Abstract
The protozoan pathogens Giardia lamblia and Cryptosporidium parvum are major causes of waterborne enteric disease throughout the world. Improved detection methods that are very sensitive and rapid are urgently needed. This is especially the case for analysis of environmental water samples in which the densities of Giardia and Cryptosporidium are very low. Primers and TaqMan probes based on the β-giardin gene of G. lamblia and the COWP gene of C. parvum were developed and used to detect DNA concentrations over a range of 7 orders of magnitude. It was possible to detect DNA to the equivalent of a single cyst of G. lamblia and one oocyst of C. parvum. A multiplex real-time PCR (qPCR) assay for simultaneous detection of G. lamblia and C. parvum resulted in comparable levels of detection. Comparison of DNA extraction methodologies to maximize DNA yield from cysts and oocysts determined that a combination of freeze-thaw, sonication, and purification using the DNeasy kit (Qiagen) provided a highly efficient method. Sampling of four environmental water bodies revealed variation in qPCR inhibitors in 2-liter concentrates. A methodology for dealing with qPCR inhibitors that involved the use of Chelex 100 and PVP 360 was developed. It was possible to detect and quantify G. lamblia in sewage using qPCR when applying the procedure for extraction of DNA from 1-liter sewage samples. Numbers obtained from the qPCR assay were comparable to those obtained with immunofluorescence microscopy. The qPCR analysis revealed both assemblage A and assemblage B genotypes of G. lamblia in the sewage. No Cryptosporidium was detected in these samples by either method.
Giardia and Cryptosporidium are protozoan parasites that cause widespread gastrointestinal illness. Ninety percent of reported outbreaks of these pathogenic protozoans occur through water, while 10% are related to food (29). Direct transmission by person-to-person contact also results in illness and is problematic for children in day care centers. Giardia is the most commonly diagnosed gastrointestinal protozoan in the world and is more frequently associated with outbreaks than Cryptosporidium. However, greater numbers of individuals are affected during outbreaks of Cryptosporidium. The Milwaukee outbreak in 1993 affected several thousand individuals served by a water treatment plant deficient for several days (15). The incidence of food-borne outbreaks due to protozoan pathogens is likely underestimated due to the difficulty of detecting low numbers of organisms, as enrichment techniques cannot be used (29). Detection of Giardia and Cryptosporidium on domestic fresh vegetables and fruits in Norway (27), a wealthy and modern country, has important implications for food safety in North America.
Waterborne outbreaks are associated with drinking water, wells, rivers, lakes, and recreational swimming pools. The reported frequencies of occurrences of contamination of surface water with Giardia and Cryptosporidium are from 60 to 96% in the United States (12, 22) and from 20 to 64% in Canada (11, 36). Their levels in surface waters are very low, ranging from 0.5 to 5,000 organisms in 100 liters of water (11, 21, 22, 23, 36). It has been proposed that action levels to prevent outbreaks with these protozoa should occur when concentrations in 100 liters of water sampled are >5 Giardia cysts (37) and 10 to 30 oocysts of Cryptosporidium (6). Detection of such low numbers of organisms is difficult and requires very sensitive techniques.
Real-time PCR (qPCR) is a technique that provides great sensitivity in detection plus quantitative results (35). Detection of the amplified product using a fluorescent probe during PCR cycling eliminates post-PCR processing and produces results within 30 to 90 min, depending on the type of real-time instrumentation that is employed.
Both Giardia and Cryptosporidium have the potential for zoonotic transmission. Genetic variability exists within species that are infective to mammals (33, 38). Major genotypes have been identified in both Giardia lamblia and Cryptosporidium parvum. G. lamblia isolates in humans and other mammals fall into two major assemblages, A and B. Similarly, C. parvum isolates can be segregated into genotype 1 (recently named Cryptosporidium hominis 20) and genotype 2 (C. parvum). Molecular methods such as qPCR allow not only sensitive detection of pathogens but also the determination of genetic variability within isolates. A difference of 1 nucleotide is distinguishable using the appropriate probe chemistry, such as a molecular beacon (19), thereby providing invaluable information as to the sources of outbreaks and individual cases, permitting rapid action to be taken.
The objective of this study was to develop qPCR primers and TaqMan probes for detection of Giardia cysts and Cryptosporidium oocysts in environmental samples. We focused on methods of extraction of DNA from cysts and oocysts and the development of methods for overcoming PCR-inhibitory substances in water samples and sewage that would be detrimental to qPCR. The detection of Cryptosporidium using qPCR has been reported (5, 8, 13, 14, 32); however, the primer-probe sequences are distinct from the ones described here. To date, there have been no reports of the use of qPCR for detection of Giardia. In addition, this is the first description of a multiplex qPCR assay for simultaneous detection of Giardia and Cryptosporidium.
MATERIALS AND METHODS
Parasites.
Live G. lamblia cysts, produced by passage of the human strain H3 of G. lamblia through Mongolian gerbils, were obtained from Waterborne Inc. (New Orleans, La.). The cysts were in phosphate-buffered saline containing antibiotics and were stored at 4°C until they were used. The WB strain was a kind gift from Mike Belosevic, University of Alberta. The Roberts-Thompson strain of Giardia muris, produced by passage in nu/nu mice, was obtained from Waterborne Inc.
Live C. parvum oocysts (IOWA strain) produced by passage in calves were obtained from Waterborne Inc. Oocysts in phosphate-buffered saline containing antibiotics were stored at 4°C and used within 3 months. The GCH1 isolate was obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergies and Infectious Diseases, NIH; C. parvum strain GCH1 oocysts were from Saul Tzipori.
DNA extraction.
DNA was extracted from cysts using the DNeasy tissue kit (Qiagen, Hilden, Germany). A modification of the animal tissue protocol was employed, adding the following sequence of steps: (i) incubation in ATL (tissue lysis buffer; Qiagen) and proteinase K for 1 h at 55°C; (ii) three freeze-thaw cycles, each cycle consisting of 2 min in liquid nitrogen followed by boiling water; and (iii) three 20-s bursts of sonication (model W-220F cell disruptor; Ultrasonics Inc.). The manufacturer's protocol was followed for purification of DNA through the column, and the DNA was eluted from the columns in sterile double-distilled water (ddH2O).
An alternative method to the use of the sonicator probe was the use of a cup horn (Sonics and Materials, Inc., Newton, Conn.). Samples in snap cap microcentrifuge tubes were placed in a floating rack and sonicated in a cup horn for 30 min at an output of 185 W and 87% amplitude. A slow flow of water was passed through the cup, maintaining the outflow temperature between 30 and 40°C.
DNA was quantified using the PicoGreen double-stranded-DNA quantitation reagent (Molecular Probes, Eugene, Oreg.). Samples were diluted in Tris-EDTA to a volume of 25 μl and added to 25 μl of picogreen Tris-EDTA solution at a 1:200 dilution. Lambda DNA (GibcoBRL, Rockville, Md.) was used to generate a standard curve. Samples were run in triplicate wells, and fluorescence was determined using the 6-carboxyfluorescein (FAM) filter set on an Mx4000 multiplex quantitative PCR system (Stratagene, La Jolla, Calif.). Regression analysis to determine concentrations of unknowns was performed using Microsoft Excel.
Oligonucleotides.
Primer-probe sets for detection of Giardia were designed against the coding region of the β-giardin gene (GenBank no. M36728) of the Portland 1 strain of G. lamblia (9). Two distinct primer-probe sets that produce 74-bp amplicons in qPCR were designed. The first primer set, P241, was based on region 222 to 296 of β-giardin, and the second set, P434, was based on region 411 to 485 (Table 1). Another primer-probe set, also in region 411 to 485 of β-giardin (Table 1), was designed based on the sequence of the H3 isolate of G. lamblia (GenBank no. AY258616).
TABLE 1.
Primer-probe sets for detection of Giardia and Cryptosporidium
| Prime-probe set | Typea | Sequence, 5′-3′ | Nucleotide positione |
|---|---|---|---|
| β-Giardin P241 | F | CATCCGCGAGGAGGTCAA | 222-239 |
| R | GCAGCCATGGTGTCGATCT | 278-296 | |
| P | FAM/AAGTCCGCCGACAACATGTACCTAACGA/BHQ-1 | 241-268 | |
| β-Giardin P434 (P1)b | F | CCTCAAGAGCCTGAACGATCTC | 411-432 |
| R | AGCTGGTCGTACATCTTCTTCCTT | 462-485 | |
| P | FAM/TTCTCCGTGGCAATGCCCGTCT/BHQ-1d | 434-455 | |
| β-Giardin P434 (H3)c | F | CCTCAAGAGCCTGAACGACCTC | 411-432 |
| R | AGCTGGTCATACATCTTCTTCCTC | 462-485 | |
| P | Cy5/TTCTCCGTGGCGATGCCTGTCT/BHQ-2d | 434-455 | |
| COWP P702 | F | CAAATTGATACCGTTTGTCCTTCTG | 583-607 |
| R | GGCATGTCGATTCTAATTCAGCT | 711-733 | |
| P | HEX/TGCCATACATTGTTGTCCTGACAAATTGAAT/BHQ-1d | 672-702 |
F, forward; R, reverse; P, TaqMan probe.
P1, Portland 1 sequence of G. lamblia β-giardin gene (assemblage A) (GenBank no. M36728).
H3, H3 sequence of G. lamblia β-giardin gene (assemblage B) (GenBank no. AY258616).
Reverse complementary sequence of nucleotides.
Relative nucleotide position in the coding sequence of the respective gene.
The gene for the Cryptosporidium oocyst wall protein (COWP) (GenBank no. AF248743) (25) was selected as the gene for designing the primer-probe set for detection of C. parvum. A 151-bp region from 583 to 733 of the coding sequence of the precursor COWP gene was targeted (Table 1). In designing the sequences, 26 partial sequences coding for the oocyst wall protein, from different isolates and species of Cryptosporidium, were examined to identify regions of the gene specific to C. parvum and to the specific genotypes 1 and 2 of C. parvum. These sequences were entered into the ClustalW program (European Bioinformatics Institute), and a multiple alignment was performed to identify unique regions of the gene. The sequences and their GenBank accession numbers are as follows: C. parvum CBAHI (AJ310765), Cryptosporidium baleyi (AF266276), Cryptosporidium sp. strain 715-dog (AF266274), Cryptosporidium felis (AF266263), Cryptosporidium sp. strain 815-bullsnake (AF266277), Cryptosporidium meleagridis (AF248742), C. meleagridis (AF266266), Cryptosporidium wrairi (AF266271), C. wrairi (U35027), C. parvum G2 (AF248743), C. parvum CPACH-1 (AJ310766), Cryptosporidium sp. strain 6-bovine (AF266273), C. parvum G2 (AF161577), Cryptosporidium sp. strain 411-mouse (AF266268), Cryptosporidium sp. strain 518-monkey (AF266272), C. parvum G1 (AF248741), C. parvum 181 (AF266265), C. parvum G1 (AF161578), Cryptosporidium sp. strain 351-ferret (AF266267), Cryptosporidium sp. strain 428-kangaroo (AF266269), Cryptosporidium sp. strain 499-pig (AF266270), Cryptosporidium serpentis (AF266275), C. serpentis (AF161580), Cryptosporidium andersoni (AF266262), Cryptosporidium muris (AF266264), and C. muris (AF161579).
All primers and TaqMan probes were designed using Primer Express versions 1.0 and 1.5 (Applied Biosystems) and were tested for specificity using an nBLAST homology search. The primers were synthesized by Sigma Genosys (Oakville, Ontario, Canada). Probes were synthesized by either Biosearch Technologies Inc. (Novato, Calif.) or Integrated DNA Technologies Inc. (Coralville, Iowa). The β-giardin probes P241 and P434 (Portland 1 sequence) were 5′ labeled with FAM (emission wavelength [λem] = 518 nm), the COWP probe P702 was 5′-hexachlorofluorescein (HEX; λem = 553 nm) labeled, and the β-giardin probe P434 for assemblage B (H3 sequence) was labeled with 1-(epsilon-carboxypentyl)-1′-ethyl-3,3,3′,3′-tetramethylindodicarbocyanine-5 (Cy5; λem = 667 nm). All probes were 3′ labeled with a nonfluorescent Black Hole Quencher (BHQ) dye: BHQ-1 for FAM and HEX and BHQ-2 for Cy5.
qPCR.
PCR was carried out using the Brilliant Quantitative PCR core reagent kit buffers (Stratagene). Each 25-μl PCR used final concentrations of 4 mM magnesium chloride, 20 mM deoxynucleoside triphosphate, 300 or 600 nM each primer, 200 nM probe, a 1:50,000 dilution of the reference dye (carboxy-x-rhodamine ([ROX]), and 2.5 U of SureStart Taq DNA polymerase. A volume of up to 5 μl of DNA was added. The PCRs were performed in an Mx4000 multiplex quantitative PCR system (Stratagene). The cycling conditions consisted of 10 min of incubation at 95°C followed by 40 cycles of alternating temperatures of 95°C for 15 s and 60°C for 1 min. Fluorescence data (three data points) were collected at the end of each cycle, and each sample was run in triplicate wells. A no-template control was included in every assay, and no cycle threshold (Ct) values were consistently obtained after 40 cycles of PCR. Bovine serum albumin (BSA) (fraction V; Sigma, St. Louis, Mo.) was added to the PCR mix to a final concentration of 20 ng/μl for evaluations of environmental samples. The PCR volume was increased to 50 μl for testing sewage extracts.
Sequencing.
The amplicons obtained from the Giardia and Cryptosporidium multiplex qPCR assay using the β-giardin P241 primer-probe set and the COWP P702 primer-probe set were separated on a 2% agarose gel. The two bands (74 and 151 bp) were excised from the gel and purified using the Mini Elute gel extraction kit (Qiagen). The purified amplicons were sequenced at the Core Facility, York University (Toronto, Ontario, Canada) by use of the dideoxy method employing an ABI Prism 377 sequencer.
Environmental water sampling, filtration, and DNA extraction.
Water samples were obtained from Erindale Pond and the Credit River, Mississauga, Ontario, Canada, and from Heart and Professor's Lakes, Brampton, Ontario, Canada, from May to September 2002. The samples were collected in 10-liter plastic carboys (Cole Palmer, Chicago, Ill.) and stored at 4°C until they were used (the same day). Samples (2 liters) were filtered through 3-μm-pore-size, 47-mm-diameter cellulose nitrate filters (Sartorius, Goettingen, Germany) in a parabolic stainless steel funnel (Gelman, Ann Arbor, Mich.) using a vacuum of 250 to 375 mm of Hg generated by a Millipore vacuum/pressure pump (115 V; 60 Hz). Upon filtration of the sample, the funnel was rinsed with ddH2O.
After filtration, the filter was removed, folded twice lengthwise with the upper surface facing out, and placed into a microcentrifuge tube. DNA was extracted directly from the filter using the DNeasy kit. Following incubation in 180 μl of ATL and 20 μl of proteinase K for 1 h at 55°C, the filter was washed with 200 μl of ATL and the wash was pooled with the initial cell lysate. The samples were then processed according to the manufacturer's instructions. DNA was eluted from the silica column using either one or two rounds of 50 μl of ddH2O.
PCR inhibitor removal.
Following filtration of 2 liters of water through the 3-μm-pore-size cellulose nitrate filter, the filter was treated with 20 ml of 0.5 M EDTA, pH 8.0, for 5 min and then washed with ddH2O. After the filter was washed with 2 volumes of ATL, Chelex 100 (Bio-Rad, Hercules, Calif.) was added to the cell lysate to a final concentration of 20%. Polyvinylpyrrolidone (PVP 360) (ICN, Aurora, Ohio) at a concentration of 8% was also added to equal a final concentration of 2%. The 8% stock solution of PVP 360 was made in ATL lysis buffer (Qiagen), incubated at 55°C for 10 min, and pipetted gently to dissolve the PVP 360. The samples were incubated for 30 min at 55°C, subjected to freeze-thaw, and centrifuged at 12,000 × g for 10 min. The supernatant was processed on a DNeasy column following the procedure recommended by the manufacturer, and DNA was eluted from the column using 50 μl of ddH2O.
Collection and processing of sewage.
Giardia and Cryptosporidium were concentrated from raw sewage obtained at the Auteuil and Fabreville wastewater treatment plants in Laval (Québec, Canada). One-liter samples of wastewater were processed by centrifugation for 30 min at 3,000 × g in 1-liter bottles in a Beckman J6 centrifuge equipped with a JS4.2 rotor. The pellets were resuspended in phosphate-buffered saline and pooled to a single volume (5.5 to 9.9 ml). Approximately 1 ml of the concentrate was used for detection of these pathogens using immunofluorescence (IF) microscopy. The remainder of the sample was sent immediately by courier to the Toronto laboratory for DNA extraction and qPCR detection of Giardia and Cryptosporidium.
DNA extraction from sewage.
Following concentration of the sewage by centrifugation, each sample (5 to 9 ml) was split among microcentrifuge tubes (1.5 ml was added to each of four to six tubes). The samples were centrifuged at 12,000 × g for 15 min, and the DNA was extracted from the pellets (∼0.5 ml of packed pellets per entire sample). The tubes were tapped to dislodge the pellets, and 360 μl of ATL was added and thoroughly mixed by vortexing. The samples were subjected to three cycles of freezing-thawing and cooled, and 40 μl of proteinase K was added per tube. The tubes were placed in a cup horn sonicator for 30 min using an output of 185 W, following which the samples were incubated for 1 h at 55°C. The tubes were centrifuged for 1 min at 12,000 × g to remove debris, and the supernatant was treated with Chelex 100 and PVP 360 to final concentrations of 20 and 2%, respectively, and incubated for 30 min at 55°C. The tubes were centrifuged for 10 min at 12,000 × g, the pellets were discarded, and the supernatants from all tubes for each sample were applied equally to two DNeasy columns (Qiagen) to separate and collect the DNA. The eluted material (50 μl [twice] of ddH2O/column) was pooled to obtain a final volume of 200 μl. The purification procedure required 3 to 4 h to perform but was necessary to ensure the removal of qPCR-inhibitory substances.
IF microscopy of sewage samples.
Approximately 1 ml of concentrated sewage was purified by Percoll-sucrose flotation (density, 1.10 g/ml). The entire 1-ml volume of the resuspended pellet was centrifuged on the gradient at 1,000 × g for 20 min. The upper layer and half of the lower layer were collected and centrifuged at 1,000 × g for 20 min and resuspended in 1 ml of water. Fifty microliters (per slide) was placed on microscope slides and dried. Cysts and oocysts were stained with anti-G. lamblia and anti-C. parvum monoclonal antibodies (Aqua-Glo G/C; Waterborne Inc.) and were counted by epifluorescence microscopy at ×400 using an Olympus B-50 research microscope. The whole surface of the slide was examined, and all characteristic structures that exhibited bright-green cyst or oocyst wall staining were counted.
RESULTS AND DISCUSSION
Simplex qPCR.
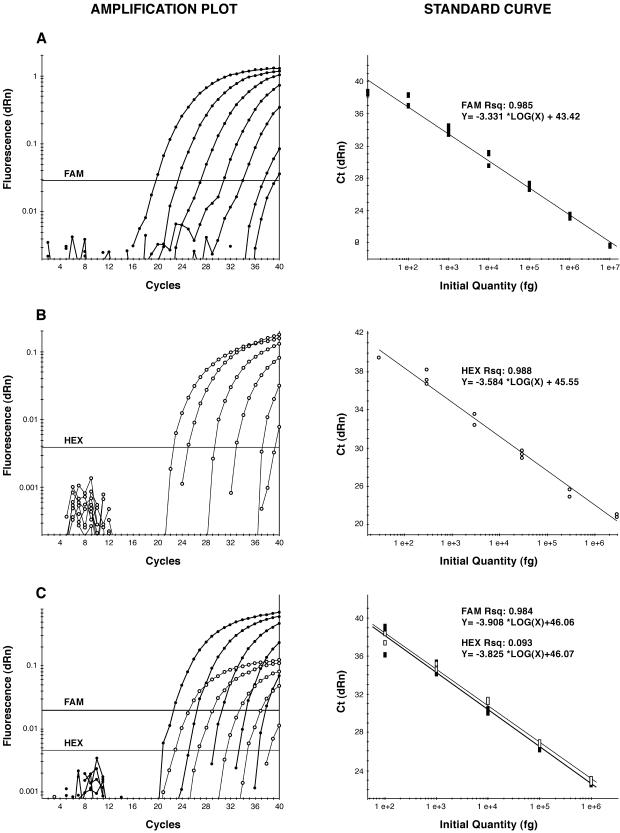
The β-giardin P241 and COWP P702 primer-probe sets provided sensitive detection of DNAs extracted from G. lamblia cysts and C. parvum oocysts, respectively. DNA was detected across a range of dilutions of 7 orders of magnitude (Fig. 1A and B). Standard curves were generated using 10-fold serial dilutions of DNA purified from cysts and oocysts, with correlation coefficients ranging from 0.984 to 0.993 and slopes of −3.331 to −3.908 (Fig. 1). Both the β-giardin and COWP genes are expressed as single-copy genes within the nucleus of each trophozoite and sporozoite, respectively (10, 31). Cysts of Giardia contain two trophozoites that have undergone multiple steps of nuclear division, resulting in 16 copies of total genetic information within each cyst (1). Each Cryptosporidium oocyst contains four nucleated sporozoites. Therefore, there are 16 copies of the β-giardin gene per Giardia cyst and 4 copies of the COWP gene per oocyst. A theoretical estimate of the DNA content of one cyst is 195 fg, and that of one oocyst is 40 fg. Results from the qPCR assay demonstrated considerable sensitivity, as seen from detection of as little as 10 fg of DNA for G. lamblia and 40 fg of C. parvum DNA, corresponding to one copy of the β-giardin gene and four copies of the COWP gene.
FIG. 1.
Simplex and multiplex detection of G. lamblia (solid symbols) and C. parvum (open symbols) using qPCR. The standard curves (right) correspond to the amplification plots (logarithmic view) on the left. (A) Tenfold serial dilutions of G. lamblia cyst DNA ranging from 10 ng to 1 fg and detection using β-giardin P241. (B) Tenfold serial dilutions of C. parvum oocyst DNA ranging from 5.7 ng to 5.7 fg and detection using the COWP primer-probe set P702. (C) Tenfold serial dilutions ranging from 1 ng to 100 fg of DNA from G. lamblia cysts and C. parvum oocysts and simultaneous detection using the β-giardin primer-probe set P241 and the COWP P702 set. The β-giardin probe was FAM labeled, and the COWP probe was HEX labeled. Horizontal lines show thresholds. dRn, baseline-corrected normalized fluorescence; RSq, R2 (indication of the fit of the standard curve to the standard data points plotted).
Our selection of targets for developing primers and probes for qPCR assays was based on molecules that are unique to these organisms and whose biology is important. The β-giardin gene of Giardia codes for a structural protein that is a component of the adhesive disk of the parasite, important in binding of trophozoites to the intestinal epithelium of their host. PCR primers based on the β-giardin gene were first described by Mahbubani and colleagues (16) and have been used for detection of Giardia in environmental water samples (17).
The COWP gene was selected because it codes for a protein that is important in maintaining the integrity of the oocyst wall, allowing the parasite to withstand harsh environmental factors until it is ingested by a new host. Based on the multiple alignments of COWP sequences from different C. parvum isolates that are available in GenBank, we designed primer-probe set P702. This primer-probe has the potential to detect Cryptosporidium that is infective to humans, including C. hominis, C. parvum, C. wrairi, and C. meleagridis. The COWP gene has been used in endpoint and restriction fragment length polymorphism analyses for genotyping Cryptosporidium (40).
Multiplex qPCR.
A multiplex qPCR assay using the β-giardin P241 (FAM-labeled) and COWP P702 (HEX-labeled) primers-probes detected G. lamblia and C. parvum (Fig. 1C) with sensitivities equivalent to that of a simplex assay (Fig. 1A and B). The amplicons generated by multiplex PCR were sequenced and proved to be identical to amplicons generated in the simplex PCR. It was observed that the addition of BSA or BLOTTO to the PCR assay mixture reduced the efficiency of the amplification of the COWP gene in the multiplex qPCR from 100 to 70%. The efficiency of amplification of the β-giardin gene was not altered.
Primer-probe specificity in qPCR.
The specificities of the primer-probe sets for Giardia and C. parvum were examined by performing qPCR assays with a panel of DNAs from related and unrelated organisms. All primer-probe sets were evaluated in qPCR with 500 pg of template DNA from the panel of organisms. The primers-probes were specific to the targets they were designed for and did not detect unrelated organisms (Table 2). The P241 primer-probe set detected G. lamblia and G. muris, the murine species of Giardia, but not C. parvum. The COWP P702 primer-probe set recognized the two C. parvum genotype 2 isolates GCH1 and IOWA but not G. lamblia. None of the primer-probe sets detected the waterborne bacteria Escherichia coli, E. coli 0157:H7, and Microcystis aeruginosa or the fungus Ophiostoma novo-ulmi.
TABLE 2.
Specificities of β-giardin and COWP probes for detection of Giardia and Cryptosporidium in qPCR
| DNA source | Detectiona
|
||
|---|---|---|---|
|
G. lamblia
|
C. parvum P702 | ||
| P241b | P434b | ||
| G. lamblia H3 | + | − | − |
| G. lamblia WB | + | + | − |
| G. muris (Roberts-Thompson) | + | − | − |
| C. parvum GCH1 | − | − | + |
| C. parvum IOWA | − | − | + |
| E. coli (ATCC 8739) | − | − | − |
| E. coli O157:H7 (ATCC 35150) | − | − | − |
| Ophiostoma novo-ulmi (VA30) | − | − | − |
| Microcystis aeruginosa (UTCC 300) | − | − | − |
+, detected; −, not detected.
Portland 1 sequence (GenBank no. M36728).
There is 3 to 5% variability in the sequence of the β-giardin gene among G. lamblia isolates (3; R. A. Guy, C. Xiao, and P. A. Horgen, unpublished data). Mismatches in the nucleotide sequences of the primers-probes were evident when the detections of different strains of G. lamblia with the probes were compared. The P241 primer-probe set detected the WB and H3 strains, corresponding to assemblages A and B of G. lamblia. However, the efficiency of amplification was lower when the mismatch was present compared to detection of a strain using the fully matched primer-probe set. G. muris was also detected using probe P241. Primer-probe set P434, designed against the sequence of the Portland 1 strain of G. lamblia, detected the WB isolate (GenBank no. AY258617; homologous to Portland 1 in the β-giardin gene region we examined) but did not detect the H3 strain of G. lamblia or G. muris (Table 2). In all cases, the matched primer-probe sets detected equivalent concentrations of the matched DNAs, demonstrating that all primer-probe sets were functional. The P434 primer-probe set was used to distinguish between assemblage A and assemblage B isolates of G. lamblia.
DNA extraction efficiency.
Maximum sensitivity for detection of pathogens in environmental samples requires not only excellent performance from primer-probe sets but also the ability to extract the entire DNA from the cysts or oocysts in a sample. Therefore, methods of DNA extraction from Giardia cysts and Cryptosporidium oocysts were evaluated and compared. The DNeasy system was selected as a preferred method because it is rapid and provides an inexpensive means of cleaning up the DNA from contaminates. A comparison was made of modifications of the DNeasy method, including freeze-thaw and sonication, to obtain the largest quantity and best quality of DNA for amplification and detection in qPCR. The PicoGreen method of dsDNA quantification (Molecular Probes) was used to minimize the numbers of cysts and oocysts that were required for these comparisons. It was determined that 1.5 × 105 cysts or oocysts was a sufficient number of cells for accurate quantification of extracted DNA.
A good yield of DNA that was suitable for qPCR was obtained using the tissue protocol of the DNeasy kit, with 1 h of incubation in ATL lysis buffer (including proteinase K) followed by separation on the silica gel column. Importantly, the addition of a three-cycle freeze-thaw step and sonication (three 20-s bursts) increased the yield two- to fivefold (Table 3). DNA concentrations (determined by PicoGreen quantification) of the freeze-thaw- and sonication-treated cysts and oocysts approached maximal theoretical estimates of the concentration that was available from 1.5 × 105 cysts or oocysts. The percent efficiencies of extraction of DNA from G. lamblia cysts were 20.14% ± 6.01% without sonication and 83.02% ± 9.86% using the sonication probe. We adopted this modified DNeasy method for preparing DNA for our standard curves and for processing environmental samples.
TABLE 3.
Concentrations of DNA extracted from cysts and oocysts following modifications to the DNeasy DNA extraction method
| DNA extraction modificationa | Concn of DNA (ng/1.5 × 105 cysts or oocysts)b
|
|
|---|---|---|
| G. lamblia cysts | C. parvum oocysts | |
| 1 h at 55°C + F/T | 5.89 ± 1.76 | 6.21 ± 0.83 |
| 1 h at 55°C + F/T + sonication (probe) | 24.28 ± 2.89 | 10.22 ± 1.52 |
| 1 h at 55°C + F/T + sonication (cup horn) | 24.62 ± 3.42 | 13.59 ± 0.64 |
| F/T + sonication (cup horn) | 29.48 ± 2.52 | 16.60 ± 1.14 |
F/T, three cycles of freeze-thaw.
Data are presented as the mean ± standard deviation of triplicate extractions per treatment group.
We also evaluated the efficacy of DNA extraction using a closed sonication system, the cup horn sonicator (Sonics and Materials Inc.). Direct lysis of cysts and oocysts in the cup horn for 30 min was as effective in releasing DNA (108.18% ± 11.49% efficiency) as was preincubation of the cells in lysis buffer for 1 h at 55°C prior to sonication (84.18% ± 11.69% efficiency) (Table 3).
qPCR-inhibitory effects of concentrated environmental water samples.
Complex matrices, such as surface water and sewage, contain numerous organic and inorganic substances with the potential to inhibit PCRs (30, 39). A study was conducted to determine the ability to use qPCR on DNA extracted from 2-liter water samples obtained from different sources. To detect the presence of inhibitors, the PCR mixtures containing the environmental water sample extracts were spiked with 500 pg of G. lamblia DNA, and DNA was detected using P241 in the qPCR assay. The Ct values from qPCR were compared to those obtained from the same concentration of DNA that was spiked into the PCR mixture containing ddH2O. We observed that DNA extracts obtained from 2-liter samples of either pond water or river water were completely inhibitory to qPCR (Table 4). The addition of BSA (final concentration, 20 ng/μl) to the PCR mixture removed the inhibitory effect (Table 4).
TABLE 4.
Effects of BSA on qPCR detection of spiked Giardia DNA in environmental water extracts
| Water samplea | Ctb
|
|
|---|---|---|
| No BSA | BSA | |
| ddH2O | 21.11 ± 0.44 | 21.17 ± 0.19 |
| Erindale Pond | ||
| 1 | None | 20.77 ± 0.21 |
| 2 | None | 20.67 ± 0.53 |
| Credit River | ||
| 1 | None | 21.08 ± 0.29 |
| 2 | None | 21.46 ± 0.17 |
1 μl from 100 μl of each sample extract was assayed.
Mean ± standard deviation of triplicate wells.
Samples from two recreational lakes were evaluated for detection of Giardia and Cryptosporidium in a multiplex assay. The addition of BSA to the PCR mixture proved sufficient to eliminate inhibition when extracts of 2-liter concentrates from Heart Lake were tested. However, extracts from Professor's Lake were completely inhibitory even in the presence of BSA. Following application of the qPCR method, these samples were analyzed on an agarose gel to determine whether inhibition was due to a lack of amplicon or to interference with fluorescence detection, possibly due to quenching of the fluorophore by inhibitory substances. No amplicon was detected in wells containing the extracts from Professor's Lake; therefore, the inhibitory substance(s) was affecting the PCR and preventing the amplification of DNA. In contrast, the Heart Lake samples revealed two bands on the agarose gel of ∼151 and 74 bp, corresponding to amplification of the COWP and β-giardin sequences.
Several chemical treatment methods were examined for removal of inhibitory substances from the Professor's Lake samples. Addition of Chelex 100 to a final concentration of 5 or 20% had no effect on reduction of inhibition. However, addition of 2% PVP 360 along with 20% Chelex 100 during the DNA extraction process effectively removed inhibitors. Amplification and detection of spiked DNAs of both G. lamblia and C. parvum was observed in the presence of extracts from Professor's Lake in the multiplex qPCR assay (Table 5). It was possible to detect the spiked DNA when 2.5 of the 50 μl of sample extract (1/20th of the total volume) was added to the qPCR. Five-microliter volumes of extract were partially inhibitory, increasing Ct values by 10 to 30%. The lower-molecular-weight material, PVP 40, was less effective at reducing inhibition than PVP 360. The use of 2.5 μl of the PVP 40-treated samples resulted in 15% inhibition, and 5 μl of sample was completely inhibitory (no Ct obtained in qPCR). Addition of Chelex 100 and PVP 360 during DNA extraction did not reduce the yield of DNA as determined by PicoGreen quantification (not shown).
TABLE 5.
Effects of Chelex 100 and PVP 360 on multiplex qPCR detection of Giardia and Cryptosporidium DNAs spiked into lake water extracts
| Water samplea | Ctb
|
|
|---|---|---|
| β-Giardin P241 | COWP P702 | |
| ddH2O | 24.88 ± 0.69 | 27.36 ± 0.40 |
| Heart Lake | ||
| Untreated | 23.98 ± 0.09 | 27.09 ± 0.35 |
| Treatedc | 24.34 ± 0.89 | 26.70 ± 0.89 |
| Professor's Lake | ||
| Untreated | None | None |
| Treated 1c | 24.89 ± 0.13 | 27.61 ± 0.19 |
| Treated 2c | 25.15 ± 0.94 | 27.99 ± 0.60 |
2.5 μl from 50 μl of each sample extract was assayed.
Mean ± standard deviation of triplicate wells.
Treated samples had 0.5 M EDTA, 20% Chelex 100, and 2% PVP 360 treatment during DNA extraction.
The chemical action of polyvinylpolypyrrolidone (PVPP; insoluble) and PVP (water soluble) in removal of inhibitors is based on the binding of phenolic groups. The formation of PVP-phenolic complexes removes molecules, such as humic acids, that contain phenolic groups (4). PVPP has been used to remove PCR inhibitors from Cryptosporidium-contaminated stool (18) and soil (4) samples. PVP 40 is a lower-molecular-weight form of PVP containing fewer reactive sites and possessing greater solubility in water than PVP 360. The lower effectiveness of PVP 40 than PVP 360 in blocking inhibition was likely due to the reduced capacity of PVP 40 to bind phenolic compounds in the samples.
Phenols in aquatic environments come from natural sources, such as biodegradation of humic substances, lignins, and tannins, and from man-made sources, such as derivatives of plastics and degradation of pesticides and herbicides (2). Concentrations of phenolics have been shown to vary between different bodies of water and also seasonally (34). In the present study, only one of the four water bodies required PVP 360 treatment to enable qPCR to be performed. Professor's Lake, a former sand and gravel quarry, is adjacent to a historic landfill site. It is possible that the landfill is a source of the phenol in the lake. An alternative source of the phenol-containing inhibitors may be herbicides applied to the extensive lawns that border the north shore of Professor's Lake.
qPCR versus IF microscopy for quantification of Giardia and Cryptosporidium in sewage.
Sewage samples were processed and examined for the presence of G. lamblia and Cryptosporidium. The extraction of DNA from 1-liter sewage samples was done by Chelex 100 and PVP 360 treatment to remove inhibitors and sonication, using a cup horn to disrupt the cyst and oocyst walls. A great deal of fibrous material was associated with the sewage, and this had the potential to trap pathogens. Within several minutes of being subjected to sonication in the cup horn, clumps that were not disrupted by vortexing were visibly disrupted, possibly releasing trapped cysts or oocysts and exposing them to the lysis reagents and sonication.
The efficiency of extraction from sewage was estimated by spiking half of one sample with 105 cysts of G. lamblia and 105 C. parvum oocysts and comparing the extraction of an equivalent number of cells in the absence of sewage. We found that the efficiency of extraction in the presence of sewage was not reduced.
A total of six sewage extracts were examined for the presence of inhibitors of qPCR by spiking the qPCR mixture containing the sewage extracts with 1 ng each of DNAs from G. lamblia and C. parvum. Inhibition was observed when 10 μl of the 200-μl sewage extract was tested in qPCR but not for 1 μl of sample (Table 6). Therefore, to quantify cysts and oocysts in these samples, 2-μl volumes of the sample extracts were used in qPCR except for sample Auteuil 1 (which inhibited PCR at 2 μl and partially at 1 μl), where 0.5 μl of the 200-μl sample was used. The multiplex assay detected Giardia in all six sewage samples but no Cryptosporidium (data not shown).
TABLE 6.
Multiplex qPCR detection of 1 ng of spiked DNA to determine the presence of inhibitors in 1-liter sewage sample extracts
| Sample | Vol added to qPCR assay (μl) | Ctb
|
|
|---|---|---|---|
| β-Giardin P241 | COWP P702 | ||
| ddH2O | 10 | 23.30 ± 0.63 | 23.88 ± 0.25 |
| Extraction control 1a | |||
| 10 | 23.23 ± 0.23 | 23.12 ± 0.29 | |
| 1 | 23.31 ± 0.49 | 23.63 ± 0.55 | |
| Extraction control 2a | |||
| 10 | 23.95 ± 0.54 | 23.30 ± 0.27 | |
| 1 | 23.31 ± 0.73 | 23.51 ± 0.41 | |
| Auteuil A1 | |||
| 10 | None | None | |
| 1 | 24.20 ± 0.31 | 24.12 ± 0.26 | |
| Auteuil A2 | |||
| 10 | 26.37 ± 0.61 | 25.00 ± 0.55 | |
| 1 | 23.51 ± 0.70 | 23.52 ± 0.46 | |
Extraction controls, all DNA extraction methods minus sewage.
Mean ± standard deviation of triplicate wells.
Quantitative analysis of G. lamblia cysts in the sewage samples was done using the P434 primer-probe set to avoid the problem of potential mismatches in the sequence of the DNA of Giardia in sewage when using the P241 primer-probe set. The primers and probe for one set of P434 were based on the Portland 1 sequence (assemblage A) of G. lamblia, and the other set was the same region of the gene but using the H3 sequence (assemblage B). We used these primer-probe sets in separate assays and summed the numbers obtained from each to obtain the total number of G. lamblia cysts present in each sample. Giardia was detected in all six samples from the Auteuil and Fabreville (Québec, Canada) treatment facilities, with cyst concentrations ranging from 2,653 to 13,408/liter of sewage (Table 7). No Ct values were obtained in the negative control samples, consisting of the extraction procedure in the absence of sewage, suggesting that Ct values obtained in the sewage samples were specific to G. lamblia. The values obtained from qPCR were similar to those obtained by enumerating cysts using IF microscopy (Table 7).
TABLE 7.
Comparison of qPCR and IF microscopy for detection of G. lamblia and Cryptosporidium in 1-liter sewage samples
| Samplea | No. of G. lamblia cysts
|
No. of C. parvum oocysts
|
||||
|---|---|---|---|---|---|---|
| qPCR
|
IF microscopy | qPCR | IF microscopy | |||
| Assemblage
|
Total | |||||
| A | B | |||||
| NC | 0 | 0 | 0 | 0 | ||
| A1 | 496 | 5,146 | 5,642 | 2,380 | 0 | 0 |
| A2 | 2,476 | 8,340 | 10,816 | 9,880 | 0 | 0 |
| A3 | 5,672 | 7,736 | 13,408 | 7,980 | 0 | 0 |
| F1 | 838 | 1,815 | 2,653 | 9,900 | 0 | 0 |
| F2 | 2,196 | 3,663 | 5,859 | 6,660 | 0 | 0 |
| F3 | 545 | 3,331 | 3,876 | 4,290 | 0 | 0 |
NC, negative control in qPCR; A, Auteuil Treatment Facility; F, Fabreville Treatment Facility.
Both assemblage A and B genotypes were detected in the samples, with the majority of cysts (58 to 91%) being of the assemblage B genotype (Table 7). Recently, Caccìo and colleagues (3) reported genotyping of G. lamblia from clinical stool samples using PCR and restriction fragment length polymorphism of the β-giardin gene. Both genotypes were detected in the stool samples, with a predominance of the assemblage A genotype. We have also detected assemblage A and B genotypes in stool; however, the majority of samples were genotype B (Guy et al., unpublished).
Interestingly, no Cryptosporidium was detected in any of the samples (Table 7), as determined by qPCR using both the multiplex assay and a simplex assay with the COWP P702 primer-probe and also using IF microscopy. The lack of detection of Cryptosporidium was not due to inhibition of either COWP amplification or fluorescence of the HEX fluorophore, because C. parvum DNA spiked into qPCR wells containing extracts from the sewage samples was detectable (Table 6). Seasonal variation in the abundance of Giardia and Cryptosporidium in sewage has been reported, with higher levels of Giardia and low to no detection of Cryptosporidium in the fall months (7), the time of year we sampled. Giardia cysts are consistently seen in sewage influent. For instance, Wallis and colleagues (36) detected Giardia in 73% of raw sewage in a cross-Canada survey. Detection of Cryptosporidium was sporadic: only 6.1% of samples contained Cryptosporidium (36), and low numbers of oocysts (<500) were observed (36). High levels of Giardia and low-level sporadic detection of Cryptosporidium has also been reported by others (7, 24, 26, 28).
The results obtained in the present study from spiking experiments revealed that there were additional qPCR-inhibitory substances in the DNA extracts from the sewage samples, as it was not possible to use larger volumes of the 200-μl sewage extract (no greater than 2 μl, and 0.5 μl in the case of the sample Auteuil 1) in the qPCR. Therefore, the quantitative evaluation limit for the qPCR method using sewage samples was 100 oocysts. Lack of detection of Cryptosporidium in the sewage suggests that oocysts were either absent or present at very low levels.
In summary, new methods for qPCR were described, and their application to pathogen detection in environmental samples, including water and sewage, was demonstrated. With the appropriate adaptation of sample extraction, qPCR is applicable to a variety of detection systems. These approaches may provide a sensitive method for detection of low numbers of pathogens, and their application would contribute to understanding the distribution and abundance of Giardia and Cryptosporidium in the environment and the risks they pose to human health.
Acknowledgments
This research was funded by NSERC Strategic and Operating grants to P. A. Horgen.
The GCH1 isolate was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH; the C. parvum strain GCH1 oocysts were from Saul Tzipori. In addition, we thank Mike Belosevic from the University of Alberta for his kind gift of the WB isolate of G. lamblia. We are very grateful to Sonics and Materials, Inc., for the use of their cup horn sonicator to test its efficiency in disrupting cyst and oocyst walls.
REFERENCES
- 1.Bernander, R., J. E. D. Palm, and S. G. Svärd. 2001. Genome ploidy in different stages of the Giardia lamblia life cycle. Cell. Microbiol. 3:55-62. [DOI] [PubMed] [Google Scholar]
- 2.Bruzzoniti, M. C., C. Sarzanini, and E. Mentasti. 2000. Preconcentration of contaminants in water analysis. J. Chromatogr. A 902:289-309. [DOI] [PubMed] [Google Scholar]
- 3.Caccìo, S. M., M. De Giacomo, and E. Pozio. 2002. Sequence analysis of the β-giardin gene and development of a polymerase chain reaction-restriction fragment length polymorphism assay to genotype Giardia duodenalis cysts from human faecal samples. Int. J. Parasitol. 32:1023-1030. [DOI] [PubMed] [Google Scholar]
- 4.Cullen, D. W., and P. R. Hirsch. 1998. Simple and rapid method for direct extraction of microbial DNA from soil for PCR. Soil Biol. Biochem. 30:983-993.
- 5.Fontaine, M., and E. Guillot. 2002. Development of a TaqMan quantitative PCR assay specific for Cryptosporidium parvum. FEMS Microbiol. Lett. 214:13-17. [DOI] [PubMed] [Google Scholar]
- 6.Hass, C. N., and J. B. Rose. 1995. Developing an action level for Cryptosporidium. J. Am. Water Works Assoc. 87:81-84.11540484 [Google Scholar]
- 7.Heitman, T. L., L. M. Frederick, J. R. Viste, N. J. Guselle, U. M. Morgan, R. C. A. Thompson, and M. E. Olson. 2002. Prevalence of Giardia and Cryptosporidium and characterization of Cryptosporidium spp. isolated from wildlife, human, and agricultural sources in the North Saskatchewan River Basin in Alberta, Canada. Can. J. Microbiol. 48:530-541. [DOI] [PubMed] [Google Scholar]
- 8.Higgins, J. A., R. Fayer, J. M. Trout, L. Xiao, A. A. Lal, S. Kerby, and M. C. Jenkins. 2001. Real-time PCR for the detection of Cryptosporidium parvum. J. Microbiol. Methods 47:323-327. [DOI] [PubMed] [Google Scholar]
- 9.Holberton, D., D. A. Baker, and J. Marshall. 1988. Segmented alpha-helical coiled-coil structure of the protein giardin from the Giardia cytoskeleton. J. Mol. Biol. 204:789-795. [DOI] [PubMed] [Google Scholar]
- 10.Holberton, D. V., and J. Marshall. 1995. Analysis of consensus sequence patterns in Giardia cytoskeleton gene promoters. Nucleic Acids Res. 23:2945-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isaac-Renton, J. L., W. Moorehead, and A. Ross,. 1996. Longitudinal studies of Giardia contamination in two community drinking water supplies: cyst levels, parasite viability, and health impact. Appl. Environ. Microbiol. 62:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeChevallier, M. W., W. D. Norton, and R. G. Lee. 1991. Occurrence of Giardia and Cryptosporidium in surface water supplies. Appl. Environ. Microbiol. 57:2610-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limor, J. R., A. A. Lal, and L. Xiao. 2002. Detection and differentiation of Cryptosporidium parasites that are pathogenic for humans by real-time PCR. J. Clin. Microbiol. 40:2335-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacDonald, L. M., K. Sargent, A. Armson, R. C. A. Thompson, and J. A. Reynoldson. 2002. The development of a real-time quantitative-PCR method for characterization of a Cryptosporidium parvum in vitro culturing system and assessment of drug efficacy. Mol. Biochem. Parasitol. 121:279-282. [DOI] [PubMed] [Google Scholar]
- 15.MacKenzie, W. R., W. L. Schell, K. A. Blair, D. G. Addiss, D. E. Peterson, N. J. Hoxie, J. J. Kazmierczak, and J. P. Davis. 1995. Massive outbreak of waterborne Cryptosporidium infection in Milwaukee, Wisconsin: recurrence of illness and risk of secondary transmission. Clin. Infect. Dis. 21:57-62. [DOI] [PubMed] [Google Scholar]
- 16.Mahbubani, M. H., A. K. Bej, M. H. Perlin, F. W. Schaeffer III, W. Jakubowski, and R. M. Atlas. 1991. Detection of Giardia cysts by using the polymerase chain reaction and distinguishing live from dead cysts. Appl. Environ. Microbiol. 57:3456-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahbubani, M. H., F. W. Schaefer III, D. D. Jones, and A. K. Bej. 1998. Detection of Giardia in environmental waters by immuno-PCR amplification methods. Curr. Microbiol. 36:107-113. [DOI] [PubMed] [Google Scholar]
- 18.McLauchlin, J., S. Pedraza-Díaz, C. Amar-Hoetzeneder, and G. L. Nichols. 1999. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J. Clin. Microbiol. 37:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mhlanga, M. M., and L. Malmberg. 2001. Using molecular beacons to detect single-nucleotide polymorphisms with real-time PCR. Methods 25:463-471. [DOI] [PubMed] [Google Scholar]
- 20.Morgan-Ryan, U. M., A. Fall, L. A. Ward, N. Hijjawi, I. Sulaiman, R. Fayer, R. C. A. Thompson, M. Olson, A. Lal, and L. Xiao. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433-440. [DOI] [PubMed] [Google Scholar]
- 21.Ong, C., W. Moorehead, A. Ross, and J. Isaac-Renton. 1996. Studies of Giardia spp. and Cryptosporidium spp. in two adjacent watersheds. Appl. Environ. Microbiol. 62:2798-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ongerath, J. E., G. D. Hunter, and F. B. DeWalle. 1995. Watershed use and Giardia cyst presence. Water Res. 29:1295-1299. [Google Scholar]
- 23.Payment, P., A. Berte, M. Prevost, B. Menard, and B. Barbeau. 2000. Occurrence of pathogenic microorganisms in the Saint Lawrence River (Canada) and comparison of health risks for populations using it as their source of drinking water. Can. J. Microbiol. 46:565-576. [PubMed] [Google Scholar]
- 24.Payment, P., R. Plante, and P. Cejka. 2001. Removal of indicator bacteria, human enteric viruses, Giardia cysts, and Cryptosporidium oocysts at a large wastewater primary treatment facility. Can. J. Microbiol. 47:188-193. [PubMed] [Google Scholar]
- 25.Pedraza-Diaz, S., C. Amar, and J. McLauchlin. 2002. The identification and characterisation of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol. Lett. 189:189-194. [DOI] [PubMed] [Google Scholar]
- 26.Rimhanen-Finne, R., P. Ronkainen, and M.-L. Hänninen. 2001. Simultaneous detection of Cryptosporidium parvum and Giardia in sewage sludge by IC-PCR. J. Appl. Microbiol. 91:1030-1035. [DOI] [PubMed] [Google Scholar]
- 27.Robertson, L. J., and B. Gjerde. 2001. Occurrence of parasites on fruits and vegetables in Norway. J. Food Prot. 64:1793-1798. [DOI] [PubMed] [Google Scholar]
- 28.Robertson, L. J., C. A. Paton, A. T. Campbell, P. G. Smith, M. H. Jackson, R. A. Gilmour, S. E. Black, D. A. Stevenson, and H. V. Smith. 2000. Giardia cysts and Cryptosporidium oocysts at sewage treatment works in Scotland, UK. Water Res. 34:2310-2322. [Google Scholar]
- 29.Rose, J. B., and T. R. Slifko. 1999. Giardia, Cryptosporidium, and Cyclospora and their impact on foods: a review. J. Food Prot. 62:1059-1070. [DOI] [PubMed] [Google Scholar]
- 30.Rossen, L., P. Nørskov, K. Holmstrøm, and O. F. Rasmussen. 1992. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 17:37-45. [DOI] [PubMed] [Google Scholar]
- 31.Spano, F., C. Puri, L. Ranucci, L. Putignani, and A. Crisanti. 1997. Cloning of the entire COWP gene of Cryptosporidium parvum and ultrastructural localization of the protein during sexual parasite development. Parasitology 114:427-437. [DOI] [PubMed] [Google Scholar]
- 32.Tanriverdi, S., A. Tanyeli, F. Balamisli, R. Koksal, F. Kilinc, X. Feng, G. Batzer, S. Tzipori, and G. Widmer. 2002. Detection and genotyping of oocysts of Cryptosporidium parvum by real-time PCR and melting curve analysis. J. Clin. Microbiol. 40:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, R. C. A. 2000. Giardiasis as a re-emerging infectious disease and its zoonotic potential. Int. J. Parasitol. 30:1259-1267. [DOI] [PubMed] [Google Scholar]
- 34.Thoss, V., M. S. Baird, M. A. Lock, and P. V. Courty. 2002. Quantifying the phenolic content of freshwaters using simple assays with different underlying reaction mechanisms. J. Environ. Monit. 4:270-275. [DOI] [PubMed] [Google Scholar]
- 35.Walker, N. J. 2002. Tech.Sight. A technique whose time has come. Science 296:557-559. [DOI] [PubMed] [Google Scholar]
- 36.Wallis, P. M., S. L. Erlandsen, J. L. Isaac-Renton, M. E. Olson, W. J. Robertson, and H. van Heulen. 1996. Prevalence of Giardia cysts and Cryptosporidium oocysts and characterization of Giardia spp. isolated from drinking water in Canada. Appl. Environ. Microbiol. 62:2789-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallis, P. M., D. Matson, M. Jones, and J. Jamieson. 2001. Application of monitoring data for Giardia and Cryptosporidium to boil water advisories. Risk Anal. 21:1077-1085. [DOI] [PubMed] [Google Scholar]
- 38.Widmer, G., J. Lin, V. Kapur, X. Feng, and M. S. Abrahamsen. 2002. Genomics and genetics of Cryptosporidium parvum: the key to understanding cryptosporidiosis. Microb. Infect. 4:1081-1090. [DOI] [PubMed] [Google Scholar]
- 39.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao, L., J. Limor, U. M. Morgan, I. M. Sulaiman, R. C. A. Thompson, and A. A. Lal. 2000. Sequence differences in the diagnostic target region of the oocyst wall protein gene of Cryptosporidium parasites. Appl. Environ. Microbiol. 66:5499-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]