB2 is identified as 2-chloro-4-fluoromuconolactone (Table 1). In line with the properties of the more stable 4-fluoromuconolactone (3, 32), this compound may become defluorinated in a nonenzymatic reaction yielding most probably 2-chloromaleylacetate. It should be noted here that out of 4-chloromuconolactone, which has never been observed thus far, only cis-dienelactone and protoanemonin have been reported to be formed (1, 16, 17, 35).
Figure 2c shows the 19F NMR results of the incubation of 2-fluoromuconate with ClcB2. A 64% decrease in the 19F NMR signal of the 2-fluoromuconate signal at −112.0 ppm is accompanied especially by the occurrence of a new resonance at −201.0 ppm with an intensity representing almost the complete decrease in intensity of 2-fluoromuconate. Based on the 1H-coupled 19F NMR splitting pattern (Fig. 2c, insert), the resonance at −201.0 ppm is assigned to 5-fluoromuconolactone, with a 2J(5F,5H) of 47.4 Hz and a 3J(5F,4H) of 25.2 Hz, being in line with what is generally observed for these types of coupling constants (3, 45). From the 19F NMR data of incubations with different ClcB2 concentrations, it was estimated that under the conditions employed, the relative rate of the ClcB2-mediated conversion of 2-fluoromuconate was about 2% of the rate with 2-chloromuconate.
FIG. 2.
19F NMR analysis of the incubation of 3-fluorocatechol by itself (a) and with the following: 1.4 μM ClcA2 (b), 0.5 μM ClcB2 (c), and 1.3 μM ClcF (d).
2-Chloro-5-fluoromuconate was a very poor substrate for ClcB2. When incubation conditions similar to those with the other fluoromuconates were used, almost no conversion was observed (Table 1).
Conversion of fluoromuconolactones by ClcF.
The two newly identified muconolactones, i.e., 2-chloro-4-fluoro-muconolactone and 5-fluoromuconolactone, were further investigated for their conversion by ClcF. A comparison of the 19F NMR spectra before and after the ClcF addition (Fig. 1c and d and 2c and d) reveals a clear decrease in the 2-chloro-4-fluoromuconolactone as well as the 5-fluoromuconolactone signal. Control incubations performed in the absence of ClcF showed that for 2-chloro-4-fluoromuconolactone, the disappearance of its 19F NMR signal occurred to similar extents in the incubations with and without added ClcF. In contrast, the 5-fluoromuconolactone incubation in the absence of ClcF showed only a 9% decrease in 5-fluoromuconolactone signal intensity, indicating that the conversion presented in Fig. 2d is dependent on the presence of ClcF and, thus, enzyme catalyzed. Figures 1d and 2d also show that the disappearance of the fluoromuconolactones is accompanied not by the occurrence of new 19F NMR signals but only by an increase of the resonance at −123.0 ppm ascribed to fluoride anions. In both cases the increase in intensity of the fluoride anion resonance matches the decrease in the amount of the fluoromuconolactone. Thus, defluorination of 5-fluoromuconolactone is catalyzed by ClcF whereas defluorination of 2-chloro-4-fluoromuconolactone occurs spontaneously due to the chemical instability of the compound under the conditions applied.
The identity of the product formed from the ClcF-mediated conversion of 5-fluoromuconolactone was determined by HPLC. To that end, 3-fluorocatechol was incubated with ClcA2, ClcB2, and ClcF, respectively (see Materials and Methods). In this way it could be established that 5-fluoromuconolactone is converted to cis-dienelactone. The latter compound was identified on the basis of its retention time, its UV absorption spectrum with a maximum at 276 nm (22), and its conversion by ClcD2.
DISCUSSION
The present paper describes the conversion of fluorinated substrates by key enzymes of the new modified ortho cleavage pathway of 3-chlorocatechol degradation in R. opacus 1cp (Fig. 3). The enzymes from 2-chlorophenol-grown R. opacus 1cp are significantly different from the corresponding enzymes induced when the same bacterial species is grown on 4-chlorophenol. ClcA2 has a high activity with unsubstituted catechol as well as with 3-chloro-, 3-methyl-, and 4-methylcatechols (21). Here we were able to show that several other halocatechols, i.e., 3-chloro-6-fluorocatechol, 3-chloro-5-fluorocatechol, 5-chloro-3-fluorocatechol, and also 3-fluorocatechol, are readily converted by ClcA2 to the corresponding fluoromuconates. ClcA2 is not active with 3,5-dichlorocatechol. Kinetic studies revealed that the latter compound acts as a strong competitive inhibitor with an inhibition constant, Ki, of 2.5 μM (21). Thus, replacement of one chlorine atom in 3,5-dichlorocatechol by a fluorine or hydrogen turns the halocatechol into a good substrate for ClcA2.
FIG. 3.
Conversion of 3-halocatechols by enzymes of the modified ortho cleavage pathway of R. opacus 1cp grown on 2-chlorophenol. The halomuconates are depicted in the 2-up orientation with the 2-halo substituents pointing upwards (30).
The difference between the enzymes from R. opacus 1cp grown on 2-chlorophenol and on 4-chlorophenol is further illustrated by the substrate specificity of ClcB2. In contrast to ClcB (3, 38), ClcB2 converts 2-fluoromuconate into 5-fluoromuconolactone. Furthermore, ClcB2 is active with 2-chloromuconate (21) and 2-chloro-4-fluoromuconate.
Studies on the catalytic and structural properties of proteobacterial muconate cycloisomerases (CatBs) and ClcBs have indicated that these Mn2+-containing enzymes share the same mechanism of lactonization but that the increased specificity towards 2-chloromuconate in ClcBs is related to the specific “2-down” orientation (Fig. 3) of the halogenated substrate in the enzyme active site (30). From mutagenesis studies it was inferred that in CatB, Ile54 and Phe327 are critically involved in creating a productive and selective binding pocket for the 2-chloromuconate substrate (30, 42). In ClcB and ClcB2 from R. opacus 1cp, all charged residues critically involved in catalysis and Mn2+ binding are strictly conserved (9, 22). However, in both Rhodococcus enzymes, the more bulky Ile54 (CatB numbering) is replaced by a proline (Pro53 in ClcB2 and Pro54 in ClcB) whereas Phe327 (CatB numbering) is conserved in ClcB2 (Phe325) but replaced by a valine (Val327) in ClcB. This suggests that in ClcB2, Pro53 could provide a larger binding pocket for 2-chloromuconate in the 2-down mode whereas in ClcB, both Pro53 and Val327 might be involved in directing the specificity towards 3-chloromuconate (3-down binding) and 2,4-dichloromuconate.
The enzymatic conversion of 2-fluoromuconate has not been reported before. From the exclusive 3,6-cycloisomerization to 5-fluoromuconolactone, it is likely that 2-fluoromuconate is bound in the 2-down mode and that Lys167 is involved in donating a proton to the exocyclic C5 carbon during the tautomerization of the enol intermediate (30). Thus, in the 2-down binding mode, ClcB2 is capable of reducing the electron-withdrawing effect of the 2-fluoro atom of 2-fluoromuconate as well as of the 2-chloro atom of 2-chloromuconate. With 2-chloro-4-fluoromuconate as a substrate, only 2-chloro-4-fluoromuconolactone was found as a cycloisomerization product. This might be somewhat surprising but fits with the fact that the 3- position of the muconolactone ring usually does not carry a substituent (16). A preferred 2-down orientation would also explain why nearly no conversion is observed with 4-chloro-2-fluoromuconate and 2-chloro-5-fluoromuconate.
Another important feature of ClcB2 is that this enzyme does not catalyze the dehalogenation of 5-halomuconolactones. For ClcB from Ralstonia eutropha, it was proposed that the lactone ring of 5-chloromuconolactone is able to rotate about the 5→4 bond, thereby bringing the acidic C4 proton next to Lys169 and promoting the formation of trans-dienelactone (30). In ClcB2, such rotation might be hindered by residues that interact with the residues in the active site. For R. eutropha ClcB, it was observed that Trp55 is located out of the active site, close to Asn276. However, in CatB from Pseudomonas putida, the equivalent Tyr59 is situated in the active site and cannot adopt the conformation seen for Trp55 because of steric interference from Lys276 (30). Interestingly, Trp55 is conserved in ClcB2 (Trp58) but the Asn276 present in R. eutropha ClcB is replaced by a lysine (Lys274). From this it is tempting to speculate that in ClcB2, Trp58 is located more into the active site, hampering rotation of the lactone ring and thus preventing dehalogenation.
5-Fluoromuconolactone, the product of the ClcB2-mediated conversion of 2-fluoromuconate, was rather stable at pH 7.2 and converted by ClcF in the same way as its chlorinated analog (22), forming cis-dienelactone. Formation of the cis-isomer discriminates the ClcF reaction from the enzymatic dehalogenation of 5-chloromuconolactone by ClcB from Pseudomonas sp. strain B13 and R. eutropha JMP134, which results in the formation of trans-dienelactone (41, 44). On the other hand, it resembles the predominant formation of cis-dienelactone from 5-chloromuconolactones by CatC from R. eutropha JMP134 (28, 29). The difference between ClcF and CatC is that the former enzyme is not able to convert nonsubstituted (+)-muconolactone (22).
Together, the data presented here confirm that the enzymes isolated from R. opacus 1cp grown on 2-chlorophenol belong to a new modified ortho cleavage pathway of 3-chlorocatechol degradation (21). All enzymes of this new pathway are only distantly related to the known chlorocatechol enzymes and appear to represent new evolutionary lines of these activities (22). New is the observation that 2-fluoromuconate is converted by ClcB2 and that ClcF is needed to convert 5-fluoromuconolactone.
Acknowledgments
This work was supported by grant NWO-RF 047-007021 for Dutch-Russian research cooperation and EC grant ICA2-CT-2000-10006.
REFERENCES
- 1.Blasco, R., R.-M. Wittich, M. Mallavarapu, K. N. Timmis, and D. H. Pieper. 1995. From xenobiotic to antibiotic, formation of protoanemonin from 4-chlorocatechol by enzymes of the 3-oxoadipate pathway. J. Biol. Chem. 270:29220-29235. [DOI] [PubMed] [Google Scholar]
- 2.Boersma, M. G., T. Y. Dinarieva, W. J. Middelhoven, W. J. H. van Berkel, J. Doran, J. Vervoort, and I. M. C. M. Rietjens. 1998. 19F nuclear magnetic resonance as a tool to investigate microbial degradation of fluorophenols to fluorocatechols and fluoromuconates. Appl. Environ. Microbiol. 64:1256-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boersma, M. G., I. P. Solyanikova, W. J. H. van Berkel, J. Vervoort, L. A. Golovleva, and I. M. C. M. Rietjens. 2001. 19F NMR metabolomics for the elucidation of microbial degradation pathways of fluorophenols. A review of previously and newly identified resonances and intermediates. J. Ind. Microbiol. Biotechnol. 26:22-34. [DOI] [PubMed] [Google Scholar]
- 4.Bondar, V. S., M. G. Boersma, E. L. Golovlev, J. Vervoort, W. J. H. van Berkel, Z. I. Finkelstein, I. P. Solyanikova, L. A. Golovleva, and I. M. C. M. Rietjens. 1998. 19F NMR study on the biodegradation of fluorophenols by various Rhodococcus species. Biodegradation 9:475-486. [DOI] [PubMed] [Google Scholar]
- 5.Bondar, V. S., M. G. Boersma, W. J. H. van Berkel, Z. I. Finkenstein, E. L. Golovlev, B. P. Baskunov, J. Vervoort, L. A. Golovleva, and I. M. C. M. Rietjens. 1999. Preferential oxidative dehalogenation upon conversion of 2-halophenols by Rhodococcus opacus 1G. FEMS Microbiol. Lett. 181:73-82. [DOI] [PubMed] [Google Scholar]
- 6.Cass, A. E. G., D. W. Ribbons, J. T. Rossiter, and S. R. Williams. 1987. Biotransformation of aromatic compounds: monitoring fluorinated analogues by NMR. FEBS Lett. 220:353-357. [DOI] [PubMed] [Google Scholar]
- 7.Dorn, E., and H.-J. Knackmuss. 1978. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1, 2-dioxygenation of catechol. Biochem. J. 174:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eulberg, D., L. A. Golovleva, and M. Schlömann. 1997. Characterization of catechol catabolic genes from Rhodococcus erythropolis 1CP. J. Bacteriol. 179:370-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eulberg, D., S. Lakner, L. A. Golovleva, and M. Schlömann. 1998. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 180:1072-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferraroni, M., R. Tarifa, A. Scozzafava, I. P. Solyanikova, M. P. Kolomytseva, L. A. Golovleva, and F. Briganti. 2003. Preliminary crystallographic analysis of 3-chlorocatechol 1,2-dioxygenase of a new modified ortho-pathway from the Gram-positive Rhodococcus opacus 1CP grown with 2-chlorophenol. Acta Crystallogr. Sect. D 59:188-190. [DOI] [PubMed] [Google Scholar]
- 11.Finkelstein, Z. I., B. P. Baskunov, M. G. Boersma, J. Vervoort, E. L. Golovlev, W. J. H. van Berkel, L. A. Golovleva, and I. M. C. M. Rietjens. 2000. Identification of fluoropyrogallols as new intermediates in the biotransformation of monofluorophenols in R. opacus 1cp. Appl. Environ. Microbiol. 66:2148-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorlatov, S. N., O. V. Maltseva, V. L. Shevchenko, and L. A. Golovleva. 1989. Degradation of chlorophenols by a culture of Rhodococcus erythropolis. Microbiology (New York) 58:647-651. (Translation of Mikrobiologiya 58:802-806.) [Google Scholar]
- 13.Held, M., W. A. Suske, A. Schmid, K.-H. Engesser, H.-P. E. Kohler, B. Witholt, and M. G. Wubbolts. 1998. Preparative scale production of 3-substituted catechols using a novel monooxygenase from Pseudomonas azelaica HBP1. J. Mol. Catal. B 5:87-93. [Google Scholar]
- 14.Helin, S., P. C. Kahn, B. L. Guha, D. G. Mallows, and A. Goldman. 1995. The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 Å resolution. J. Mol. Biol. 254:918-941. [DOI] [PubMed] [Google Scholar]
- 15.Katz, B. A., D. Ollis, and H. W. Wyckoff. 1985. Low resolution crystal structure of muconolactone isomerase. A decamer with a 5-fold symmetry axis. J. Mol. Biol. 184:311-318. [DOI] [PubMed] [Google Scholar]
- 16.Kaulmann, U., S. R. Kaschabek, and M. Schlömann. 2001. Mechanism of chloride elimination from 3-chloro- and 2, 4-dichloro-cis, cis-muconate: new insight obtained from analysis of muconate cycloisomerase variant CatB-K169A. J. Bacteriol. 183:4551-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhm, A. E., M. Schlömann, H.-J. Knackmuss, and D. H. Pieper. 1990. Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP134. Biochem. J. 266:877-883. [PMC free article] [PubMed] [Google Scholar]
- 18.Maltseva, O. V., I. P. Solyanikova, and L. A. Golovleva. 1994. Chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP. Kinetic and immunochemical comparison with analogous enzymes from Gram-negative strains. Eur. J. Biochem. 226:1053-1061. [DOI] [PubMed] [Google Scholar]
- 19.Maltseva, O. V., I. P. Solyanikova, L. A. Golovleva, M. Schlömann, and H.-J. Knackmuss. 1994. Dienelactone hydrolase from Rhodococcus erythropolis 1CP: purification and properties. Arch. Microbiol. 162:368-374. [Google Scholar]
- 20.Moiseeva, O. V., E. V. Linko, B. P. Baskunov, and L. A. Golovleva. 1999. Degradation of 2-chlorophenol and 3-chlorobenzoate by Rhodococcus opacus 1cp. Microbiology (New York) 68:400-405. [Google Scholar]
- 21.Moiseeva, O. V., O. V. Belova, I. P. Solyanikova, M. Schlömann, and L. A. Golovleva. 2001. Enzymes of a new modified ortho-pathway utilizing 2-chlorophenol in Rhodococcus opacus 1CP. Biochemistry (Moscow) 66:548-555. [DOI] [PubMed] [Google Scholar]
- 22.Moiseeva, O. V., I. P. Solyanikova, S. R. Kaschabek, J. Gröning, M. Thiel, L. A. Golovleva, and M. Schlömann. 2002. A new modified ortho cleavage pathway of 3-chlorocatechol degradation by Rhodococcus opacus 1cp: genetic and biochemical evidence. J. Bacteriol. 184:5282-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neujahr, H. Y., and K. G. Kjellén. 1978. Phenol hydroxylase from yeast. Reaction with phenol derivatives. J. Biol. Chem. 253:8835-8841. [PubMed] [Google Scholar]
- 24.Peelen, S., I. M. C. M. Rietjens, M. G. Boersma, and J. Vervoort. 1995. Conversion of phenol derivatives to hydroxylated products by phenol hydroxylase from Trichosporon cutaneum. A comparison of regioselectivity and rate of conversion with calculated molecular orbital substrate characteristics. Eur. J. Biochem. 227:284-291. [DOI] [PubMed] [Google Scholar]
- 25.Pieper, D. H., K. Stadler-Fritzsche, K.-H. Engesser, and H.-J. Knackmuss. 1993. Metabolism of 2-chloro-4-methylphenoxyacetate by Alcaligenes eutrophus JMP134. Arch. Microbiol. 160:169-178. [DOI] [PubMed] [Google Scholar]
- 26.Pieper, D. H., K. Stadler-Fritzsche, H.-J. Knackmuss, and K. N. Timmis. 1995. Formation of dimethylmuconolactones from dimethylphenols by Alcaligenes eutrophus JMP134. Appl. Environ. Microbiol. 61:2159-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potrawfke, T., J. Armengaud, and R.-M. Wittich. 2001. Chlorocatechols substituted at positions 4 and 5 are substrates of the broad-spectrum chlorocatechol 1,2-dioxygenase of Pseudomonas chlororaphis RW71. J. Bacteriol. 183:997-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prucha, M., A. Peterseim, K. N. Timmis, and D. H. Pieper. 1996. Muconolactone isomerase of the 3-oxoadipate pathway catalyzes dechlorination of 5-chloro-substituted muconolactones. Eur. J. Biochem. 237:350-356. [DOI] [PubMed] [Google Scholar]
- 29.Prucha, M., V. Wray, and D. H. Pieper. 1996. Metabolism of 5-chlorosubstituted muconolactones. Eur. J. Biochem. 237:357-366. [DOI] [PubMed] [Google Scholar]
- 30.Schell, U., S. Helin, T. Kajander, M. Schlömann, and A. Goldman. 1999. Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates. Proteins Struct. Funct. Genet. 34:125-136. [PubMed] [Google Scholar]
- 31.Schlömann, M. 1994. Evolution of chlorocatechol catabolic pathways. Conclusions to be drawn from comparisons of lactone hydrolases. Biodegradation 5:301-321. [DOI] [PubMed] [Google Scholar]
- 32.Schlömann, M., P. Fischer, E. Schmidt, and H.-J. Knackmuss. 1990. Enzymatic formation, stability, and spontaneous reactions of 4-fluoromuconolactone, a metabolite of the bacterial degradation of 4-fluorobenzoate. J. Bacteriol. 172:5119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlömann, M., E. Schmidt, and H.-J. Knackmuss. 1990. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J. Bacteriol. 172:5112-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt, E., G. Remberg, and H.-J. Knackmuss. 1980. Chemical structure and biodegradability of halogenated aromatic compounds. Halogenated muconic acids as intermediates. Biochem. J. 192:331-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, E., and H.-J. Knackmuss. 1980. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem. J. 192:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schreiber, A., M. Hellwig, E. Dorn, W. Reineke, and H.-J. Knackmuss. 1980. Critical reactions in fluorobenzoic acid degradation by Pseudomonas sp. B13. Appl. Environ. Microbiol. 39:58-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sejlitz, T., and H. Y. Neujahr. 1987. Phenol hydroxylase from yeast. A model for phenol binding and improved purification procedure. Eur. J. Biochem. 170:343-349. [DOI] [PubMed] [Google Scholar]
- 38.Solyanikova, I. P., O. V. Maltseva, M. D. Vollmer, L. A. Golovleva, and M. Schlömann. 1995. Characterization of muconate and chloromuconate cycloisomerase from Rhodococcus erythropolis 1CP: indications for functionally convergent evolution among bacterial cycloisomerases. J. Bacteriol. 177:2821-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suske, W. A., M. Held, A. Schmid, T. Fleischmann, M. G. Wubbolts, and H.-P. E. Kohler. 1997. Purification and characterization of 2-hydroxybiphenyl 3-monooxygenase, a novel NADH-dependent, FAD-containing aromatic hydroxylase from Pseudomonas azelaica HBP1. J. Biol. Chem. 272:24257-24265. [DOI] [PubMed] [Google Scholar]
- 40.Suske, W. A., W. J. H. van Berkel, and H.-P. E. Kohler. 1999. Catalytic mechanism of 2-hydroxybiphenyl 3-monooxygenase, a flavoprotein from Pseudomonas azelaica HBP1. J. Biol. Chem. 274:33355-33365. [DOI] [PubMed] [Google Scholar]
- 41.Vollmer, M. D., P. Fischer, H.-J. Knackmuss, and M. Schlömann. 1994. Inability of muconate cycloisomerases to cause dehalogenation during conversion of 2-chloro-cis, cis-muconate. J. Bacteriol. 176:4366-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vollmer, M. D., H. Hoier, H.-J. Hecht, U. Schell, J. Gröning, A. Goldman, and M. Schlömann. 1998. Substrate specificity of and product formation by muconate cycloisomerases: an analysis of wild-type enzymes and engineered variants. Appl. Environ. Microbiol. 64:3290-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vollmer, M. D., U. Schell, V. Seibert, S. Lakner, and M. Schlömann. 1999. Substrate specificities of the chloromuconate cycloisomerases from Pseudomonas sp. B13, Ralstonia eutropha JMP134 and Pseudomonas sp. P51. Appl. Microbiol. Biotechnol. 51:598-605. [DOI] [PubMed] [Google Scholar]
- 44.Vollmer, M. D., and M. Schlömann. 1995. Conversion of 2-chloro-cis, cis-muconate and its metabolites 2-chloro- and 5-chloromuconolactone by chloromuconate cycloisomerases of pJP4 and pAC27. J. Bacteriol. 177:2938-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wray, V. 1983. Fluorine-19 nuclear magnetic resonance spectroscopy, p. 149-191. In G. A. Webb (ed.), Annual Reports on NMR Spectroscopy, vol. 14. Academic Press Inc., London, United Kingdom.