Abstract
Constructed wetlands have been recognized as a removal treatment option for high concentrations of contaminants in agricultural waste before land application. The goal of this study was to characterize microbial composition in two constructed wetlands designed to remove contaminants from dairy washwater. Water samples were collected weekly for 11 months from two wetlands to determine the efficiency of the treatment system in removal of chemical contaminants and total and fecal coliforms. The reduction by the treatment was greatest for biological oxygen demand, suspended solids, chemical oxygen demand, nitrate, and coliforms. There was only moderate removal of total nitrogen and phosphorus. Changes in the total bacterial community and ammonia-oxidizing bacterial composition were examined by using denaturing gradient gel electrophoresis (DGGE) and sequencing of PCR-amplified fragments of the gene carrying the α subunit of the ammonia monooxygenase gene (amoA) recovered from soil samples and DGGE bands. DGGE analysis of wetlands and manure samples revealed that the total bacterial community composition was dominated by bacteria from phylogenetic clusters related to Bacillus, Clostridium, Mycoplasma, Eubacterium, and Proteobacteria originally retrieved from the gastrointestinal tracts of mammals. The population of ammonia-oxidizing bacteria showed a higher percentage of Nitrosospira-like sequences from the wetland samples, while a higher percentage of Nitrosomonas-like sequences from manure, feces, raw washwater, and facultative pond was found. These results show that the wetland system is a natural process dependent upon the development of healthy microbial communities for optimal wastewater treatment.
Although natural wetlands have existed for ages across the globe, the use of constructed or artificial wetlands built for the improvement of water quality is a relatively new concept of the last two decades. Wastewater from intensive agricultural activities (cattle feedlots and dairies) typically have significantly higher concentrations of organic matter and nutrients than treated municipal effluent. The high pollutant loads being generated pose particular problems and challenges for the dairy industry, since high concentrations of nutrients can contribute to water management problems if wastes are allowed to discharge directly into receiving waters. For this reason agricultural wastes must be treated prior to disposal. Constructed wetlands in association with stabilization ponds have been suggested as a potential treatment option prior to land application. The extent of water treatment in natural or constructed wetlands depends upon the wetland design, microbial community, and types of plants involved. The bulk of the water quality improvement in natural and constructed wetlands is due to bacteria (2).
It has long been recognized that certain microbial groups in animal waste are responsible for breaking down various organic compounds and for the suppression of pathogens in waste. However, very little work has been done on the functional composition of the different bacterial groups in a subsurface flow wetland. Gersberg et al. (2) explored many of the biological parameters that affected the performance of horizontal subsurface flow wetlands. The authors discussed the important roles of aquatic plants, such as bulrushes, in translocating oxygen from the upper parts of the plant to the ends of the roots. They concluded that nitrifying bacterial communities may develop around this microaerobic zone and convert ammonium to nitrate. As the water flows into the anaerobic zone, the denitrifying bacteria convert nitrate to nitrogen gas. Therefore, an aerobic and anaerobic regime develops that contains complementary microbial communities. These communities can nitrify ammonium-laden water under aerobic conditions or convert the nitrate to nitrogen gas by denitrifying bacteria under anaerobic conditions. One advantage of this system over a surface flow system is that the water is being exposed to a far greater surface area of bacterial biofilms, resulting in a higher level of nutrient removal over a shorter time period.
The diversity of microorganisms in the wetland environment may be critical for the proper functioning and maintenance of the system. One such group of bacteria involved in different biological and chemical transformations of organic compounds in wastewater are the chemolithotrophic ammonia-oxidizing bacteria. These bacteria are responsible for the first, rate-limiting step in nitrification in which ammonium (NH4) is transformed to nitrite (NO2) and to nitrate (NO3) during natural nitrogen cycling (7, 15). Oved et al. (13) studied the effects of urban sewage effluent on the community composition and function of ammonia-oxidizing bacteria in soil by using denaturing gradient gel electrophoresis (DGGE) of PCR-amplified fragments of the amoA gene. They found a significant and consistent shift in the population composition of ammonia-oxidizing bacteria in soil irrigated with effluent dominated by Nitrosomonas-like populations, while noneffluent soil was dominated by Nitrosospira-like populations.
The use of the functional gene carrying the α subunit of amoA has been shown to be a good molecular marker for ammonia-oxidizing bacteria in soil (18, 22). The functional gene has been shown to detect only Nitrosospira in agricultural soils. More diverse species within this genus were determined by using the amoA gene rather than the 16S rRNA gene (4). Other investigators have also demonstrated the successful use of a specific set of PCR primers to amplify a fragment of amoA from a variety of pure cultures of ammonia-oxidizing bacteria from environmental samples (3, 9, 17, 18, 22).
This study examined the composition of the general bacterial population and ammonia-oxidizing bacteria in manure, feces, and dairy washwater prior to and after treatment in subsurface wetlands. The first objective was to characterize microbial communities in constructed wetland wastewater and to determine how the composition of the community may influence the final wastewater effluent quality. The second objective was to determine the effectiveness of the wetland treatment technology in reducing contaminants in dairy waste effluent. Present dairy washwater management practices involve storing washwater in ponds or spraying raw washwater onto crops and/or disposal land. Storage in lagoons affects the groundwater quality through the percolation of washwater, contributing significant amounts of nutrients to the groundwater. During a significant storm event, overflow from the storage lagoons also may occur, posing a threat to surface water quality from the high levels of organic material and pathogens present in washwater.
MATERIALS AND METHODS
Wetland design and sampling regime.
The wetland design consisted of two subsurface horizontal flow beds (60 m by 10 m by 1 m) operating in parallel and a raw and facultative pond for central collection of the washwater prior to treatment. Coarse and finer gravel were placed up to a depth of 1 m. The gravel particles were graded across the length of the flow bed. The first 6 m of the wetlands contained coarse and fine gravel to act as a sink for particulate matter. The next 6 m contained a bed of reeds (Phragmites communus) with a shallow dense root system. The root system acts as a physical filter to remove suspended matter. The remaining 48 m of the wetland contained bulrush (Scirpus validus).
Wetland 1 was an end-loading design. The washwater entered through multiple inlets onto the coarse gravel, reeds, and bulrush and then drained into the collection box at the end of the basin. Wetland 2 was a side-loading basin. Washwater entered through multiple inlets along both sides of the wetland and passed through a narrow gravel bed containing reeds and bulrush before being collected through a perforated pipe along the center of the wetlands that drained into the collection box. The combination of open gravel and specific plants was used for the initial removal of suspended solids and subsequent nitrification of the wastewater in the microaerobic zone surrounding the roots of the bulrushes. The overall design of the wetlands was based on studies carried out by Gersberg et al. (2), with some modifications for higher biochemical oxygen demand (BOD), nutrient levels, and suspended solids.
Monthly samples were collected from six locations: wastewater lagoon, facultative pond, and wetlands 1 and 2 influents and effluents. Fresh cow and calf fecal samples and manure samples were collected for microbial analysis. Manure samples were taken from piles of fecal materials that had been deposited for about 2 weeks. Weekly water samples were collected for chemical analysis and determination of the quality of the final effluent water. All samples were collected between December 2000 and September 2001. Weekly samples were analyzed for total Kjeldahl nitrogen (TKN), ammonium nitrogen (NH4-N), nitrate-nitrogen (NO3-N), BOD, total suspended solids (TSS), orthophosphate (PO4-P), pH, potassium (K+), sodium (Na+), chloride (Cl−), total dissolved solids (TDS), chemical oxygen demand (COD), and fecal and total coliform bacteria. All analyses were done with protocols from the Standard Methods of the American Public Health Association (1).
DNA extraction and purification from soil and effluent.
Total bacterial DNA was extracted from 500 mg of fecal or manure samples and from 250 mg of pellets of concentrated effluent sample centrifuged at 3,000 × g for 10 min. DNA was extracted by using UltraClean fecal and water DNA kits (MO BIO, Inc., Solana Beach, Calif.) according to the manufacturer's protocol with slight modifications.
PCR primers and DGGE analysis of total bacterial community.
PCR was performed with about 50 ng of template DNA with the primers PRB 338f and PRUN 518r, located at the V3 region of the 16S rRNA genes of bacterioplankton (14) to assess bacterial community diversity. PCR mixtures for the bacterial 16S rRNA PCR DGGE sequence amplification contained 10 pmol of each primer, Ready-To-Go PCR beads from Amersham-Pharmacia Biotech (Piscataway, N.J.), and sterile distilled water in a final volume of 25 μl; PCR conditions used were those described by Ibekwe et al. (5).
DGGE was performed with 8% (wt/vol) acrylamide gels containing a linear chemical gradient ranging from 30 to 70% denaturant with 100% defined as 7 M urea and 40% formamide. Gels were run for 3 h at 200 V with the Dcode Universal Mutation System (Bio-Rad Laboratories, Hercules, Calif.). DNA was visualized after ethidium bromide staining by UV transillumination and was photographed with a Polaroid camera. Major bands were excised for identification of bacterial species. Bands were placed into sterilized vials with 20 μl of sterilized, distilled water and were stored overnight at 4°C to allow the DNA to passively diffuse out of the gel strips. Ten microliters of eluted DNA was used as the DNA template with the eubacteria primers. DNA was cloned by using the pGEM-T Easy vector system (Promega, Madison, Wis.) and was transformed into Escherichia coli JM109. Isolation of plasmids from E. coli was performed by using the Qiagen plasmid mini kit (Valencia, Calif.). The purified plasmids were sequenced with the ABI PRISM dye terminator cycle sequencing kit with AmpliTaq DNA polymerase, FS (Applied Biosystems, Foster City, Calif.).
DGGE and sequence analyses of ammonia oxidizers.
The diversity of the ammonia-oxidizing bacteria in the samples was performed with amoA primers targeting a partial stretch of the genes that encode the active-site polypeptide of ammonia monooxygenase (18). Products with the expected size of 491 bp were excised from the gel and were purified with a QiaexII gel extraction kit (Qiagen). Purified fragments were cloned as described above. A second amplification was done by a seminested PCR using the amoA primers for DGGE analysis. For the second assay, a GC clamp (5′ CCGCCGCGCGGCGGGCGGGGCGGGGGCACGGGG-3′) was attached to the forward primer to increase the DGGE gel separation (11). PCRs contained Ready-To-Go PCR beads, 50 pmol of each primer, template DNA, and sterile distilled water to a total volume of 50 μl. The reaction conditions were 94°C for 2 min, 30 cycles of 92°C for 60 s, 60°C for 60 s, and 68°C for 45 s, followed by a final single extension at 72°C for 5 min. DGGE was performed as described above, except a linear chemical gradient ranging from 30 to 60% denaturant was used. The DNA was visualized, the major bands were excised, and the DNA was diffused as described above. A total of 10 μl of eluted DNA was used as template with amoA primers without the GC clamp for reamplification. The products were purified with the QIAquick PCR purification kit and were cloned as described previously.
Statistical analysis and analysis of DGGE bands.
Data analyses of nutrient concentrations were performed by using SAS (20). Analysis of variances, means, and standard deviations for the individual nutrient in triplicate samples were determined (P = 0.05) to compare the concentration of each nutrient in each sample over the sampling period.
DNA fingerprints obtained from the 16S rRNA banding patterns on the DGGE gels were photographed and digitized by using an ImageMaster Labscan (Amersham-Pharmacia Biotech, Uppsala, Sweden). The lanes were normalized to contain the same amount of total signal after background subtraction. The gel images were straightened and aligned by using ImageMaster 1D Elite 3.01 (Amersham-Pharmacia Biotech) and were analyzed to give a densitometric curve for each gel. Band positions were converted to Rf values between 0 and 1, and profile similarity was calculated by determining Dice's coefficient for the total number of lane patterns. Cluster analyses of the lane patterns were constructed by using the Minitab statistical software (version 13; State College, Pa).
Phylogenetic analysis.
Sequence analyses were done by using the BLAST database (National Center for Biotechnology Information; www.ncbi.nlm.nih.gov). Partial amoA gene sequences were aligned with parts from the complete amoA gene sequences of ammonia-oxidizing bacteria obtained from the BLAST gene bank. Sequence alignments were performed by using the PILEUP program from the University of California-Riverside Genetics Computer Group (GCG programs). Matrices of evolutionary distances were computed by using the Phylip program with the Jukes-Cantor model (8). Phylogenetic trees were constructed and checked by bootstrap analysis (100 data sets) by using the program SEQBOOT. Bootstrap values represented the frequency of resampling that supported a specific branching pattern.
RESULTS
Wastewater constituents.
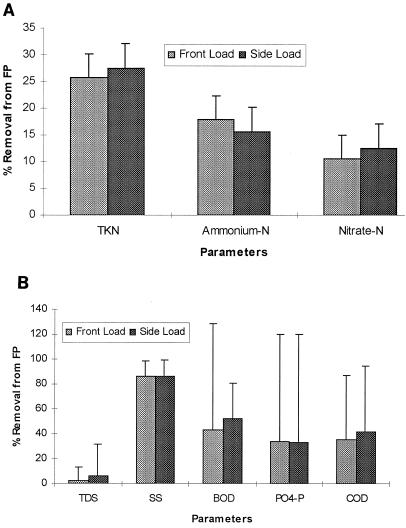
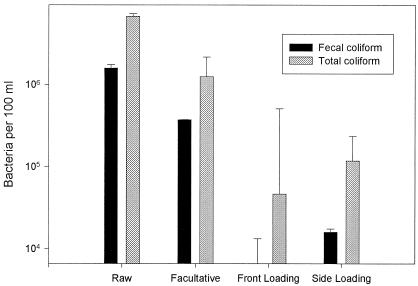
Wastewater samples from wetlands were analyzed for TKN, PO4-P, K, NH4-N, NO3-N, BOD, TDS, pH, TSS, Na+, Cl−, COD, and coliform bacteria. The TKN levels in raw washwater were 150 to 250 mg liter−1 with ammonium levels of 100 to 200 mg liter−1 and were reduced 25 and 16% through wetlands treatment, respectively (Fig. 1A). The total nitrogen in the raw washwater was removed on an average of 25%. Although the BOD of the raw washwater ranged from 50 to 350 mg liter−1 and steadily increased as the ambient temperature increased in the spring and summer, reductions through wetlands treatment remained constant at an average of 73% (Fig. 1B). The suspended solids ranged widely (100 to 1,100 mg liter−1) in the raw water, while levels in wetlands effluent were consistently lower, with an average drop of 91%. COD data were collected as well in order to determine the amount of oxidizable organic and inorganic matter in the raw and treated washwater. There was a 38% drop in COD and a 33% decrease in orthophosphate (Fig. 1B). The pH remained close to neutral (range of 7 to 8) throughout the system. The removal efficiencies for TSS, BOD, and total and fecal coliform bacteria were similar to those described by Gersberg et al. (2) in wetlands treatment of domestic wastewater. Total and fecal coliform concentrations followed the same trend of total E. coli from the same wetlands (6). Wetlands treatment achieved a 2 log (99%) decrease in total coliform bacteria and a 3 log (99.9%) decrease in fecal coliform bacteria between the raw washwater pond and the average wetland effluent (Fig. 2). This is significant both from the standpoint of surface runoff as well as potential airborne pathogens released during the spray irrigation of raw washwater on disposal lands. There were no significant differences in the treatment efficiencies between the front-loading and side-loading wetland systems, as illustrated by the removal rates noted in the water quality above (Fig. 1 and 2). However, lower removal efficiencies were achieved for total nitrogen and phosphate, which may be attributable to higher levels in the influent to the wetland system. The greatest removal seen from the wetlands treatment was for BOD, suspended solids, COD, and coliforms.
FIG. 1.
Percent removal of major chemical components in wetland. Analyses were done on weekly samples. Results are presented as an average removal rate for the 11 months that samples were collected. FP, facultative pond.
FIG. 2.
Changes in fecal and total coliform concentrations in and out of wetlands. Values shown here are based on the geometric mean from four samples averaged from each of the sampling months. Concentration of bacteria was determined from 100 ml of samples taken at different locations in the wetlands.
Except for a few variations, there was little change between the influent and effluent concentrations of the conservative ions (Na+, Cl−, K+; data not shown). These ions are conservative constituents that vary little, if any, due to biological activity. Because of their conservative nature, they are useful tools for determining retention and travel times of the washwater through the wetlands system. Residence time is the average length of time washwater remains in the wetlands from the time of introduction of the raw water to the discharge of treated effluent. Residence time is inversely related to the flow velocity, such that higher flow velocities correspond to a shorter residence time, all other factors being equal. Actual flow velocity through the wetlands may vary due to a number of factors, including the flow rate of washwater into the wetlands, permeability of the gravel and amount of biofilm growth on the gravel, density of plant roots, and deposition of sediment in the gravel pore space.
Values for the conservative ions determined for wetland influent and treated effluent were used in cross-correlation analyses to calculate the residence time in the wetlands (12). Based on the variation in concentrations of these constituents, the residence time was estimated at approximately 7 days. Cross-correlation analyses were possible based on the variations or spikes in the concentrations of ions that pulsed through the wetlands. The variations in ion concentrations were observable in both the wetlands influent and effluent, allowing for estimation of the residence time based on the time difference from when the spike occurred at the influent and at the effluent.
Bacterial community composition in manure and wetlands.
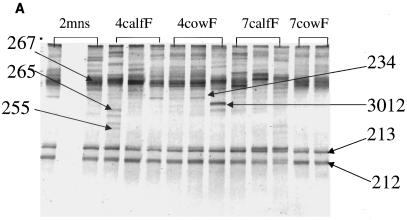
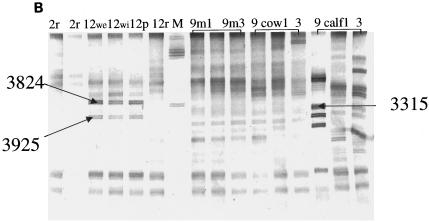
Analysis of the bacterial community composition by PCR-DGGE was performed on all manure, fecal, and wetland samples collected from December 2000 to September 2001. Banding patterns for the 16S rRNA DGGE-PCR amplicons are presented in Fig. 3A to D. Gels for other sampling times showed similar DGGE patterns (data not shown), while DGGE banding patterns from cow and calf feces were significantly different from each other. The gel images consist of Fig. 3A and part of 3B for fecal and manure samples. The number of bands per lane varied from 10 to 22. Some differences were noted in band position, intensity, and number of bands present in the fecal bacterial DGGE patterns throughout the collection period. Each calf or cow sample had its own unique profile, indicating variation within and between animals, as well as sampling time. However, the DGGE analyses from manure samples demonstrated relatively stable banding patterns throughout the collection period (Fig. 3A and B). Two bands (212 and 213) were common in all samples. The Image Master ID was used to create a database set for these samples. The set contained a total of 46 unique bands based on a synthetic band from the software as the standard band. The bands were found in different combinations among the DGGE banding patterns. Occurrence of different bands from the four gels showed that 24% of the bands occurred in more than 88% of the samples. Pattern comparisons were made among the four gels by using Dice's coefficient. From the database analysis, the Dice values indicated that all samples shared a large portion of the band set (data not shown), while a few had unique bands as indicated by the banding patterns shown in Fig. 3A to D.
FIG. 3.
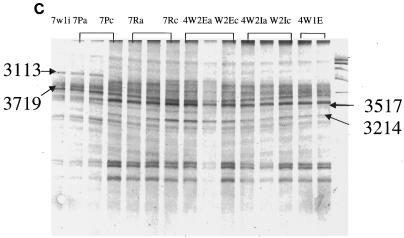
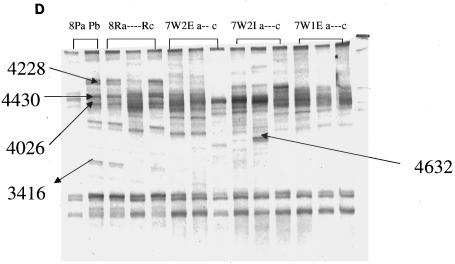
DGGE analysis of 16S rRNA fragments of total bacterial population from fecal and manure samples. (A) Gel image from samples collected from manure in February (2mns), calf feces in April (4calfF), cow feces in April (4cowF), calf feces in July (7calfF), and cow feces in July (7cowF). (B) Gel image from samples collected from raw pond in February (2r), collected in December from wetland effluent (we), wetland influent (wi), facultative pond (p), and raw pond (r). Manure (9m1-9m3) and fecal samples were also collected in September from cow (9 cow1-3) and from calf (9 calf 1-3). (C) Numbers before letters represent months that samples were taken; W1 and W2, wetlands 1 and 2; I, influent; E, effluent. P and R represent facultative pond and raw pond, respectively. (D) Numbers before letters represent months that samples were taken. Amplified products were separated on a gradient gel of 30 to 70% denaturant. All labeled bands were excised from the gel, reamplified, and subjected to sequence analysis. These reamplification products were cloned and screened as described in the text.
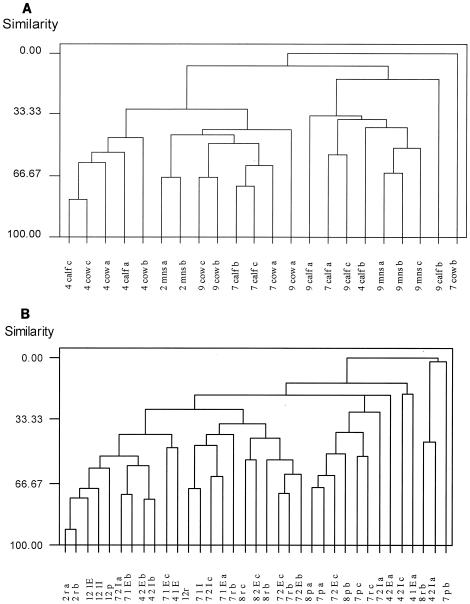
Since distinct banding patterns were observed within different sources of fecal samples and manure and also in sampling times, cluster analysis was performed by using the UPGMA (unweighted pair-group method using arithmetic average) algorithm. Figure 4A is an example of the cluster pattern formed when all fecal samples collected from cows and calves were analyzed. The samples separated into two clusters based on time, and within each cluster based on age. The manure samples separated into two different clusters based on time. The results show individual pattern stability and agree with the work of Zoetendal et al. (23), who reported individual pattern stability after analysis of four human fecal samples over a 6-month period using temperature gradient gel electrophoresis to determine bacterial population profiles. The relative stability and individuality of the patterns indicated that each cow or calf harbored a specific and unique fecal bacterial community. Zoetendal et al. (23) concluded after examination of several unrelated individuals of different ages and different dietary preferences that the reasons for pattern uniqueness were likely to be found in host factors. The calves in the present experiment were about 2 months old and were fed the same diet, yet banding patterns were still unique for each calf. In another study, Simpson et al. (21) demonstrated the stability of DGGE banding profiles from fecal samples collected from piglets under different diets. The DGGE banding profiles indicated that each individual maintained a unique fecal bacterial population that was stable over time, suggesting a strong host influence. Individual DGGE patterns could be separated into distinct time-dependent clusters.
FIG. 4.
Cluster analysis of microbial communities generated by the analysis of DGGE 16S rRNA PCR patterns representing the genetic similarity of the microbial community profiles obtained by PCR-DGGE. Data in panel A are derived from the gels shown in Fig. 3A and B (lanes 9m-9 calf1 3). Symbols are as shown in gels. (B) Data in panel B are derived from the gels shown in Fig. 3B (lanes 2r-12r), C, and D.
PCR-amplifiable DNA was recovered from all wetland samples as well as from the corresponding facultative pond and raw pond. Samples collected from different sampling sites during the year showed little variation in banding patterns when analyzed by PCR-DGGE (Fig. 3C and D). In all, 15 to 23 bands of various intensities were detected per sample, with about 12 bands shared among all samples. The most obvious bands in these samples were bands 212, 213, 3214, 3517, and 3719, and they were present in all the wetland samples. Clustering of the profiles revealed that all profiles were similar, with no clear distinction between time and sampling locations (Fig. 4B). One possible explanation may be that from April until September the microbial community composition in the wetlands had been well established.
Analysis of predominant bacterial species by PCR-DGGE.
The analysis of predominant bacterial species was carried out with fecal, manure, and wetland samples collected during the study. Bands selected for analysis are shown in Fig. 3A to D. Table 1 shows the prominent bands recovered from the DGGE gel. In Fig. 3A and B all fecal and manure samples generated very complex and unique banding patterns, except for the manure samples that showed very stable banding patterns. In addition to the two prominent bands (212 and 213) derived from fecal and manure samples and present in all other samples, two other bands (4026 and 4228) derived from the raw waste and facultative ponds, respectively, were also found in all samples. The derived sequences from these bands confirmed 212 to be 100% similar to salt marsh clone LCP-89 and 213 to be 100% similar to uncultured Caulerpa texifolia bacterium. Bands 4026 and 4228 derived from the wetlands were 100% similar to uncultured delta proteobacterium and uncultured bacterium MK17, respectively. Most of the excised bands produced legible DNA sequences; the only exception was band 267. This rather diffuse band may have contained DNA from more than one bacteria species. The majority of the DGGE bands showed the highest levels of identity to clones recovered from the gastrointestinal tract. Most of the strains from this study had 99 to 100% similarity levels with sequences from the database (Table 1). Most of the prominent bands retrieved from the fecal and wetland samples (234, 255, 265, and 267) were 99 or 100% similar to bacterial sequences identified by Leser et al. (10) from pig gastrointestinal tract. Sequence identity of most of the bands in Table 1 showed that most of the clones were closely associated with gastrointestinal bacteria from different species or bacteria from sludge or wastewater environments.
TABLE 1.
Sequence analysis of bands excised from DGGE gels derived from bacterial 16S rRNA extracted from feces, manure, and wastewater from wetland samples
| Band (sample source) | Related bacterial sequence | % Similarity | Sample present | Accession no. |
|---|---|---|---|---|
| 212 (calf feces) | Saltmarsh clone LCP-89 | 100 | All | AF286032 |
| 213 (manure) | Uncultured Caulerpa texifolia bacterium | 100 | All | AF259615 |
| 234 (cow feces) | Uncultured bacterium p-2173-s959-3 | 100 | Manure and feces | AF371881 |
| 255 (calf feces) | Uncultured bacterium p-1628-c5 | 100 | Manure and feces | AF317920 |
| 265 (manure) | Uncultured bacterium p-43-a5 | 99 | Manure and feces | AF371601 |
| 267 (calf feces) | Uncultured bacterium p-43-a5 | 99 | Manure and feces | AF371601 |
| 3012 (cow feces) | Unidentified rumen bacterium 30-2 | 100 | Manure and feces | AF018566 |
| 3113 (wetland 1 influent) | E. coli | 99 | Wetland | AY078065 |
| 3214 (facultative pond) | Catenibacterium mitsuokai | 100 | Wetland | AB030225 |
| 3315 (calf feces) | Uncultured bacterium GOUTA14 | 100 | Manure and feces | AY050585 |
| 3416 (facultative pond) | Mycoplasma caviae | 100 | Wetland | AF221111 |
| 3517 (wetland 2 influent) | Uncultured bacterium | 100 | Wetland | AF339858 |
| 3719 (wetland 2 influent) | Uncultured Bay of Fundy bacterium | 100 | Wetland | AF323555 |
| 3824 (raw waste pond) | Uncultured marine bacterium | 100 | Wetland | AJ298382 |
| 3925 (tacultative pond) | Unidentified eubacterium | 100 | Wetland | AF010082 |
| 4026 (raw waste pond) | Uncultured delta proteobacterium | 100 | All | AF369724 |
| 4228 (facultative pond) | Uncultured bacterium MK17 | 100 | All | AF087080 |
| 4430 (facultative pond) | Uncultured bacterium p-478-02 | 100 | Wetland | AF371691 |
| 4632 (wetland 2 influent) | Uncultured delta proteobacterium | 100 | Wetland | AF354151 |
Ammonia-oxidizing bacteria composition in fecal and wetland samples by DGGE of amoA genes.
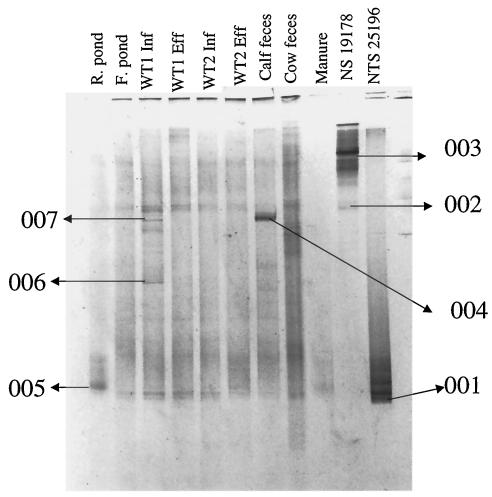
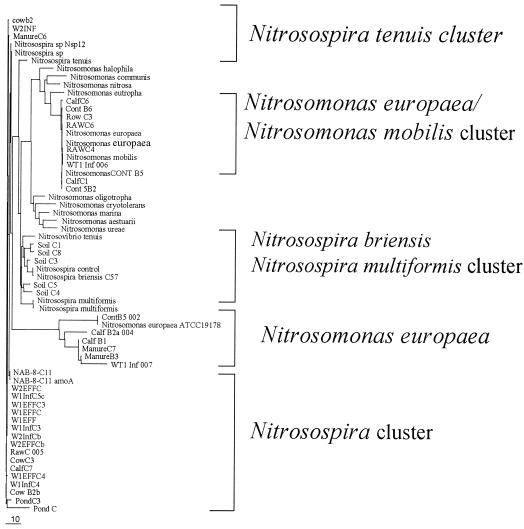
The roles of some members of the wetland and fecal bacterial community structure were further explored by examining the functional gene encoding the α subunit of ammonia monooxygenase (amoA). Results obtained from the DGGE analysis of the ammonia-oxidizing bacteria were very reproducible across the sampling points throughout the year (data not shown). DNA over the sampling time was pooled, and a total of nine samples and two positive controls were used for the DGGE analysis (Fig. 5). DGGE patterns from the sampling points showed four distinct dominant banding patterns from the nine samples and the controls. The two control samples migrated to the opposite end of the gel, with Nitrosomonas sp. strain ATCC 19178 migrating to the top portion of the gel and Nitrosospira sp. strain ATCC 25196 migrating to the lower portion of the gel. Sequence analysis also revealed that the bands in the lower third of the DGGE gel were phylogenetically related to Nitrosospira species, while the bands in the upper third of the gel were phylogenetically related to Nitrosomonas species (Fig. 5 and 6). However, most of the bands in the DGGE gel phylogenetically clustered with Nitrosomonas sp. strain ATCC 19178 (002, 004, and 007). These results are in agreement with previous results from DGGE analysis (13) using the same primers. Two out of three Nitrosomonas-like sequences obtained from the wetland samples were grouped into one cluster with Nitrosomonas europaea. The only Nitrosomonas-like sequence which did not cluster within this group was band 006, obtained from the wetland influent, which grouped with the N. mobilis cluster (Fig. 6). All sequences related to Nitrosomonas obtained from the wetland and manure samples belonged to the N. europaea-N. mobilis cluster (Fig. 6). None of the samples obtained from the wetlands and manure clustered with the Nitrosospira control sample (Fig. 6). Band 005 clustered with the soil isolate Nitrosospira sp. strain NAB-8-C11.
FIG. 5.
DGGE analysis of amoA fragments obtained from feces, manure, and wetland samples. Similar samples were pooled from all sampling dates (December 2000 to September 2001) for analysis. Nine samples were used for the final analysis plus the control samples for Nitrosomonas sp. strain ATCC (NS 19178) and Nitrosospira sp. strain ATCC (NTS 25196). F and R represent facultative pond and raw pond, respectively; WT1 and WT2, wetland 1 and 2, respectively; Inf, influent; Eff, effluent.
FIG. 6.
Phylogenetic tree constructed for partial amoA gene sequences and aligned by the GCG program from the University of California, Riverside, genetic group. The tree was produced by using a neighbor-joining algorithm. The tree shows the relationship between ammonia-oxidizing bacteria from wetland, manure, and fecal samples and sequences obtained from agricultural soils. The bar indicates an estimated 10% sequence divergence. The cluster nomenclature is that of Purkhold et al. (17).
Ammonia-oxidizing bacteria sequences from fecal and wetland samples by cloning of amoA genes.
Cloned beta-subclass ammonia-oxidizing bacteria composition was performed in all the wetland, manure, and fecal samples pooled from all the samples collected throughout the sampling period. The amoA PCR products (primers amoA-1F and amoA-2R) retrieved from the samples were used for the generation of amoA libraries. A total of 30 clones were randomly selected and sequenced. Phylogenetic analysis demonstrated that all clones contained amoA sequences affiliated with the beta subclass ammonia-oxidizing bacteria (Fig. 6). Nitrosospira-related sequences were detected in all the samples analyzed in this study, whereas Nitrosomonas-related sequences were detected in manure, feces, and the raw wastewater holding pond, and fewer sequences were detected in the wetland samples. Most of these samples clustered with N. europaea and N. mobilis. (Fig. 6). As seen in Fig. 6, the Nitrosomonas-related sequences from the environmental samples clustered into two groups closely related to N. europaea ATCC 19178, N. europaea, and N. mobilis.
The analyses of Nitrosospira-related sequences showed that the majority of the wetland samples were phylogenetically related to Nitrosospira. These samples clustered into two main groups, with most of them closely related to uncultured bacterium NAB-8-C11. The second group clustered with Nitrosospira sp. strain Nsp12 and Nitrosospira tenuis. To determine how closely related the wetland and agricultural soil clones were, five soil clones from eastern Washington state soils (4) were included in the analysis. The results showed that none of the wetland Nitrosospira sequences were in the same cluster as the sequences from agricultural soils (Fig. 6). All the sequences from agricultural soil were closely related to Nitrosospira briensis and Nitrosospira multiformis, which are known to be some of the most common soil ammonia-oxidizing bacteria (16). The Nitrosospira-like sequences identified in the wetlands, manure, and fecal samples in this study were clearly different from the Nitrosospira-like sequences identified in the agricultural soils under different management regimes.
DISCUSSION
One of the major objectives for the establishment of constructed wetlands is to use the final effluent for irrigation and/or for disposal into other bodies of water. Data from this study showed that there is a diverse community of ammonia-oxidizing bacteria that may be involved in the nitrification process in the wetlands, ultimately influencing the final effluent water quality. In this study, the application of total bacterial DGGE was used to study the diversity of dominant bacterial populations in fecal, manure, and wetland samples. The predominant ammonia-oxidizing bacteria populations identified in this study, Nitrosospira-like species and Nitrosomonas-like species, were readily separated by DGGE analyses. All analyzed amoA bands which migrated to the lower third of the DGGE gel were determined to be phylogenetically related to Nitrosospira species, while all amoA bands migrating to the upper third of the gel were phylogenetically associated with Nitrosomonas species. The cloning experiment showed high diversity of ammonia-oxidizing bacteria in the wetlands, including most of the effluent and influent samples from the facultative pond, raw pond manure, and fecal samples. Clearly, there is a significant gradient of ammonia-oxidizing bacteria diversity in the wetland system. This may be due to the stratification that exists in unmixed manure, raw, and pond samples. As the waste is degraded it changes in composition as it passes through the wetland system. The waste entering the raw and facultative ponds contains a high amount of organic matter, and as the waste is degraded, the level of organic matter will be reduced and, concomitantly, oxygen will be consumed. In the presence of high levels of organic matter, heterotrophs can outcompete autotrophic ammonia oxidizers for oxygen (16) and ammonia (19). As the organic matter declines, the number of heterotrophs decreases, and consequently, ammonia-oxidizing bacteria can proliferate. These results are in agreement with the work of Rowan et al. (19), who showed higher diversity of ammonia-oxidizing bacteria at the base of the secondary filter bed than in the primary filter bed. Ammonia-oxidizing bacteria were not quantified in this study, but the data suggest that the wetlands remove more ammonium than the facultative pond and ammonium concentration decreased significantly from the wetland influent to the wetland effluent (Fig. 1A). This was reflected in the extent of ammonia-oxidizing bacteria diversity within the wetlands. The high diversity of ammonia-oxidizing bacteria in this system does not necessarily establish a direct link to ammonium removal, since actual quantification of ammonia-oxidizing bacteria in the system was not determined.
The wetlands had greater ammonia-oxidizing bacteria diversity (especially Nitrosospira sp.) than the other components studied. It has been suggested that the level of ammonia-oxidizing bacteria diversity within a wastewater treatment facility has a major influence on process stability (19); the greater the diversity, the more stable the process. These authors concluded that a treatment plant with greater diversity would cope better with changing conditions. A reduction in the numbers of one organism may not mean process failure, as other organisms better adapted to the new conditions proliferate, resulting in a more functionally stable system. The results from this study support this notion, since a more diverse population of both Nitrosomonas and Nitrosospira was identified, and Nitrosospira was inherently more diversified in the wetlands. If diversity does play a major role in wastewater treatment in the constructed wetlands, the engineering of wetlands to have higher diversity may make processes such as nitrification and denitrification more efficient.
In this study, the wetland effluent is more suitable for on-site reuse and will reduce the amount of contaminants entering groundwater supplies as a result of percolation of washwater stored in ponds and sprayed on disposal lands. In addition, the removal of solids in the irrigation water prevents the formation of a hardened manure crust on the disposal surface, which reduces surface runoff. On-site reuse of the wetland-treated washwater through spray irrigation on disposal lands is optimized by the reduction of organic loads and solids in washwater, allowing more water of higher quality to be used for the irrigation of pastures as it has been adopted in this farm. Improved washwater quality also reduces the buildup of solids in irrigation lines and decreases the need for pump and line service by the dairies.
The removal of the main pollutants from the dairy washwater will have a beneficial effect on the surface and groundwater in the Chino Basin. In turn, this process will benefit the quality of water leading into the Santa Ana River and the Orange County groundwater basin. The wetland project will serve as an innovative model for waste management for the dairy industry and other confined-animal facilities. Providing a cost-effective, low-maintenance process that can be independently built and managed, wetland treatment systems throughout the Chino basin will have a significant long-term effect on the quality of the ground and surface water supply.
.
Acknowledgments
We thank Katherine O'Connor, project director of the Chino dairy wetlands, and the members of the staff of the Orange County Water District who were instrumental in the construction of the wetlands, especially the Technical Advisory Committee.
This research was supported by the 206 Manure and By-Product Utilization Project of the U.S. Department of Agriculture—Agricultural Research Service.
REFERENCES
- 1.American Public Health Association. 1995. Standard methods for the examination of the examination of water and wastewater, 19th ed. American Public Health Association, Washington, D.C.
- 2.Gersberg, R. M., B. V. Elkins, S. R. Lyon, and C. R. Goldman. 1986. Role of aquatic plants in wastewater treatment by artificial wetlands. Water Res. 20:363-368. [Google Scholar]
- 3.Horz, H. P., J. H. Rotthauwe, T. Lukow, and W. Liesack. 2000. Identification of major subgroups of ammonia-oxidizing bacteria in environmental samples by T-RFLP analysis of amoA PCR products. J. Microbiol. Methods 39:197-204. [DOI] [PubMed] [Google Scholar]
- 4.Ibekwe, A. M., A. C. Kennedy, P. S. Frohne, S. K. Papiernik, C.-H. Yang, and D. E. Crowley. 2002. Microbial diversity along a transect of agronomic zones. FEMS Microbiol. Ecol. 39:183-191. [DOI] [PubMed] [Google Scholar]
- 5.Ibekwe, A. M., S. K. Papiernik, J. Gan, S. R. Yates, C.-H. Yang, and D. E. Crowley. 2001. Impact of fumigants on soil microbial communities. Appl. Environ. Microbiol. 67:3245-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibekwe, A. M., P. M. Watt, C. M. Grieve, V. K. Sharma, and S. R. Lyon. 2002. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands. Appl. Environ. Microbiol. 68:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jetten, M. S., S. Logemann, G. Muyzer, L. A. Robertson, S. de Vries, M. C. van Loosdrecht, and J. G. Kuenen. 1997. Novel principles in the microbial conversion of nitrogen compounds. Antonie Leeuwenhoek 71:75-93. [DOI] [PubMed] [Google Scholar]
- 8.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 2113-2132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 9.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Møller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orange County Water District. 2002. Dairy wetlands washwater treatment demonstration project. Final report. Orange County Water District, Fountain Valley, Calif.
- 13.Oved, T., A. Shaviv, T. Goldrath, R. T. Mandelbaum, and D. Minz. 2001. Influence of effluent irrigation on community composition and function of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 67:3426-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Øvreås, L., L. Forney, F. L. Daae, and T. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Sælenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul, E. A., and F. E. Clark. 1989. Soil microbiology and biochemistry. Academic Press, Inc., San Diego, Calif.
- 16.Prosser, J. I. 1986. Nitrification. IRL Press, Oxford, United Kingdom.
- 17.Purhkold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implication for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowan, A. K., J. R. Snapeb, D. Fearnsidec, M. R. Barerd, T. P. Curtise, and I. M. Head. Composition and diversity of ammonia-oxidising bacterial communities in wastewater treatment reactors of different design treating identical wastewater. FEMS Microbiol. Ecol., in press. [DOI] [PubMed]
- 20.SAS Institute. 1988. Users guide: statistic version 6. Statistical Analytical Institute, Cary, N.C.
- 21.Simpson, J. M., V. J. McCracken, H. R. Gaskins, and R. I. Mackie. 2000. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Appl. Environ. Microbiol. 66:4705-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephen, J. R., Y. J. Chang, S. J. Macnaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil beta-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoetendal, E., A. Akkermans, and W. deVos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]