Abstract
A real-time reverse transcription-PCR method targeting the rbcL gene was developed for the detection and quantitation of the Florida red tide organism, Karenia brevis. The assay was sensitive to less than 1 cell per reaction, did not detect rbcL from 38 nontarget taxa, and accurately quantitated K. brevis organisms in red tide samples from around Florida. These studies have resulted in a sensitive and specific method for K. brevis detection in the marine environment.
Karenia brevis (Davis cf. Hansen & Moestrup = Gymnodinium breve) is an unarmored, non-peridinin-containing dinoflagellate that grows to ca. 20 to 40 μm in diameter. The organism is positively phototactic (3), is negatively geotactic (8), swims at a speed of ca. 1 m h−1 (12) and is thought to be an obligate photoautotroph (1). K. brevis is the causative agent of the recurring red tide blooms (21 of 22 years from 1975 to 1997) observed in the Gulf of Mexico and off the southeastern Atlantic coast of the United States (14), which have been reported since the Spanish conquests (5). Lipophilic brevetoxins (9) produced by K. brevis can result in massive fish kills and have been implicated in the mortality of 700 bottlenose dolphins off the east coast of the United States in 1987 (6) and the mysterious deaths of 149 Florida manatees in 1995 and 1996 (15). In cases of human exposure, brevetoxin can cause respiratory distress by inhalation and food poisoning by consumption of tainted shellfish.
Current methods for the detection of K. brevis depend on microscopy or pigment analysis, methods which are time-consuming and require a considerable amount of expertise and skill (10). Isolation of dinoflagellates and cultivation from environmental samples to confirm identity may take months. Consequently, rapid molecular methods to detect K. brevis in the environment are needed. To this end, we have been investigating the ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) large-subunit gene (rbcL) as a potential molecular marker for this organism. RuBisCO is the primary carbon-fixing enzyme in photoautotrophic organisms. K. brevis and the other fucoxanthin-containing dinoflagellates have a form ID rbcL enzyme, and genetic evidence suggests that they contain plastids of haptophyte origin acquired through tertiary endosymbiosis (7, 13).
As rbcL is highly expressed in viable cells and mRNA levels can be orders of magnitude greater than those of DNA, the mRNA was targeted for this study. As RNA is rapidly degraded in the environment, an RNA target will give an indication of a viable population compared to what is detected by DNA-based methods, which may detect dead cells as well.
To obtain sequence data, a PCR primer set was designed with sequence data from Karenia mikimotoi (GenBank accession no. ABO34635) (13) by modifying existing chromophyte rbcL primers (11) in order to amplify a 554-bp region (approximately one-third) of Karenia's rbcL gene (forward primer, GATGATGARAAYATTAACTC; reverse primer, ATTTGTCCCGCATTGATTCCT [International Union of Pure and Applied Chemistry degeneracy symbols were used]).
Cultures of K. brevis were provided courtesy of Karen Steidinger of the Florida Fish and Wildlife Conservation Commission's Florida Marine Research Institute. Strains were isolated by her lab from the following locations around the Florida coast: Apalachicola, Charlotte Harbor, Mexico Beach, Jacksonville, and Piney Island. Strains used in this analysis were named for their isolation location and the plate well into which they were isolated. Several nontarget algal strains of diverse lineage were obtained from either the Provasoli-Guillard Center for Culture of Marine Phytoplankton (CCMP; West Boothbay Harbor, Maine) or from the Steidinger lab (see Table 1). All strains were under a 12-h-light-12-h-dark light regimen at 26 μmol s−1 m−2 and were incubated at 20 or 14°C in F/2 medium (4), which was modified for each strain's needs according to CCMP's directions.
TABLE 1.
Positive and negative controls for amplification by real-time RT-PCR
| Species | Strain or clone | Detection by real-time PCR | Species | Strain or clone | Detection by real-time PCR | |
|---|---|---|---|---|---|---|
| Positive controls | ||||||
| K. brevis | Apalachicola B5 | + | ||||
| K. brevis | Apalachicola C6 | + | ||||
| K. brevis | Charlotte Harbor A2 | + | ||||
| K. brevis | Charlotte Harbor C2 | + | ||||
| K. brevis | Mexico Beach B3 | + | ||||
| K. brevis | Mexico Beach C5 | + | ||||
| K. brevis | Jacksonville C3 | + | ||||
| K. brevis | Piney Island A3 | + | ||||
| K. brevis | Piney Island B4 | + | ||||
| K. brevis | Wilson | + | ||||
| Negative controls | ||||||
| Dinoflagellates | ||||||
| K. mikimotoi | CCMP430 | − | ||||
| Amphidinium carterae | CCMP1314 | − | ||||
| Akashiwo sanguinea | CCMP1321 | − | ||||
| Alexandrium tamarense | CCMP1493 | − | ||||
| Glenodinium foliacrum | NAa | − | ||||
| Gymnodinium catenatum | CCMP1937 | − | ||||
| Gyrodinium sp. | NA | − | ||||
| Kryptoperidinium foliaceum | NA | − | ||||
| Lingulodinium polyedra | CCMP1738 | − | ||||
| Prorocentrum micans | NA | − | ||||
| Scrippsiella trochoidea | NA | − | ||||
| Scrippsiella precaria | NA | − | ||||
| Diatoms | ||||||
| Phaeodactylum tricornutum | CCMP1327 | − | ||||
| Cylindrotheca sp. | ST6CH2 clone | − | ||||
| Skeletonema sp. | ST4CH31 clone | − | ||||
| Skeletonema sp. | ST4CH14 clone | − | ||||
| Raphidophyte Heterosigma akashiwo | NA | − | ||||
| Praisinophytes | ||||||
| Tetraselmis sp. | 850001 | − | ||||
| Tetraselmis sp. | CCMP961 | − | ||||
| Unidentified species | CCMP1536 | − | ||||
| Prymnesiophytes | ||||||
| Isochrysis sp. | 3C | − | ||||
| Pavlova lutheri | CCMP1325 | − | ||||
| Prymnesium parvum | NA | − | ||||
| Unidentified species | ST8CH26 clone | − | ||||
| Unidentified species | ST1CH3 clone | − | ||||
| Chlorophytes | ||||||
| Clamydomonas euryale | CCMP219 | − | ||||
| Unidentified species | ST5SY7 clone | − | ||||
| Chlamydomonas sp. | ST2SY2 clone | − | ||||
| Pycnococcus sp. | ST6SY8 clone | − | ||||
| Cyanophytes | ||||||
| Synechococcus sp. | CCMP836 | − | ||||
| Synechococcus sp. | WH7803 | − | ||||
| Synechococcus sp. | ST2SY26 clone | − | ||||
| Prochlorococcus sp. | ST2SY33 clone | − | ||||
| Trichodesmium sp. | ST8SY15 clone | − | ||||
| Trebouxiophyte Chlorella autotrophica | CCMP243 | − | ||||
| Coscinodicsophyte Thatassiosira pseudonana | CCMP1335 | − | ||||
| Eustigmatophytes | ||||||
| Nannochloropsis sp. | ST3CH27 clone | − | ||||
| Nannochloropsis sp. | ST1CH4 clone | − | ||||
| Xanthophyte Heterococcus sp. | ST6CH33 clone | − |
NA, not available.
K. brevis cells were harvested by centrifugation (10 min at 5,000 × g), and the DNA was extracted by a modified phenol-chloroform method (2). PCR amplification was conducted with final concentrations of 1 μM for the primers, 3 mM for MgCl2, 0.4 mM for each deoxynucleoside triphosphate, and 2.5 U of Taq polymerase (Promega Corp., Madison, Wis.). Cycling conditions were 40 repetitions of 95°C for 1 min, 50°C for 1 min, and 72°C for 1.5 min, with a final extension step at 72°C for 15 min. Amplification was confirmed by agarose gel electrophoresis. PCR amplicons were purified with a QIAquick PCR purification kit (QIAGEN, Valencia, Calif.) and ligated into the pCR II vector, and TOP10 cells were transformed according to the manufacturer's instructions (Invitrogen Corp., Carlsbad, Calif.). Transformants were plated onto 2XYT plates containing 50 μg (each) of kanamycin and ampicillin per ml. White colonies were screened for insert size by PCR amplification. Positive clones were grown in 2XYT broth with antibiotics, and plasmid DNA was extracted with a Wizard Plus SV miniprep spin kit (Promega Corp.). Clones from nontarget species from our rbcL clone library were also grown and extracted as described above. Sequencing of the 554-bp K. brevis and K. mikimotoi rbcL insert was performed at the DNA Sequencing Core laboratory at the University of Florida.
One of the sequenced clones carrying the 554-bp insert from K. brevis APC6 (clone 15) was selected for use in sensitivity testing. Nontarget environmental rbcL clones (from the same region of the gene) were obtained from the Gulf of Mexico during a previous study to initially test specificity (see Table 1). Based on the direction of the insert, the vector was linearized by digesting the plasmid with either HindIII or EcoRV and a sense transcript was made by in vitro transcription using the T7 or SP6 promoter site. The transcripts were purified with a QIAGEN RNeasy RNA extraction kit, with the DNase digestion step being performed according to the manufacturer's instructions. These transcripts were quantified with a Ribogreen RNA quantification kit according to the manufacturer's instructions (Molecular Probes, Inc., Eugene, Oreg.), mixed 1:1 with an RNA storage buffer (8 M guanidinium isothiocyanate, 80 mM Tris-HCl [pH 8.5], 24 mM MgCl2, 140 mM KCl), aliquoted, and frozen at −80°C. The K. brevis APC6 clone 15 transcript was used to generate real-time reverse transcription (RT)-PCR standard curves, while the others were used to test the specificity of the primer-probe set.
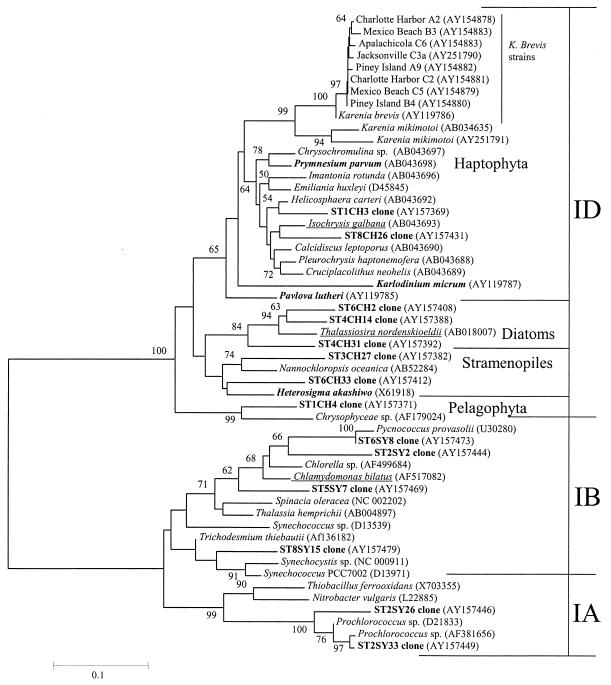
Sequences for phylogenetic comparison were obtained from GenBank. During the course of this study, a sequence for K. brevis appeared in GenBank (16). Sequences were aligned and analyzed using the KODON software package, version 1.0 (Applied Maths, Inc., Austin, Tex.), which uses a Clustal W alignment method. Phylogenetic and molecular evolutionary analysis was conducted using MEGA2 software (version 2.1; S. Kumar, K. Tamura, I. B. Jakobsen, and M. Nei, Arizona State University, Tempe, 2001) using both nucleotide and deduced amino acid sequence data. All nontarget strains, their representative accession numbers, and their relationships based on deduced amino acid residues are shown in Fig. 1.
FIG. 1.
Neighbor-joining phylogenetic tree based on deduced amino acid sequences with a Poisson distance correction showing relationships between form I rbcL sequences from K. brevis and other phytoplankton species, as well as clones obtained on a cruise to the Mississippi River plume in the Gulf of Mexico. Boldface taxa were tested by real-time RT-PCR as nontarget controls. There were many taxa tested as nontarget strains whose rbcL sequences were not available in GenBank, and closest sequenced representatives are underlined.
Sequence data from the K. brevis rbcL clones showed a short (91-bp) region that was markedly different from K. mikimotoi's rbcL sequence. This portion of the rbcL gene of K. brevis was selected as the target for a primer and probe set for the TaqMan Taq nuclease assay. A primer set and an internal fluorogenic probe were designed to amplify and detect the 91-bp region (forward primer, TGAAACGTTATTGGGTCTGT; reverse primer, AGGTACACACTTTCGTAAACTA; internal probe, FAM [6-carboxyfluorescein]-TTAACCTTAGTCTCGGGTA-TAMRA [6-carboxytetramethylrhodamine]). For real-time RT-PCR, 5 μl of the target was added to 45 μl of a one-step RT-PCR mixture prepared from 2×RT-PCR TaqMan master mix (Applied Biosystems, Foster City, Calif.) containing each primer at a concentration of 1 μM, 2 mM MgCl, and a 0.5 μM concentration of the probe. Cycling conditions were as follows: a precycling reverse transcription step of 45°C for 30 min; an initial denaturation step of 95°C for 10 min; and then 40 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. Reaction mixtures were run in the Applied Biosystems 7700 sequence detection system and analyzed with their supplied software.
Cell counts for all cultured algal strains (including K. brevis) were carried out by filtering 1 ml of culture onto 0.22-μm-pore-size black polycarbonate Poretics filters (Osmonics Inc., Minnetonka, Minn.). Cells were counted by using epifluorescence microscopy on an Olympus BX-60 microscope with the 20× objective and blue excitation (filter set U-MNIB).
RNA from the culture was extracted using the QIAGEN RNeasy spin kit with the following modifications. Culture samples (1 ml) were filtered onto a 0.45-μm-pore-size HVpolyvinylidene difluoride filter (Millipore Durapore). The filters were placed into 2-ml screw-cap microcentrifuge tubes containing 750 μl of RLT lysis buffer (QIAGEN) with 2-mercaptoethanol (10 μl ml−1). The filters were incubated for 10 min at room temperature, 500 μl was removed into a 1.5-ml microcentrifuge tube, and RNA extraction continued according to the manufacturer's instructions (QIAGEN). The extracted RNA was quantified using a Ribogreen RNA quantification kit according to the manufacturer's instructions. All nontarget algal strains were tested for amplification with the real-time primer-probe set with 10 pg of nontarget RNA per reaction mixture.
Field samples were collected by the Florida Marine Research Institute from several locations at several different times in Collier County (west coast of Florida; collected 28 March, 2 and 9 April, and 2 May 2003) and from the Indian River lagoon (east coast of Florida; collected 13 December 2002) during both bloom and nonbloom events. Algae in field samples were counted by microscopy by the Florida Fish and Wildlife Conservation Commission prior to our receiving them. Algae from field samples were extracted as described above, but 10 to 20 ml was extracted and 5 μl of the extract was added to the RT-PCR mixture.
The TaqMan probe-based RT-PCR assay (91-bp amplicon) yielded only positive results with K. brevis strains (Table 1). All other dinoflagellates (including K. mikimotoi) and algal strains resulted in no amplification. All strains tested were present in sufficient concentrations to allow for amplification based on the lowest detectable concentration of K. brevis.
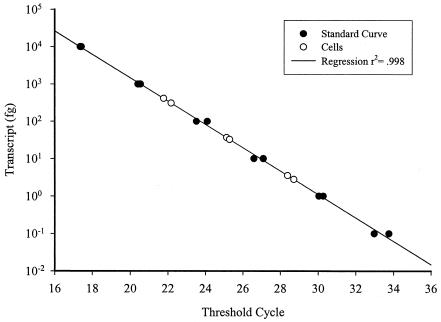
Standard curves derived by using the in vitro transcript from APC6 clone 15 showed sensitivity over a range of concentrations spanning 7 orders of magnitude, ranging from 0.1 fg to 1,000 pg, as shown in Fig. 2. Standard curves using whole-cell extracts from K. brevis culture were sensitive to as little as 1 pg of total RNA (less than 1 cell per reaction, based on cell counts and dilution).
FIG. 2.
Real-time RT-PCR standard curve generated from the APC6 clone 15 transcript showing the linearity of the method, covering 7 orders of magnitude (filled circles [trendline]). Also shown are amplification results from K. brevis cellular extracts corresponding to 100 cells, 10 cells, and 1 cell per reaction (open circles).
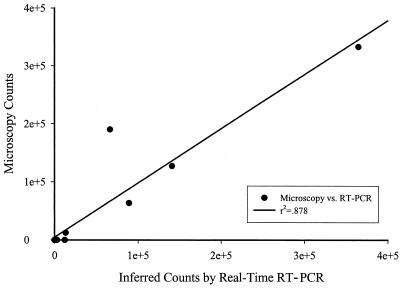
Red tide bloom and nonbloom samples from around Florida were analyzed for K. brevis using this method. From the west coast, 15 samples were analyzed; 11 were nonbloom and 4 were moderate to high bloom. The two samples from the east coast were composed of one bloom and one nonbloom. Microscopy counts of the nonbloom samples were below the detection limit of 333 cells liter−1. Counts inferred by RT-PCR were mostly 0.0 cell liter−1 or below the detection limit by microscopy (7 of 12 samples).Of the remaining five nonbloom samples, three gave a result of approximately 1,000 cells liter−1, one indicated 3,000 cells liter−1, and one indicated 12,000 cells liter−1. The last sample's result may be due to contamination of the sample. For the bloom samples, all but one indicated that cell density was very close to that of the microscopy counts, and one sample indicated approximately one-third the density by microscopy. As this method targets mRNA, it is possible that the cells in the last sample were not producing high levels of transcript or that they were no longer viable. Figure 3 summarizes this comparison of cell densities for these field samples as enumerated by microscopy and inferred from real-time RT-PCR. A good correlation (r2 = 0.878) was observed between the results of both methods for field samples.
FIG. 3.
Comparison of microscopy cell counts and real-time RT-PCR-inferred cell counts from natural bloom samples.
Using the TaqMan probe, we were able to amplify and detect a wide range of concentrations of K. brevis to the exclusion of all nontarget DNA and RNA tested, with a detection limit of less than 100 cells liter−1 when 20 ml of seawater is extracted. When larger volumes are filtered, lower detection limits should be attainable. The dynamic range over which this technique is effective covers the range of natural K. brevis blooms in the environment. When an environment contains <1,000 cells liter−1 (as determined by microscopy cell counts), K. brevis is considered to be present but poses no risk of adverse health effects or shellfish contamination. Samples with >1,000 cells liter−1 are considered to have a very low level bloom, carrying a slight risk of respiratory irritation. At concentrations of >5,000 cells liter−1 shellfish harvesting is closed. The highest level of a bloom has been reached when there are >106 cells liter−1. A bloom of this magnitude can result in massive fish kills, respiratory distress in humans, and discoloration of the water and can affect the health of marine mammals such as dolphins and manatees.
This method represents the first molecular detection strategy for K. brevis, and it is well suited for the detection and monitoring of red tide blooms caused by K. brevis in the Gulf of Mexico and the southern Atlantic coast of the United States. Although diel regulation of rbcL in K. brevis has not been characterized, this assay may provide an easy and relatively rapid procedure that might be employed as an alternative to the more difficult and time-consuming methods currently used by red tide monitoring and management programs in Florida and other states affected by K. brevis.
Acknowledgments
This work was supported by a grant from the NOAA Florida ECOHAB program.
We thank Karen Steidinger and Bill Richardson for supplying many of the cultures used in this study and Earnest Truby for supplying counted bloom samples.
REFERENCES
- 1.Aldrich, D. V. 1962. Photoautotrophy in Gymnodinium breve. Science 137:988-990. [DOI] [PubMed] [Google Scholar]
- 2.Fuhrman, J. A., D. E. Comeau, Å. Hagstrom, and A. M. Chan. 1988. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl. Environ. Microbiol. 54:1426-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geesey, M. E., and P. A. Tester. 1993. Gymnodinium breve: ubiquitous in Gulf of Mexico waters, p. 251-256. In T. J. S. Smayda and Shimizu (ed.), Toxic phytoplankton blooms in the sea: Proceedings of the Fifth International Conference on Toxic Marine Phytoplankton. Elsevier Science Publishing, Inc., New York, N.Y.
- 4.Guillard, R. R., and J. H. Ryther. 1962. Studies of marine planktonic diatoms. 1. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can. J. Microbiol. 8:229-239. [DOI] [PubMed] [Google Scholar]
- 5.Gunther, G., R. H. Williams, C. C. Davis, and F. G. W. Smith. 1947. Catastrophic mass mortality of marine animals and coincident phytoplankton bloom on the west coast of Florida. Ecol. Monogr. 18:311-324. [Google Scholar]
- 6.Hersh, S. H. 1989. Why the dolphins die. Sea Front. July-August:246-248. [Google Scholar]
- 7.Ishida, K., and B. R. Green. 2002. Second- and third-hand chloroplasts in dinoflagellates: phylogeny of oxygen-evolving enhancer 1 (PsbO) protein reveals replacement of a nuclear-encoded plastid gene by that of a haptophyte tertiary endosymbiont. Proc. Natl. Acad. Sci. USA. 99:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamykowski, D., E. J. Milligan, and R. E. Reed. 1998. Relationships between geotaxis/phototaxis and diel vertical migration in autotrophic dinoflagellates. J. Plankton Res. 20:1781-1796. [Google Scholar]
- 9.Lee, M. S., G. Qin, K. Nakanishi, and M. G. Zagorski. 1989. Biosynthesis studies of brevetoxins, potent neurotoxins produced by the dinoflagellate Gymnodinium breve. J. Am. Chem. Soc. 111:6234-6241. [Google Scholar]
- 10.Millie, D. F., O. M. Schofield, G. J. Kirkpatrick, G. Hohnsen, P. A. Tester, and B. T. Vinyard. 1997. Detection of harmful algal blooms using photopigments and absorption signatures: a case study of the Florida red tide dinoflagellate, Gymnodinium breve. Limnol. Oceanogr. 42:1240-1251. [Google Scholar]
- 11.Paul, J. H., A. Alfreider, and B. Wawrik. 2000. Micro- and macrodiversity in rbcL sequences in ambient phytoplankton populations from the southeastern Gulf of Mexico. Mar. Ecol. Prog. Ser. 198:9-18. [Google Scholar]
- 12.Steidinger, K. A., and E. A. Joyce, Jr. 1973. Florida red tides. State Fla. Dep. Nat. Resour. Educat. Ser. 17:1-26. [Google Scholar]
- 13.Takishita, K., K. Nakano, and A. Uchida. 2000. Origin of the plastid in the anomalously pigmented dinoflagellate Gymnodinium mikimotoi (Gymnodiniales, Dinophyta) as inferred from phylogenetic analysis based on the gene encoding the large subunit of form I-type RuBisCo. Phycol. Res. 48:85-89. [Google Scholar]
- 14.Tester, P. A., and K. A. Steidinger. 1997. Gymnodinium breve red tide blooms: initiation, transport, and consequences of surface circulation. Limnol. Oceanogr. 42:1039-1051. [Google Scholar]
- 15.Trainer, V. L., and D. G. Baden. 1999. High affinity binding of red tide neurotoxins to marine mammal brain. Aquat. Toxicol. 46:139-148. [Google Scholar]
- 16.Yoon, H. S., J. D. Hackett, and D. Battacharya. 2002. A single origin of the peridinin- and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proc. Natl. Acad. Sci. USA. 99:11724-11729. [DOI] [PMC free article] [PubMed] [Google Scholar]