Abstract
Current velocity affected the architecture and dynamics of natural, multiphyla, and cross-trophic level biofilms from a forested piedmont stream. We monitored the development and activity of biofilms in streamside flumes operated under two flow regimes (slow [0.065 m s−1] and fast [0.23 m s−1]) by combined confocal laser scanning microscopy with cryosectioning to observe biofilm structure and composition. Biofilm growth started as bacterial microcolonies embedded in extracellular polymeric substances and transformed into ripple-like structures and ultimately conspicuous quasihexagonal networks. These structures were particularly pronounced in biofilms grown under slow current velocities and were characterized by the prominence of pennate diatoms oriented along their long axes to form the hexagons. Microstructural heterogeneity was dynamic, and biofilms that developed under slower velocities were thicker and had larger surface sinuosity and higher areal densities than their counterparts exposed to higher velocities. Surface sinuosity and biofilm fragmentation increased with thickness, and these changes likely reduced resistance to the mass transfer of solutes from the water column into the biofilms. Nevertheless, estimates of dissolved organic carbon uptake and microbial growth suggested that internal cycling of carbon was more important in thick biofilms grown in slow flow conditions. High-pressure liquid chromatography-pulsed amperometric detection analyses of exopolysaccharides documented a temporal shift in monosaccharide composition as the glucose levels decreased and the levels of rhamnose, galactose, mannose, xylose, and arabinose increased. We attribute this change in chemical composition to the accumulation of diatoms and increased incorporation of detrital particles in mature biofilms.
An understanding of the structure-function relationships in microbial biofilms is central to interpret and predict biofilm impacts on the habitat where they form (46). Our current perception of biofilm structure results from the interplay of mathematical modeling and experimental studies. At least three conceptual models have been derived from this research (57): (i) planar, homogeneous biofilms are typified in dental plaque exposed to extremely high substrate concentrations in the saliva; (ii) heterogeneous, mosaic biofilms consisting of stacks of bacterial colonies embedded in extracellular polymeric substances (EPS) and appearing as columns surrounded by the bulk liquid have been described from drinking water distribution systems (21); and (iii) biofilms with complex three-dimensional structures with channel networks through which liquid flow occurs have been described from studies by using confocal laser-scanning microscopy (CLSM), microelectrodes, and fluorescent particles (10, 23, 45). A unifying thread of these conceptual models is that they attempt to explain biofilm structure as a function of substrate concentration and hydrodynamics (47, 50, 57). Additionally, the biofilms described in these models were derived from nuisance growths that colonize medical and industrial systems, where sunlight is absent and communities are largely bacterial monocultures. In contrast, biofilms from natural ecosystems exposed to sunlight include greater trophic (phototrophs and heterotrophs) and phylogenetic (eucaryotes and procaryotes) diversity and complexity.
Although some of the earliest biofilm research was related to rock surfaces in stream ecosystems (14, 27, 28, 43), our current knowledge of lotic biofilms lags far behind their medical and industrial counterparts. However, biofilms represent the dominant mode of microbial life in streams (12, 14), where they can drive ecosystem metabolism, nutrient cycling, and biodegradation. Stream biofilms are functionally (42) and phylogenetically (2, 29) diverse communities and possess highly complex EPS chemistry (33, 34, 58). Flow is an inherent feature of stream ecosystems that influences function, as well as structure, and yet studies of flow effects in streams primarily have focused on the composition and shape of diatom communities (3, 4). Most of our current understanding of the relationships between flow and structure-function coupling of biofilms is largely derived from bacterial biofilms manipulated in laboratory-scale reactors (see, for example, references 30, 38, and 56). Thus, there is a need to relate hydrodynamics to the structure and function of stream biofilms and the resultant impacts on biological and chemical processes in stream ecosystems.
We report here on the nascent architecture and dynamics of natural, microbial biofilms in a forested piedmont stream. Biofilms were cultivated under two different current velocities in streamside flumes to incorporate the natural dynamics of stream water chemistry, particle behavior, and hydrodynamics into our experimental design. We describe biofilm development from bulk biomass measurements, we used CLSM, along with cryosectioning, to explore the spatial organization of bacterial and algal cells, bulk exopolysaccharides, and particles within biofilms, and we analyzed the monosaccharide composition of the EPS.
MATERIALS AND METHODS
Cultivation of biofilms.
Biofilms were grown in four streamside flumes (length, 30 m; width, 0.30 m; depth, 0.30 m) adjacent to a forested reach of White Clay Creek (WCC; Stroud Water Research Center, Pa.). Flumes were continuously fed in a once-through mode with streamwater from a header tank to ensure constant flow. WCC nutrient concentrations typically range from 3 to 4 mg of NO3-N liter−1 and 10 to 60 μg of PO4-P liter−1 (35). All flumes were packed with the same clean, commercially available cobble and pea gravel (median grain size, 20.9 ± 0.8 mm) and unglazed ceramic coupons (1 by 1.5 by 0.3 cm) (6). Flume slopes were adjusted to yield duplicate open channel flow velocities of 0.065 ± 0.003 and 0.23 ± 0.005 m s−1. The average water temperature was 18.4°C and was not affected by water residence time in the flumes.
Microbial biomass and exopolysaccharides.
Bacterial abundance was estimated by epifluorescence direct microscopic counts (17). Briefly, preserved biofilms (2.5% formaldehyde) were treated with 0.1 mM sterile tetrasodium pyrophosphate (54) for 10 min, and cells were removed by sonication (75 W, 60 s). Aliquot samples (1 ml) were transferred into sterile vials containing 1 ml of tetrasodium pyrophosphate, thoroughly vortexed, and then sonicated (75 W, 30 s) again. Samples were then mixed with sterile glycerol (30% final concentration), vortexed, and spun to pellet particles that likely cause background fluorescence (2). After staining with propidium iodide, bacterial cells in the supernatant were enumerated on a black 0.2-μm-pore-size GTBP Millipore filter in 10 to 30 randomly selected fields to account for 300 to 500 cells. Two filters were counted per ceramic coupon. Chlorophyll a was assayed to provide an estimate of algal biomass (44). Bulk organic and inorganic biofilm accumulations were derived from dry and ash masses (450°C, 5 h).
Carbohydrate analyses were performed on bulk EPS harvested from biofilms by extraction in 50 mM EDTA (53) on a rotary shaker. The extract was filtered (0.2-μm pore size) to remove particles and cells, and EPS in the filtrate was precipitated with ice-cold ethanol (99% [vol/vol]) and held at −20°C for 24 h. The polymeric fraction was pelleted by centrifugation (1,500 × g, 30 min), and the supernatant was discarded. The pellet was redissolved in 2 ml of deionized water, and EPS was washed a second time in ice-cold ethanol. The resulting pellet was lyophilized and subsampled. One aliquot was assayed for total carbohydrates by the sulfuric acid-phenol method (11), and another aliquot (0.5 ml) was digested (100°C, 5 h) with 4 M trifluoroacetic acid in precombusted glass ampoules that were flushed with nitrogen and analyzed for monosaccharides by high-pressure liquid chromatography (HPLC)-pulsed amperometric detection (PAD) (7). Bulk EPS carbohydrates, bacterial abundance, and chlorophyll a were measured on each sampling date, whereas monosaccharides were determined on days 3 and 5 (young biofilms) and days 18 and 21 (mature biofilms) of the biofilm development.
DOC uptake.
To estimate biofilm functional dynamics, we monitored the flume-scale uptake of dissolved organic carbon (DOC) during biofilm growth. DOC concentrations in the flume influent and effluent waters were measured with an OI-700 organic carbon analyzer (20). DOC uptake was estimated from mass balance calculations and related to the flume bed surface area.
Cryosections and CLSM.
Developmental patterns were derived from the CLSM investigation of the biofilm xy plane from ceramic coupons that were randomly sampled for structural analyses, carefully removed from the flumes, and transferred into microchambers (Fisher Scientific) containing streamwater (0.2-μm pore size filtered). Within 30 min of sampling, bacterial cells were stained with the fluorescent nucleic acid stain SYTO 13 (Molecular Probes, Inc., Eugene, Oreg.) at a concentration of 20 μg ml−1 (10 min, room temperature). Microbial EPS was stained with the lectin probe Concanavalin A (ConA)-Texas red (Molecular Probes), which purportedly binds to terminal α-d-glucose and α-d-mannose, although ConA might bind to non-EPS targets or nonspecifically to the biofilm matrix (19). We stained EPS with a lectin concentration of 5 mg ml−1 (10 min at room temperature). Excess staining solutions were removed by gently rinsing the samples with filtered (0.2-μm pore size) streamwater. These samples were used either for direct CLSM analysis or cryoembedding. Cryoembedding was performed by embedding samples in Tissue-Tek O.C.T. compound (Miles, Inc., Elkhart, Ind.) for 15 min and then freezing them on dry ice (18, 60). Once completely frozen, the biofilm was removed from the ceramic coupon, turned over, placed back on dry ice, and reembedded in O.C.T. compound. Embedded biofilms were cut into 6-μm sections (yz plane) with a Leica (Germany) cryostat. Cryosections were transferred onto poly-l-lysine-coated slides, allowed to air dry, and subsequently mounted in Citifluor medium (Citifluor, Canterbury, United Kingdom). Sections were immediately analyzed with a Zeiss LSM 510 scanning confocal microscope (Carl Zeiss, Jena, Germany) equipped with an ArKr laser (488 and 568 nm) and an HeNe laser (633 nm). For routine imaging, a ×10 Plan Neo-Fluar lens was used to cover cryosections over a length of 922 μm. For high magnifications, we used ×40 Plan Neo-Fluar (NA 1.3, oil immersion) and a ×63 C-Apochromat (NA 1.2, water immersion) objective lenses. The algal autofluorescence was recorded in the far-red channel, and its overlay with Conavalin-Texas red resulted in a purple appearance.
Image acquisition and analysis.
Digitized cryosections (16 bits pseudocolor TIFF files, 1,024 by 1,024 pixels) were imported from the LSM 510 into the GIS and mapping software Arc-View 2. 2 (ESRI, Inc.) for image analysis. Each imported image layer (red-green-blue [RGB]) was converted to a unique grid in a Cartesian coordinate system. We applied a fixed threshold to all images (15). Thresholding results in a matrix with “zero values” in positions where the numbers of RGB pixels are <6 and “one values” where the numbers of RGB pixels are between 6 and 255. One values represent positions containing microbial cells, ConA-EPS, or particles; zero values represent positions with voids. After thresholding, we removed background noise by using the Spatial Analyst (version 1.1; ESRI). Biomass pixels that were not connected to adjacent biomass clusters and that contained <50 RGB pixels were set to the zero value. These images were used for the quantification of microstructural heterogeneity in the xz plane. Cryosections proved appropriate for quantitative analyses of biofilm structure (31).
Microstructural parameters.
Microstructural parameters estimated from these images included the average thickness and roughness of the biofilm. Roughness was determined as the deviation from 200 to 250 individual thickness measurements per section (cf. reference 31). Biofilm porosity was determined as the ratio of the void surface area (zero value RGB pixel) to the total cross-sectional surface area (sum of zero and one value RGB pixels). The sinuosity of the biofilm surface front was determined as the ratio between the number of connected RGB pixels of the biofilm surface between two given points and the corresponding linear distance between these two points. This parameter is analogous to surface enlargement, which is thought to affect mass transport (40). Biofilm fragmentation was determined as the ratio of the perimeter and the surface area of biofilm clusters. Biofilm fragmentation is analogous to the average diffusion distance suggested as a measure of the distance over which a substrate molecule must diffuse from the void to bacterial cells within a cluster (59). The biofilm areal density was calculated from average thickness and total mass estimates of biofilms (39).
Statistical analyses.
Hydraulic parameters and DOC uptake are presented as the mean of duplicate flume-scale measurements performed at each sampling date. Biofilm biomass parameters were determined from two to three ceramic coupons per flume per sample date. Biofilm microarchitecture parameters were determined from duplicate coupons collected from a single flume representing each flow treatment on eight occasions. Because low sample numbers contribute to uneven variation, we used the nonparametric Mann-Whitney Wilcoxon procedure to test for flow effects on biofilm structures (13). Correlations between biofilm microarchitecture parameters were explored by using the Pearson r coefficient. All values are given as mean ± the standard deviation (SD).
RESULTS
Hydraulic characteristics of the flumes.
The two flow regimes resulted in a 13-fold difference in shear stress and a 4-fold difference in Reynolds numbers (Table 1). The Reynolds numbers for open channels were indicative of transitional and fully turbulent flow.
TABLE 1.
Hydraulic characteristics of the experimental flumes
| Parameter | Mean ± SD (n = 14) for:
|
|
|---|---|---|
| Slow flow | Fast flow | |
| Flow velocity (m s−1) | 0.065 ± 0.011 | 0.23 ± 0.02 |
| Shear stress (N m−2) | 1.07 ± 0.26 | 13.31 ± 0.48 |
| Reynolds (Ref) | 1869 ± 710 | 7559 ± 254 |
Biomass development and flume-scale dynamics.
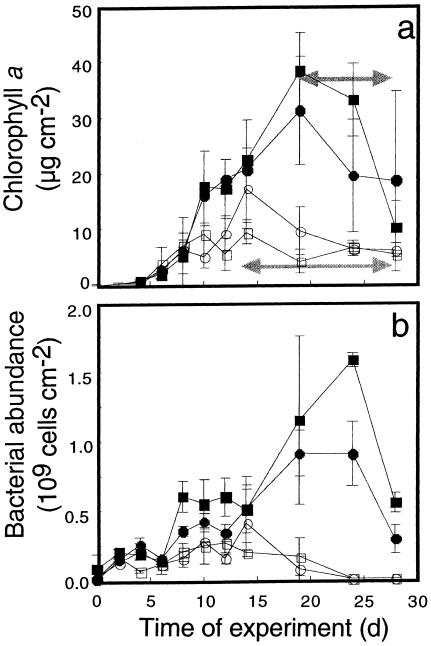
Flow significantly affected bulk microbial biomass, with 3.4-fold-higher bacterial abundance and 2.5-fold-higher chlorophyll a concentrations in the slow flow treatments from approximately day 10 onward (Fig. 1). After an initial 3- to 5-day lag phase, chlorophyll a and bacterial abundance steadily increased and then decreased due to massive biofilm detachment and grazing by invertebrates, as revealed by visual inspection. The development of long filamentous streamers (up to 6 cm long) contributed to large within-flume variations of mature biofilms.
FIG. 1.
Developmental dynamics of microbial biomass as indicated by chlorophyll a (a) and bacterial abundance (b). Closed symbols denote slow-flow treatments, and open symbols denote fast-flow treatments. Horizontal arrows indicate periods of increased biofilm detachment and grazing. The results are averages ± the SDs of two to three coupons for each experimental flume.
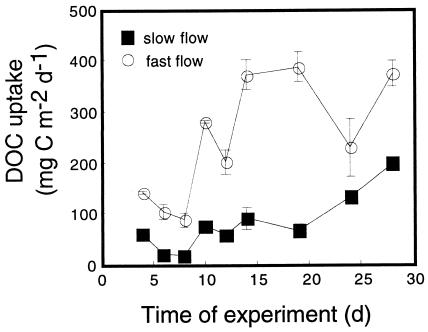
The trajectory of flume-scale uptake of DOC followed the broad pattern of biomass development (Fig. 2). DOC uptake, measured as a decline in DOC concentrations between the header tank inflow and the flume outflow, occurred consistently when the influent waters were pumped from the stream at baseflow and had a DOC concentration of ≤2.4 mg of C liter−1. Outflow concentrations from the treatments were very similar (average difference of 0.028 mg of C liter−1 over a range of values from 1.49 to 2.29 mg of C liter−1) between treatments. However, when the uptake values were multiplied by the discharge to obtain estimates of the uptake flux, average rates were 3.9 times higher in the fast-flow treatment. The percentages of influent DOC removed within the flumes gradually increased with biofilm development from initial values of 2 to 6% from days 3 to 7, to 7 to 11% from days 9 to 17, and finally to 32% when last measured on day 27. The 32% DOC removal represented a difference of 0.479 mg of C liter−1 from a starting concentration of 1.49 mg of C liter−1.
FIG. 2.
Temporal dynamics of DOC uptake from the streamwater. The results are averages ± the SDs from duplicate flow treatments.
Exopolysaccharides.
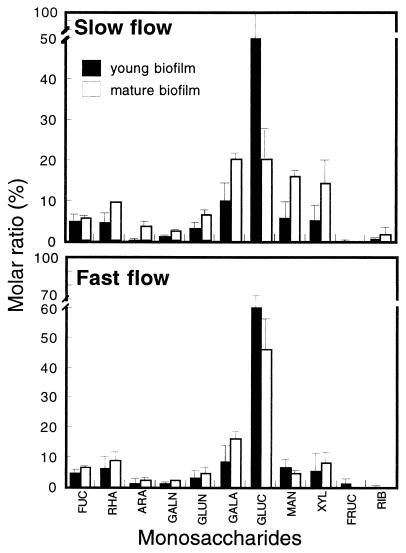
HPLC-PAD analyses revealed glucose as the dominant monomer in all biofilms, followed by galactose and mannose (Fig. 3). Young biofilms showed similar monomer compositions in both flow treatments. The exopolysaccharide fingerprints of biofilms under the slow-flow treatment changed with biofilm development as follows: the mole percent value for glucose decreased 3.2 times; the mole percent values for rhamnose, galactose, mannose, and xylose increased 2 to 2.8 times; and the mole percent value for arabinose increased 17 times (Fig. 3). The average levels of exopolysaccharides (as glucose [cf. reference 11]) were significantly higher in the fast-flow treatments and, when combined with the significantly lower bacterial abundance, yielded a nearly threefold-higher exopolysaccharide/cell ratio in the fast-flow treatments (Table 2).
FIG. 3.
Monosaccharide composition of young and mature biofilms from slow- and fast-flow treatments. FUC, fucose; RHA, rhamnose, ARA, arabinose; GALN, galactosamine; GLUN, glucosamine; GALA, galactose; GLUC, glucose; MAN, mannose; XYL, xylose; FRUC, fructose; RIB, ribose. Molar ratio is the ratio of the number of moles of the monomer to the total number of moles of saccharides; data are averages ± the SDs from duplicate coupons.
TABLE 2.
Effect of flow velocity on bulk exopolysaccharides
| Parameter | Mean result ± SD (n = 11) for:
|
Pa | |
|---|---|---|---|
| Slow flow | Fast flow | ||
| Bacterial abundance (109 cells cm−2) | 0.47 ± 0.37 | 0.14 ± 0.09 | 0.003 |
| Exopolysaccharides (μg of glucose cm−2) | 1.30 ± 1.40 | 1.14 ± 1.40 | NS |
| Exopolysaccharides cell−1 (fg of glucose cell−1) | 2.98 ± 3.32 | 9.36 ± 7.78 | 0.007 |
The Mann-Whitney Wilcoxon procedure was used to test for significance of flow effects. NS, not significant.
Structural development of the xy plane.
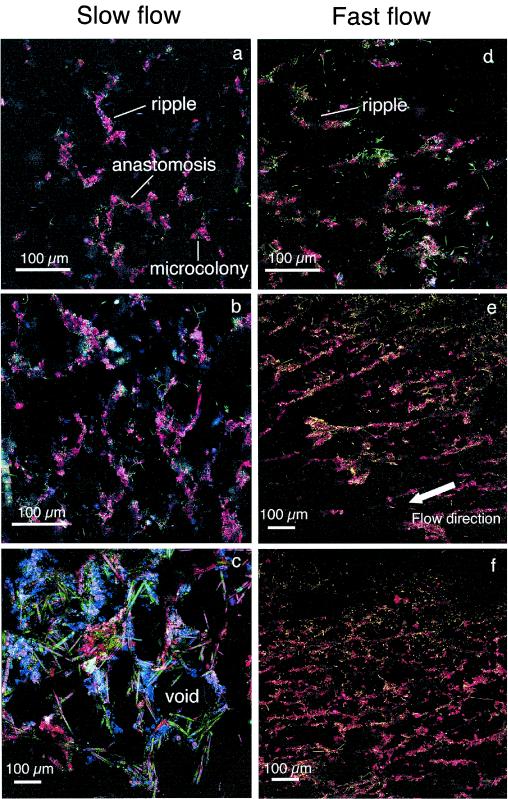
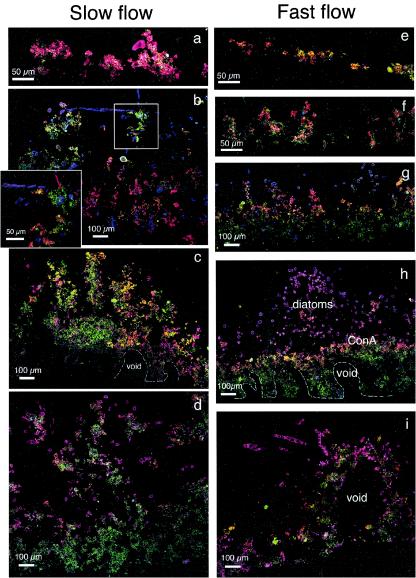
The progression of biofilm growth began with bacterial colonization, which by day 4 consisted of isolated microcolonies and bacterial cells scattered over the entire substratum and the first appearance of ripple-like structures with distinct ConA staining of the EPS (Fig. 4a and d). At this developmental stage, long filamentous bacteria were prominent in the fast flow treatment (Fig. 4d). By day 8, anastomoses developed between ripples, along with a first appearance of diatoms and amorphous material (Fig. 4b and e). The overall orientation of these initial structures was parallel to the main flow direction. This was particularly pronounced in the fast-flow treatments. By day 21, a prominent network of quasihexagonal structures developed from the basal pattern, especially in those biofilms grown in slow flow (Fig. 4c). Elongated pennate diatoms (Syndera ulna) formed the sides of the hexagons around a central void and amorphous material accumulated around the diatoms. Pores or voids defined by the quasihexagonal configuration of biomass had an average diameter of 160 ± 36 μm (n = 14). At the same time, biofilms from fast-flow treatments consisted of a dense network of conspicuous ripples with a high degree of anastomosis, creating central voids that were similar to those in the slower flow but without the same depth and layering (Fig. 4f).
FIG. 4.
CLSM images showing the structural dynamics and formation of quasihexagonal structures in stream biofilms (xy plane). (a, b, and c) Images of 4-, 12-, and 21-day-old biofilms, respectively, from slow-flow treatments; (d, e, and f) images of 6-, 12-, and 24-day-old biofilms, respectively, from fast-flow treatments.
Structural development of the xz plane.
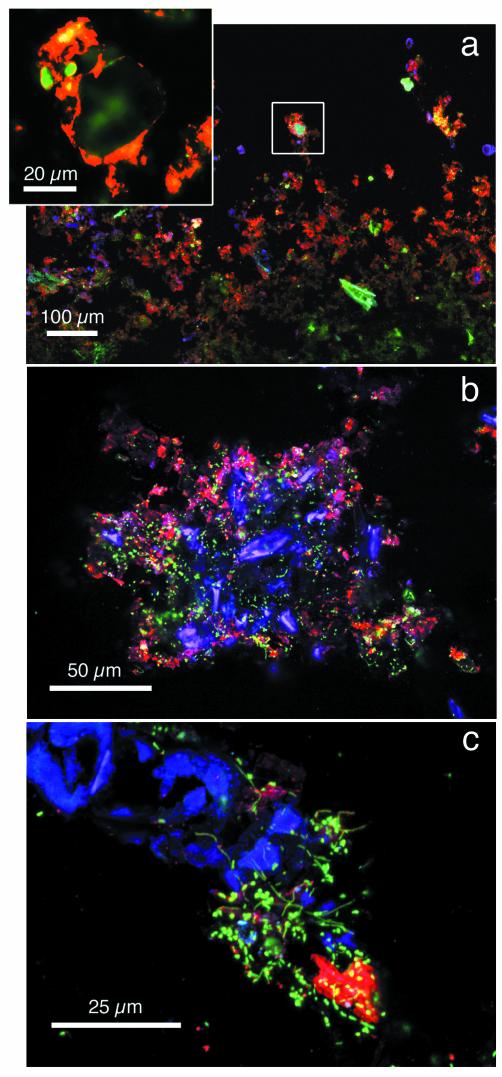
Low-magnification CLSM analysis of cryosections generally supported the observed xy patterns of living biofilms, with loose but copious clusters of ConA-stained EPS (Fig. 5a, e, and f). Virtually no algal cells were detectable in these 3- to 5-day-old biofilms predominantly composed of bacteria. With continued growth, a distinct base layer and a canopy developed. The base layer was generally recognized by low ConA-stained EPS and the presence of accumulated particles and bacterial cells (Fig. 5c, d, g, and h) arrayed around arcade-like void spaces (Fig. 5c and h). Initially, the canopy consisted of finger-like protuberances (Fig. 5c) and, as biofilms developed, algal cells became the dominant canopy component (Fig. 5d, h, and i). A cross-section of a quasihexagonal space with a long algal cell bridging the gap shows this pattern (Fig. 5b). Depending on the developmental stage of biofilms, valleys between the canopy protuberances can extend into the biofilm base to form extensive channel networks (Fig. 5c and d). In some cases we observed a conspicuous stratification of ConA-stained EPS layers and diatoms (Fig. 5g and h).
FIG. 5.
CLSM micrographs of cryosections showing the structural development of the biofilm xz plane in slow flows (a to d) and fast flows (e to i). (a) Coalescing microcolonies of a 4-day-old largely bacterial biofilm; (b) a relatively compact biofilm (12 days) with finger-like protuberances and the first appearance of algal cells; (c) mature biofilm (17 days) with elongated algal cells bridging a possibly quasihexagonal void; (d) 24-day-old biofilm with distinct structural differentiation into a base layer and a canopy; (e and f) images showing initial microcolonies with filamentous bacterial cells in initial biofilms (at 3 and 6 days, respectively); (g and h) images showing the structural differentiation into a base layer and a diatom-rich canopy (g) and clear microstratification with a ConA-EPS-rich layer (h); (i) cross-section of ridge-like structure with void, possibly resulting from metazoan burrowing. Note the arcade-like void system within the base layer of mature biofilms (c and h).
With higher magnification, we were able to observe that mineral particles coated with ConA-stained EPS were the major component of biofilm microlayers (Fig. 6a). Diatoms constituted a central building block of the filamentous streamers that also included abundant bacterial cells and noticeable peripheral ConA-stained EPS (Fig. 6b). High magnification revealed large bacteria densely colonizing a broken algal cell and its exudates, as visualized by ConA-stained EPS (Fig. 6c), suggesting an autotrophic-heterotrophic coupling.
FIG. 6.
CLSM images showing the incorporation of ConA-EPS-coated particles in a mature biofilm (a), a cross-section of the distal part of a streamer of a 14-day-old biofilm (b), and an example of the close coupling between an algal cell and bacteria (c).
Quantification of biofilm microstructures.
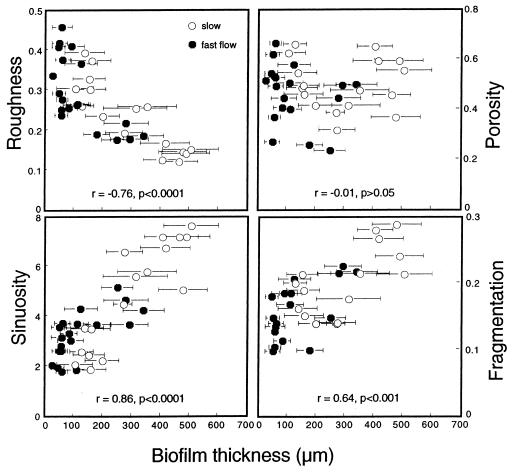
Biofilms grown in slow-flow treatments were thicker and had higher average sinuosity and areal density and yet had less roughness than biofilms from fast-flow treatments (Table 3). Biofilm fragmentation and porosity were not affected by flow. Surface roughness decreased, whereas both sinuosity and fragmentation increased, with biofilm thickness (Fig. 7). Fragmentation did not correlate with biofilm thickness.
TABLE 3.
Microstructural biofilm parameters for slow (n = 18)- and fast (n = 18)-flow treatments
| Parametera | Mean result ± SD for:
|
Pb | |
|---|---|---|---|
| Slow flow | Fast flow | ||
| Thickness (μm) | 295 ± 143 | 123 ± 97 | <0.001 |
| Roughness | 0.22 ± 0.09 | 0.29 ± 0.09 | <0.05 |
| Sinuosity | 4.78 ± 2.09 | 3.22 ± 0.96 | <0.05 |
| Porosity | 0.50 ± 0.10 | 0.46 ± 0.12 | NS |
| Fragmentation | 0.20 ± 0.05 | 0.17 ± 0.05 | NS |
| Areal density (g cm−3) | 0.03 ± 0.02 | 0.01 ± 0.02 | <0.05 |
Roughness, sinuosity, porosity, and fragmentation are dimensionless indices.
The effects of flow treatment were tested with the Mann-Whitney Wilcoxon test. NS, not significant.
FIG. 7.
Relationships between biofilm thickness and roughness, sinuosity, porosity, and fragmentation. Error bars indicate the SDs from 200 to 250 single-thickness measurements. See the text for further explanation.
DISCUSSION
Structural development.
The development of ripple-like or ridge-like structures that we have documented are similar to biofilm features observed in other systems. For example, migratory ripples were described from mixed-species bacterial biofilms cultivated in turbulent flow (48). Ridges aligned in the main flow direction that ultimately developed into dense parallel structures were observed in biofilms grown on polycarbonate slides within rotating annular reactors fed by stream water (31).
Quasihexagonal structures similar to those we observed as anastomoses between parallel ridges evolved into a basal pattern have been described for biofilms from wastewater (36) and marine sulfate-reducing bacteria (61). This mode of biofilm growth thus seems to be more common than hitherto thought, and yet the mechanisms underlying this pattern remain largely speculative. This growth form may be a response to the hydrodynamics of the flume microenvironments and facilitate the competition for a substratum during initial biofilm formation as described by Picioreanu (41), for instance. Alternatively, bacterial cells that initiate biofilm growth are believed to skip over crevices to avoid zones of reduced mass transfer (32), and this could potentially lead to biomass growth clustered around pores in the substratum. However, close CLSM inspection of the coupon texture revealed that the distribution of pores did not agree with biomass patterns.
The increased physical stability of quasihexagonal structures is another possible cause of this biofilm architecture. Stoodley et al. (49) described biofilm rheology as viscoelastic and demonstrated structural deformation of bacterial biofilms grown in turbulent flow. The quasihexagonal patterns fortified by pennate diatoms as major building blocks could prevent biofilms from detrimental deformations and heavy erosion when faced with changes in current velocity in stream ecosystems. Furthermore, pores or voids within the interior of these quasihexagons may facilitate advective solute transport as envisioned by current biofilm models (see, for example, references 8 and 9).
CLSM has become a powerful tool for studying the spatial organization of biofilms. However, where limited penetration depth of the confocal laser beam, along with photobleaching of fluorescent probes, becomes an issue, sectioning of the specimen prior to CLSM can provide better images and information on the spatial organization (26, 55). The combination of cryosections with CLSM yielded xz images of stream biofilms with high-quality resolution that could not have been achieved with confocal optical sections and subsequent reconstruction (cf. reference 33). Our CLSM analyses of cryosections conveyed excellent insight into the vertical growth of biofilms and underscore the importance of organic and inorganic particles in stream biofilms.
Upper layers of initial microcolonies coalesced through growth, which resulted in the development of voids within the base layer of the biofilm. Algal cells extended away from the basal layer to form the biofilm canopy, along with inorganic particles and organic debris. In some instances we found a microstratification of particles coated with EPS and diatoms, which probably reflects particle dynamics in the stream water. Particles can increase biofilm surface area and transport attached bacteria to biofilms, but they may also affect mass transfer by increasing the diffusional resistance within biofilms. Organic particles can furthermore constitute a significant carbon source for heterotrophs. The combination of these factors certainly contributes to the shell-like growth with elevated bacterial abundances and activities close to the biofilm surface observed in various systems (see, for example, reference 33) and derived from multidimensional modeling (40). On the other hand, bacterial growth in streams can also increase particle deposition velocity through enhanced filtration and adhesion [T. J. Battin, L. A. Kaplan, and J. D. Newbold, Eos. Trans. AGU, no. 82(47), Fall Meet. Suppl., abstr. H12G-02, 2001]. Once accumulated and aggregated within biofilms, particles can be exported from the ecosystem through detachment of entire biomass clusters.
We found disparate developmental patterns of surface roughness and sinuosity in the stream biofilms. Roughness decreased with biofilm maturity in our stream systems, whereas it has been shown to increase in Pseudomonas aeruginosa biofilms (39) and multidimensional modeling (41). However, laboratory-grown biofilms are not exposed to particles and do not include photoautotrophs or heterotrophic grazers. In streams, biofilm structural development is the net result of growth and detachment of microbial biomass, hydrodynamics, substrate availability, and grazing (cf. reference 25). Among other things, enhanced particle filtration and accumulation within the heterogeneous canopies that were particularly well developed in biofilms from our slow-flow treatments could also explain the higher areal density of these communities.
The increase of surface sinuosity with biofilm thickness and hence with maturation agrees with modeling results by Picioreanu et al. (40, 41). These authors showed that the width of the valley defined by two neighboring protuberances (i.e., sinuosity) determines advective transport to the biofilm rather than surface roughness. Thus, we postulate that sinuosity increases with biofilm thickness to assure the mass transfer of substrates necessary to satisfy microbial demand. Considering the shell-like growth of biofilms, enhanced transport of particles and substrates between neighboring protuberances would thus successively reduce surface roughness. Similarly, fragmentation increased with biofilm thickness, and this likely enhanced solute transport within biofilms by increasing the channel network and thus reducing the average diffusive pathway for substrate molecules (59).
Exopolysaccharides.
The monosaccharide composition we described from the stream biofilms generally agrees with other gas chromatography- or HPLC-derived carbohydrate fingerprints from intertidal sediments (52) or terrestrial biofilms (51). Glucose is always the dominant monomer, followed by mannose, rhamnose, galactose, or xylose. Noninvasive lectin-binding assays support the frequent occurrence of these monomers and further illustrate their overwhelming spatial heterogeneity (33, 34, 58). Patterns of ConA-stained EPS in the present study also support the HPLC-PAD-derived patterns of glucose and mannose as dominant carbohydrates in the stream biofilms.
Our results are the first to show a relative shift of the monosaccharide composition during biofilm development. Whether this change is attributable to a shift of the community structure remains unknown. For instance, diatoms, which are well known to produce copious amounts of EPS (16), became particularly abundant in mature biofilms from slow-flow treatments. Alternatively, increased incorporation of allochthonous EPS (e.g., via particles) could also shift the polysaccharide fingerprint. No lysis control was included in our EPS extraction, and so we cannot assess the contribution of intracellular polysaccharides to our exopolysaccharide estimates.
We did find higher exopolysaccharide/cell ratios in biofilms growing in a fast flow. This agrees with findings from Pseudomonas fluorescens biofilms grown in flow cell reactors (38) and from biofilms collected from a high-gradient glacial stream (2). Elevated exopolysaccharide/cell ratios should improve adhesion under elevated shear stress and thus reflect a functional response to avoid detachment through erosion and sloughing.
Biofilm function.
Our results on DOC uptake indicate that biofilms grown in fast flow rely more on organic substrates from the water column, a finding that agrees with the uptake of experimentally injected glucose (Battin et al., Eos. Trans. AGU abstr. H12G-02, 2001). We attribute this difference between flow treatments to the overall lower diffusive resistance in the fast-flow treatments. In fact, the external boundary layer is thinner in a fast flow, and the internal diffusive resistance is lower due to reduced thickness and areal density. The converse effects isolate slow-flow biofilms from external supply, and heterotrophs within those biofilms predominantly rely on internal energy sources. Algal exudates (see Fig. 6c) and DOC immobilized within the EPS matrix figure among these internal sources (see, for example, reference 1). Close coupling between heterotrophs and autotrophs is thus believed to be more important in thicker biofilms, a notion that agrees with findings from microbial mats (see, for example, reference 22). This implies elevated internal cycling—or close spatial coupling of carbon production and consumption—within these slow-flow communities, leading to reduced downstream cycling.
Conclusion.
Performing the present study in large-scale streamside flumes helped place our biofilm research into an ecosystem context (sensu Palmer and White [37]) since the flumes received sunlight, natural concentrations of solutes and particles, and simulated hydrodynamics that occur at the water-sediment interface. These conditions allowed us to observe the dynamics of biofilm structure and function at a level of complexity that could not be obtained with, for instance, more artificial, laboratory-scale rotating annular bioreactors (24, 33). We found that stream microbial biofilms grown under quasinatural conditions were highly dynamic and complex communities composed of organisms that cross trophic levels and represent multiple phyla. An open system such as a stream provides a broad repertoire of structural (inorganic and organic particles) and functional (procaryotic and eucaryotic phototrophs and heterotrophs) building blocks or components for biofilms. This expansive view of microbial biofilms complicates our ability to probe the evolutionary, molecular, and genetic bases for biofilm formation, such as the description of genes involved in the maintenance of biofilm architecture in a bacterial monoculture (5). Alternatively, it opens up possibilities for observing interactions that are common in natural environments, at a scale that is relevant to organic matter dynamics and biogeochemical processes in stream ecosystems.
Acknowledgments
We thank Sherman Roberts and Mike Gentile for technical assistance, Kirk Czymmek for CLSM support, and Stefan Würtz and two anonymous reviewers for constructive comments.
This research was supported by a FWF Erwin Schrödinger grant (J1879-BIO) to T.J.B., U.S. National Science Foundation grant DEB-9904047 to L.A.K., and the Pennswood No. 2 Fund for Environmental Research.
REFERENCES
- 1.Battin, T. J., A. Butturini, and F. Sabater. 1999. Immobilization and metabolism of dissolved organic carbon by natural sediment biofilms in a Mediterranean and temperate stream. Aquat. Microb. Ecol. 19:297-305. [Google Scholar]
- 2.Battin, T. J., A. Wille, B. Sattler, and R. Psenner. 2001. Phylogenetic and functional heterogeneity of sediment biofilms along environmental gradients in a glacial stream. Appl. Environ. Microbiol. 67:799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggs, B. J. F., D. G. Goring, and V. I. Nikora. 1998. Subsidy and stress responses of stream periphyton to gradients in water velocity as a function of community growth form. J. Phycol. 34:598-607. [Google Scholar]
- 4.Blenkinsopp, S. A., and M. A. Lock. 1994. The impact of storm-flow on river biofilm architecture. J. Phycol. 5:807-818. [Google Scholar]
- 5.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardinale, B. J., M. A. Palmer, C. M. Swan, S. Brooks, and N. LeRoy-Poff. 2002. The influence of substrate heterogeneity on biofilm metabolism in a stream ecosystem. Ecology 83:412-422. [Google Scholar]
- 7.Cheng, X., and L. A. Kaplan. 2001. Improved analysis of dissolved carbohydrates in stream water with HPLC-PAD. Anal. Chem. 73:458-461. [DOI] [PubMed] [Google Scholar]
- 8.Costerton J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W. 2000. Biofilms in the new millenium: musings from a peak in Xanadu, p. 329-344. In D. Allison, P. Gilbert, H. M. Lappin-Scott, and M. Wilson (ed.), Community structure and co-operation in biofilms: SGM symposium 59. Cambridge University Press, Cambridge, United Kingdom.
- 10.De Beer, D., P. Stoodley, F. Roe, and Z. Lewandowski. 1994. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol. Bioeng. 43:1131-1138. [DOI] [PubMed] [Google Scholar]
- 11.Dubois, M. G., K. A. Hamilton, J. K. Rebers, and R. Smith. 1956. Colorimetric method for the determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 12.Fischer, H., and M. Pusch. 2001. Comparison of bacterial production in sediments, epiphyton, and the pelegiac zone of a lowland river. Freshwater Biol. 46:1335-1348. [Google Scholar]
- 13.Fowler, J., and L. Cohen. 1990. Practical statistics for field biology. Open University Press, Philadelphia, Pa.
- 14.Geesey, G. G., R. Mutch, J. W. Costerton, and R. B. Green. 1978. Sessile bacteria: an important component of the microbial population in small mountain streams. Limnol. Oceanogr. 23:1214-1223. [Google Scholar]
- 15.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. Erboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 16.Hoagland, K. D., J. R. Rosowski, M. R. Gretz, and S. C. Roemer. 1993. Diatom extracellular polymeric substances: function, fine structure, chemistry, and physiology. J. Phycol. 29:537-566. [Google Scholar]
- 17.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by epifluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, C. T., G. A. McFeters, and P. S. Stewart. 1996. Evaluation of physiological staining, cryoembedding, and autofluorescence quenching techniques on fouling biofilms. Biofouling 9:269-277. [Google Scholar]
- 19.Johnson, A. R., M. Hausner, A. Schnell, and S. Würtz. 2000. Evaluation of fluorescently labeled lectins for noninvasive localization of extracellular polymeric substances in Sphingomonas biofilms. Appl. Environ. Microbiol. 66:3487-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan, L. A. 1994. A field and laboratory procedure to collect, process, and preserve freshwater samples for dissolved organic carbon analysis. Limnol. Oceanogr. 39:1470-1476. [Google Scholar]
- 21.Keevil, C. W., and J. T. Walker. 1992. Normaski DIC microscopy and image analysis of biofilms. BINARY 4:93-95. [Google Scholar]
- 22.Kuehl, M., R. N. Glud, H. Plough, and N. B. Ramsing. 1996. Microenvironmental control of photosynthesis and photosynthesis coupled respiration in an epilithic cyanobacterial biofilm. J. Phycol. 32:799-812. [Google Scholar]
- 23.Lawrence, J. R., D. R. Korber, B. D. Hoyle, B. D. Costerton, and D. E. Caldwell. 1991. Optical sectioning of microbial biofilms. J. Bacteriol. 173:6558-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence, J. R., G. D. W. Swerhone, and T. T. Neu. 2000. A simple rotating annular reactor for replicated biofilm studies. J. Microbiol. Methods 42:215-224. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence, J. R., B. Scharf, G. Packroff, and T. T. Neu. 2002. Microscale evaluation of the effects of grazing by invertebrates with contrasting feeding modes on river biofilm architecture and composition. Microb. Ecol. 43:199-207. [DOI] [PubMed] [Google Scholar]
- 26.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography: a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lock, M. A., R. R. Wallace, J. W. Costerton, R. M. Ventullo, and S. E. Charlton. 1984. River epilithon: toward a structural-functional model. Oikos. 42:10-22. [Google Scholar]
- 28.Madsen, B. L. 1972. Detritus on stones in small streams. Proceedings of the international biological program-UNESCO symposium on detritus and its role in aquatic ecosystems. Mem. Inst. Ital. Idrobiol. Suppl. 29:385-403. [Google Scholar]
- 29.Manz, W., T. R. Wendt-Potthoff, T. R. Neu, U. Szewzyk, and J. R. Lawrence. 1999. Phylogenetic composition, spatial structure, and dynamics of lotic bacterial biofilms investigated by fluorescent in situ hybridization and confocal laser scanning microscopy. Microb. Ecol. 37:225-237. [DOI] [PubMed] [Google Scholar]
- 30.Melo, L., and M. J. Vieira. 1999. Physical stability and biological activity of biofilms under turbulent flow and low substrate concentration. Bioproc. Eng. 20:363-368. [Google Scholar]
- 31.Murga, R., P. S. Stewart, and D. Daly. 1995. Quantitative analysis of biofilm thickness variability. Biotechnol. Bioeng. 45:503-510. [DOI] [PubMed] [Google Scholar]
- 32.Murray, J. L. S., and P. A. Jumars. 2002. Clonal fitness of attached bacteria predicted by analog modeling. Bioscience 52:343-355. [Google Scholar]
- 33.Neu, T. R., and J. R. Lawrence. 1997. Development and structure of microbial biofilms in river water studied by confocal laser scanning microscopy. FEMS Microb. Ecol. 24:11-25. [Google Scholar]
- 34.Neu, T. R., G. D. W. Swerhone, and J. R. Lawrence. 2001. Assessment of lectin-binding analysis for in situ detection of glycoconjugates in biofilm systems. Microbiology 147:299-313. [DOI] [PubMed] [Google Scholar]
- 35.Newbold, J. D., T. L. Bott, L. A. Kaplan, B. W. Sweeney, and R. L. Vannote. 1997. Organic matter dynamics in WCC, Pennsylvania, USA. J. N. Am. Benthol. Soc. 16:46-50. [Google Scholar]
- 36.Okabe, S., H. Kuroda, and Y. Watanabe. 1998. Significance of biofilm structure on transport of inert particulates into biofilms. Water Sci. Technol. 38:163-170.
- 37.Palmer, R. J., and D. C. White. 1997. Developmental biology of biofilms: implications for treatment and control. Trends Microbiol. 5:435-440. [DOI] [PubMed] [Google Scholar]
- 38.Pereira, M. A., M. Kuehn, S. Wuertz, T. Neu, and L. F. Melo. 2002. Effect of flow regime on the architecture of a Pseudomonas fluorescens biofilm. Biotechnol. Bioeng. 78:164-171. [DOI] [PubMed] [Google Scholar]
- 39.Peyton, B. M. 1996. Effects of shear stress and substrate loading rate on Pseudomonas aeruginosa biofilm thickness and density. Water Res. 30:29-36. [Google Scholar]
- 40.Picioreanu, C., M. C. M. Van Loosdrecht, and J. J. Heijnen. 1998. Mathematical modeling of biofilm structure with a hybrid differential-discrete cellular automaton approach. Biotechnol. Bioeng. 58:101-116. [DOI] [PubMed] [Google Scholar]
- 41.Picioreanu, C., M. C. M. Van Loosdrecht, and J. J. Heijnen. 2000. A theoretical study on the effect of surface roughness on mass transport and transformation in biofilms. Biotechnol. Bioeng. 68:355-369. [DOI] [PubMed] [Google Scholar]
- 42.Romani, A. M., and S. Sabater. 2001. Structure and activity of rock and sand biofilms in a Mediterranean stream. Ecology 82:3232-3245. [Google Scholar]
- 43.Rounick, J. S., and M. J. Winterbourne. 1983. The formation, structure and utilization of stone surface organic layers in two New Zealand streams. Freshwater Biol. 13:57-72. [Google Scholar]
- 44.Steinman, A. D., and G. A. Lamberti. 1996. Biomass and pigments of benthic algae, p. 295-314. In F. R. Hauer and G. A. Lamberti (ed.), Methods in stream ecology. Academic Press, Inc., New York, N.Y.
- 45.Stoodley, P., D. Debeer, and Z. Lewandowski. 1994. Liquid flow in biofilm systems. Appl. Environ. Microbiol. 60:2711-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoodley, P., J. D. Boyle, I. Doods, and H. M. Lappin-Scott. 1997. Consensus model of biofilm structure, p. 1-9. In J. W. T. Wimpenny, P. Handley, P. Gilbert, H. M. Lappin-Scott, and M. Jones (ed.), Biofilms: community interactions and control. Biolins, Cardiff, United Kingdom.
- 47.Stoodley, P., J. D. Boyle, D. De Beer, and H. M. Lappin-Scott. 1999. Evolving perspectives of biofilm structure. Biofouling 14:75-90. [Google Scholar]
- 48.Stoodley, P., Z. Lewandowski, J. D. Boyle, and H. M. Lappin-Scott. 1999. The formation of migratory ripples in a mixed species bacterial biofilm growing in turbulent flow. Environ. Microbiol. 1:447-455. [DOI] [PubMed] [Google Scholar]
- 49.Stoodley, P., Z. Lewandowski, J. D. Boyle, and H. M. Lappin-Scott. 1999. Structural deformation of bacterial biofilms caused by short term fluctuations in flow velocity: an in situ demonstration of biofilm viscoelasticity. Biotechnol. Bioeng. 65:83-92. [PubMed] [Google Scholar]
- 50.Stoodley, P., L. Hall-Stoodley, J. D. Boyle, F. Jøorgensen, and H. M. Lappin-Scott. 2000. Environmental and genetic factors influencing biofilm structure, p. 53-64. In D. Allison, P. Gilbert, H. M. Lappin-Scott, and M. Wilson (ed.), Community structure and co-operation in biofilms: SGM symposium 59. Cambridge University Press, Cambridge, United Kingdom.
- 51.Sutherland, I. W. 1995. A natural terrestrial biofilm. J. Indust. Microbiol. 17:281-283. [Google Scholar]
- 52.Taylor, I. S., D. M. Paterson, and A. Mehlert. 1999. The quantitative variability and monosaccharide composition of sediment carbohydrates associated with intertidal diatom assemblages. Biogeochemistry 45:303-327. [Google Scholar]
- 53.Underwood, G. J. C., D. M. Paterson, and R. J. Parkes. 1995. The measurement of microbial carbohydrate exopolymers from interstitial sediments. Limnol. Oceanogr. 40:1243-1253. [Google Scholar]
- 54.Velji, M. I., and L. J. Albright. 1985. Microscopic enumeration of attached marine bacteria of seawater, marine sediment, fecal matter, and kelp blade samples following pyrophosphate and ultrasound treatments. Can. J. Microbiol. 32:121-126. [Google Scholar]
- 55.Wagner, M., P. Hutzler, and R. Amann. 1998. Three-dimensional analysis of complex microbial communities by combining confocal laser scanning microscopy and fluorescence in situ hybridization, p. 467-486. In M. H. F. Wilkinson and F. Schut (ed.), Digital analysis of microbes: imaging, morphometry, fluorometry, and motility techniques and applications. John Wiley & Sons, Ltd., London, England.
- 56.Wijeyekoon, S., T. Mino, H. Satoh, and T. Matsuo. 2000. Growth and novel structural features of tubular biofilms produced under different hydrodynamic conditions. Water Sci. Technol. 41:129-138.11381983 [Google Scholar]
- 57.Wimpenny, J., W. Manz, and U. Szewzyk. 2000. Heterogeneity in biofilms. FEMS Microbiol. Rev. 24:661-671. [DOI] [PubMed] [Google Scholar]
- 58.Wolfaardt, G. M., J. R. Lawrence, R. D. Robarts, and D. E. Caldwell. 1998. In situ characterization of biofilm exopolymers involved in the accumulation of chlorinated organics. Microb. Ecol. 35:213-233. [DOI] [PubMed] [Google Scholar]
- 59.Yang, X. M., H. Beyenal, G. Harkin, and Z. Lewandowski. 2000. Quantifying biofilm structure using image analysis. J. Microbiol. Methods 39:109-119. [DOI] [PubMed] [Google Scholar]
- 60.Yu, F. P., G. M. Callis, P. S. Stewart, T. Griebe, and G. A. McFeters. 1994. Cryosectioning of biofilms for microscopic examination. Biofouling 8:85-91. [Google Scholar]
- 61.Zhang, T., J. D. Gu, and H. H. P. Fang. 1999. Microbial distribution in the marine biofilm: proceedings of the IWAQ/IWSA conference on biofilm systems. IWAQ/IWSA, New York, N.Y.