Abstract
Experiments were conducted to determine if a novel pmoA-like gene (pmoA2) recently discovered in the methane-oxidizing bacterium Methylocystis strain SC2 (P. F. Dunfield, M. Tchawa Yimga, S. D. Dedysh, U. Berger, W. Liesack, and J. Heyer, FEMS Microbiol. Ecol. 41:17-26, 2002) is present in other methane-oxidizing bacteria (MOB), and if it is expressed. A newly developed primer combination (pmoA206f-pmoA703b) allowed a differential detection of pmoA1 and pmoA2. By using this primer combination, we identified pmoA2 in a wide range of type II MOB of the Methylosinus-Methylocystis group. However, screening by PCR and by Southern hybridization using a newly developed pmoA2-specific oligonucleotide probe also showed that closely related type II MOB, exhibiting 16S rRNA gene sequence identities of higher than 97%, may or may not harbor pmoA2. No pmoA2 was detected in five type I MOB tested: Methylococcus capsulatus strain Bath, Methylocaldum strain E10A, Methylobacter luteus, Methylomicrobium album, and Methylomonas strain D1a. In comparative sequence analyses, all pmoA2-like sequences formed a coherent cluster clearly distinct from pmoA1 sequences of type I and type II MOB, and from amoA sequences of the Nitrosomonas-Nitrosospira group. Phylogenetic analysis using the paml model suggested that pmoA2 is subject to strong purifying selection and therefore has an important cellular function. We probed total RNA extracts of Methylocystis strain SC2 for gene expression of pmoA. A strong signal was observed for pmoA1 in Northern hybridization, while the results obtained for pmoA2 were ambiguous. However, reverse transcription-PCR confirmed that pmoA2 was expressed, albeit at lower level than pmoA1. This provided experimental evidence that the gene product of pmoA2 may be a functionally active enzyme.
Methane-oxidizing bacteria (MOB) are able to utilize methane (CH4) as their sole source of carbon and energy for growth (13). Their ability to oxidize CH4 released at the interface of methanogenic environments and to act as sink for atmospheric CH4 makes these organisms key players in balancing the global CH4 budget and mitigating global warming due to CH4 (4, 28). Phylogenies based on 16S rRNA genes show that MOB form distinct lineages in the Gammaproteobacteria (type I MOB) and Alphaproteobacteria (type II MOB) (2, 5, 6, 13, 16).
The first step in CH4 oxidation, the conversion of methane to methanol, is carried out by a methane monooxygenase (MMO). This enzyme exists in two forms, a particulate, membrane-associated form (pMMO) and a soluble form (sMMO). The two forms of enzyme differ in their structures, kinetic properties, and ranges of substrates they utilize (26). Only a restricted number of MOB species harbor sMMO, while almost all MOB possess pMMO. The only MOB lacking pMMO is Methylocella palustris (5).
Cloning and sequence analysis of genes encoding pMMO revealed three consecutive open reading frames (pmoC, pmoA, and pmoB) in both type I (30, 31) and type II MOB (11). The pmoA gene, which encodes the 27-kDa subunit (PmoA) of pMMO, has been shown to be evolutionarily highly conserved among methanotrophs (17). The type I MOB Methylococcus capsulatus strain Bath and Methylomicrobium album strain BG8 (30, 31), as well as the type II species Methylosinus trichosporium strain OB3b and Methylocystis sp. strain M (11) contain duplicate pmoA gene copies. The sequences of these duplicate gene copies have been shown to be nearly identical.
However, we recently demonstrated that the type II MOB Methylocystis strain SC2 contains two very different pmoA-like genes (9). The first gene (pmoA1 or conventional pmoA) exhibited very high sequence homology to pmoA genes of other type II MOB (even identical amino acid sequence to PmoA of some other Methylocystis strains). The second gene (pmoA2 or novel pmoA) possessed only 73% identity with pmoA1 at the nucleotide level and 68.5% identity at the amino acid level. PmoA2 of Methylocystis strain SC2 was closely related to the deduced amino acid sequence of a pmoA-like gene retrieved in a previous study by cultivation-independent methods from rice field soil (86.3% identity) (19), indicating that the presence of multiple, diverse pmoA gene copies might not be unique to strain SC2. This was also demonstrated by the detection of a novel pmoA2-like copy in M. trichosporium strain KS21 (9).
Our study aimed to assess whether the presence of multiple, diverse pmoA gene copies is a common genotypic trait among methanotrophs or a phenomenon restricted to a few methanotroph strains. We developed PCR primers and hybridization probes for differential detection of pmoA1 and pmoA2 in single MOB genospecies. PCR-based screening by terminal restriction fragment length polymorphism (T-RFLP) analysis, Southern hybridization and comparative sequence analysis revealed that the presence of multiple, diverse pmoA genes in single genospecies is common among type II MOB, but not type I MOB. We also show that, contrary to previous results (9), pmoA2 of Methylocystis strain SC2 is expressed under standard growth conditions, although the level of expression is clearly lower than that of pmoA1.
MATERIALS AND METHODS
MOB strains.
The 32 strains used in this study are listed in Table 1. The conditions used for growth of the MOB were adapted from Heyer et al. (15, 16) or Leadbetter and Forster (22) and depended upon whether the cultures were intended for screening of pmoA2 by PCR amplification or Southern blot analysis. Cultures to be used for PCR-based screening were grown, as described by Heyer et al. (16), on agar plates of mineral salts medium incubated at 30°C in closed glass chambers containing a gas mixture of 20% (vol/vol) CH4, 5% CO2, and 75% air. For Southern or Northern hybridization experiments, MOB were grown in liquid culture of medium 10 for M. trichosporium strain OB3b and Methylocystis strain SC2 or in medium 10 containing NaNO3 instead of NH4Cl as the nitrogen source for M. capsulatus strain Bath and Methylomonas strain D1a. For growth of Methylomonas strain D1a, a supplement of vitamin solution was added to final concentrations of 1 μg/liter for vitamin B6; 0.2 μg/liter for biotin and folic acid; 0.5 μg/liter for vitamin B1, vitamin B2, and dl-Ca-pantothenate; and 0.01 μg/liter for vitamin B12. Liquid cultures were grown for 3 to 5 days at 30°C under a headspace of 20% (vol/vol) CH4, 5% CO2, and 75% air. Cells were pelleted at 4,000 × g for 20 min at 4°C and washed once with TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]). The cell biomass was immediately used for extraction of DNA.
TABLE 1.
List of methanotrophic strains screened for the presence of pmoA2
| Strain designation | Taxonomic assignment | T-RFs at tempa:
|
Detection of pmoA2 | Source or reference | |
|---|---|---|---|---|---|
| 60°C | 66°C | ||||
| SM16 | Methylocystis sp. | 245, 438 | 438 | + | 16 |
| LR1 | Methylocystis sp. | 245, 438 | 438 | + | 8 |
| B2/7 | Methylocystis sp. | 245, 438 | 438 | + | 16 |
| 62/12 | Methylocystis sp. | 245, 438 | 438 | + | 16 |
| KS9 | Methylocystis sp. | 245, 350 | 350 | + | 16 |
| O14c | Methylocystis sp. | 245 | 245 | − | 16 |
| IMET 10484 | Methylocystis sp. | 245, 350 | 350 | + | 16 |
| 21/1 | Methylocystis sp. | 245 | 245 | − | 16 |
| IMET 10499 | Methylocystis sp. | 245, 350 | 350 | + | 16 |
| F10V2a | Methylocystis sp. | 115, 245 | 115 | + | 16 |
| KS12 | Methylocystis sp. | 209 | 209 | − | 16 |
| IMET 10486 | Methylocystis sp. | 245, 438 | 438 | + | 16 |
| SC2 | Methylocystis sp. | 245, 438 | 438 | + | 16 |
| IMET 10491T | Methylocystis echinoides | 245, 438 | 438 | + | 16 |
| Pi5/4 | Methylocystis sp. | 245 | 245 | − | 16 |
| 81 | Methylocystis parvus | 245, 350 | 350 | + | 16 |
| SC8 | Methylosinus sporium | 245, 350 | 350 | + | 16 |
| SK13 | Methylosinus sporium | 245, 350 | 350 | + | 16 |
| 20/3 | Methylosinus sporium | 245, 159 | 159 | + | 16 |
| H1b | Methylosinus sporium | 245, 350 | 350 | + | 16 |
| SM6 | Methylosinus trichosporium | 245, 350 | 350 | + | 16 |
| H5 | Methylosinus trichosporium | 245 | NDb | − | 16 |
| M23 | Methylosinus trichosporium | 245 | 245 | − | 16 |
| 39/3 | Methylosinus trichosporium | 245 | 245 | − | 16 |
| NCIMB 11131T, OB3b | Methylosinus trichosporium | 245 | ND | − | NCIMBc |
| SC10 | Methylosinus trichosporium | 130, 350 | 350 | + | 16 |
| KS21 | Methylosinus trichosporium | 245, 279 | 279 | + | 16 |
| NCIMB 11914T | Methylobacter luteus | ND | ND | − | NCIMB |
| NCIMB 11123T, BG8 | Methylomicrobium album | ND | ND | − | NCIMB |
| NCIMB 11853, Bath | Methylococcus capsulatus | ND | ND | − | NCIMB |
| D1ad | Methylomonas sp. | ND | ND | − | This study |
| E10ad | Methylocaldum sp. | ND | ND | − | This study |
T-RFs observed in differential T-RFLP analysis. PCR at an annealing temperature of 60°C enabled simultaneous detection of both pmoA1 and pmoA2, while PCR at 66°C led to the specific PCR amplification of pmoA2 (if present).
ND, not detected.
NCIMB, National Collections of Industrial and Marine Bacteria Ltd.
Methylomonas strain D1a and Methylocaldum strain E10a are novel isolates of type I MOB. Their taxonomic assignment is based on comparative 16S rRNA gene sequence analysis. Further details of the two strains will be published elsewhere.
DNA extraction.
DNA extraction for PCR-based studies was performed by a procedure based on mechanical agitation in a FastPrep FP120 cell disrupter (Savant, Holbrook, N.Y.) of 2-ml screw-cap reaction vessels filled with a mixture of culture, 0.1-mm-diameter silica-zirconium beads and phosphate buffer (pH 8.0) containing 2% sodium dodecyl sulfate (SDS) (14).
Cells grown in liquid culture (∼1 g of fresh biomass) were suspended in 10 ml of TE buffer to which 25 mg of lysozyme and 20 μl of proteinase K (25 mg/ml) were added. The suspension was placed in a 37°C water bath for 2 h, after which a 10% (wt/vol) solution of SDS was added to a final concentration of 1%, followed by a 1.5-h incubation at 37°C. After centrifugation for 15 min at 4°C, the supernatant was collected, and 1 ml of 5 M potassium acetate (pH 7.5) was added. The suspension was centrifuged for 15 min at 4°C, and the resulting supernatant was transferred to a new vessel. Total nucleic acids were extracted from the supernatant twice with 1 volume of chloroform-isoamyl alcohol (24:1 [vol/vol]) and then precipitated from the aqueous phase with 1 volume of isopropanol. The nucleic acids were resuspended in 5 ml of TE buffer.
For removal of coextracted RNA, 30 μl of 100-mg/ml RNase A and 20 μl of 100,000-U/ml RNase T1 were added, followed by incubation for 2 h at 37°C. The RNase treatment was stopped by extraction three times with 1 volume of chloroform-isoamyl alcohol (24:1 [vol/vol]). The DNA was precipitated from the aqueous phase with 1 volume of isopropanol and then resuspended in 100 μl of TE buffer. The amount of DNA extracted was estimated by electrophoresis of aliquots on a 1% agarose gel and comparison to a PstI digest of phage λ DNA.
Extraction of total RNA from Methylocystis strain SC2.
Total nucleic acids were extracted as described above for strains grown in liquid culture, except that solutions for extracting RNA were prepared with diethyl pyrocarbonate (DEPC)-treated deionized water. For the removal of coextracted DNA, 1 volume of TMC buffer (10 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 0.1 mM CsCl2) (7) and 20 μl (5 U) of RNase-free DNase (Promega, Madison, Wis.) were added, followed by incubation for 2 h at 37°C. The reaction was stopped by extraction three times with 1 volume of chloroform-isoamyl alcohol (24:1 [vol/vol]). The total RNA was precipitated from the aqueous phase with 1 volume of isopropanol and resuspended in 100 ml of DEPC-pretreated TE buffer. The integrity of the 16S and 23S rRNA fragments was checked by electrophoresis on a 1.2% agarose gel and comparison to an rRNA standard from Escherichia coli (Roche Diagnostics).
Differential detection of pmoA1 and pmoA2.
The pmoA-based diversity present in a single strain was assessed by determining the number and the size of terminal restriction fragments (T-RFs) observed in restriction digests of pmoA genes PCR-amplified with the primer combination pmoA206f-pmoA703b at two different annealing temperatures (Table 2). For T-RFLP analysis, the 5′ primer (pmoA206f) was labeled with the dye 5-carboxyfluorescein. The reaction mixture contained 1 μl of template DNA, 7.5 μl of 10× reaction buffer, 1.5 mM Mg2+, 150 μM (each) deoxynucleoside triphosphate (dNTP), 0.125 μM (each) primer, and 2.5 U of Taq DNA polymerase (Promega, Mannheim, Germany). The thermal PCR profile was as follows: initial denaturation for 3 min at 94°C followed by 32 cycles consisting of denaturation at 94°C for 60s, primer annealing at 60 or 66°C for 60 s, and elongation at 72°C for 60s. The final extension step was extended to 7 min. Aliquots of the amplicons (10 μl) were checked by electrophoresis on a 1% agarose gel.
TABLE 2.
Oligonucleotide primers and probes used in this study
| Namea | Target | Strategy | Gene positionsb | Sequence (5′→3′)c | Reference |
|---|---|---|---|---|---|
| A189f | pmoA | PCR, RT-PCR | 172-189 | GGNGACTGGGACTTCTGG | 17 |
| A682b | pmoA | PCR, RT-PCR | 702-685 | GAASGCNGAGAAGAASGC | 17 |
| pmoA206f | pmoA1 and pmoA2d | PCR, RT-PCR | 172-206 | GGNGACTGGGACTTCTGGATCGACTTCAAGGATCG | This study |
| pmoA703b | pmoA1 and pmoA2d | PCR | 702-668 | GAASGCNGAGAAGAASGCGGCGACCGGAACGACGT | This study |
| pmoA636b | pmoA2 | S/N blot,e RT-PCR | 635-610 | ATCATGCGGATGTATTCMGGSGTGCC | This study |
| A593b | pmoA1 (strain SC2) | S/N blot, RT-PCR | 615-593 | CATCGACGTGCGGACGAAGTGGA | 9 |
| Eub9f | 16S rRNA gene | PCR | 9-27 | GAGTTTGATCMTGGCTCAG | 21 |
| Eub1492b | 16S rRNA gene | PCR | 1512-1492 | ACGGYTACCTTGTTACGACTT | 34 |
All (including labeled) oligonucleotides were purchased from Metabion (Martinsried, Germany).
Numbering for pmoA refers to the pmo gene sequence of Methylococcus capsulatus strain Bath (30). Numbering for 16S rDNA refers to the 16S rRNA gene sequence of E. coli (3).
N = A,T,C, or G; M = C or A; R = A or G; Y = C or T; and S = G or C.
The use of the primer combination pmoA206f and pmoA703b in PCR at an annealing temperature of 60°C enabled simultaneous detection of both pmoA1 and pmoA2, while use at 66°C led to the specific PCR amplification of pmoA2 (if present).
S/N blot, Southern or Northern blot.
The digestion of purified amplicons (100 ng) and separation of the restriction digests on an ABI 373A automated sequencer (Perkin-Elmer Applied Biosystems, Weiterstadt, Germany) were carried out as described previously (24).
For direct sequencing of amplicons obtained at an annealing temperature of 66°C, PCR was carried out with primer pmoA206f without fluorescence label. Amplicons were purified by using the Qiagen PCR purification kit (Qiagen, Hilden, Germany) according to the recommendations of the supplier. Both strands of the PCR products were analyzed by using the dye terminator sequencing chemistry of PE Applied Biosystems and either pmoA206f or pmoA703b as the sequencing primer.
Southern hybridization.
Aliquots of genomic DNA (10 μg) were digested overnight at 37°C with 50 U of restriction enzyme PstI, EcoRI, or XhoI according to the protocols recommended by the supplier (Promega, Madison, Wis.). The restricted DNA was precipitated with ethanol, dried in a vacuum desiccator (Savant), resuspended in 20 μl of TE buffer, and separated on a 0.8% agarose gel at 25 V for 12 h. Southern blotting was carried out according to the procedure described in the DIG Application Manual for Filter Hybridization (Roche Diagnostics GmbH, Mannheim, Germany, 2000).
The digoxigenin (DIG)-labeled oligonucleotide probe pmoA636b (Table 2) was used for specific detection of pmoA2. A pmo gene probe was used for simultaneous detection of both pmoA1 and pmoA2. This probe was based on a mixture of DIG-labeled amplicons of the different strains tested. PCR was carried out with either primer combination A189f-A682b (16) or pmoA206f-pmoA703b and an annealing temperature of 60°C. For direct labeling during PCR, DIG-dUTP was mixed with dTTP in the proportional relationship of 1:3.
Both oligonucleotide probe pmoA636b and the pmo gene probe were applied under the same stringency conditions. Hybridizations were carried out overnight at 42°C with a standard hybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% N-lauroylsarcosine, 0.02% SDS, 1% blocking solution). Membranes were washed in 0.5× SCC at 60°C for 30 min, and hybridization signals were detected on a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) after reaction with the ECF substrate for Western blotting (Amersham, Piscataway, N.J.).
Northern hybridization.
Approximately 60 μg of total RNA from Methylocystis strain SC2 was first separated on a 1% formaldehyde agarose gel at 60 V for 3 h in presence of 1× MOPS (morpholinepropanesulfonic acid) buffer (10× MOPS buffer is 200 mM MOPS, 50 mM sodium acetate, 20 mM EDTA [pH 7.0]). Northern blotting was carried out according to the procedure outlined in the DIG Application Manual for Filter Hybridization (Roche Molecular Diagnostics). Blotting was performed overnight on a Hybond-N-membrane (Amersham). The DNA was UV cross-linked using a UV Stratalinker 2400 (Stratagene, La Jolla, Calif.) at 266 nm for 3 min. The DIG-labeled oligonucleotide probes A593b and pmoA636b were used for specific detection of pmoA1 and pmoA2, respectively (Table 2). Hybridizations were performed overnight at 40°C. Washing and visualization were carried out as described above for Southern hybridization.
RT-PCR.
Copy DNA (cDNA) of mRNA from pmoA genes expressed in Methylocystis strain SC2 was synthesized with a Qiagen Omniscript kit according to the instructions of the manufacturer. The reverse transcription (RT) was carried out in a total volume of 20 μl at 37°C for 30 min. The reaction mixture contained 2 μg of total RNA from strain SC2, 0.5 mM (each) dNTP, reverse transcriptase buffer, 10 U of RNase inhibitor, 1.0 μM primer A682b (Table 2), and 4 U of Omniscript reverse transcriptase. Aliquots (1 μl) of the cDNA solution were used for subsequent PCR amplification. The primer combinations A189f-A593b and pmoA206f-pmoA636b were used to specifically amplify cDNA from either pmoA1 or pmoA2. The reaction cocktail contained 1 μl of RT product, 10 μl of 10× reaction buffer, 1.5 mM Mg2+, 200 μM each deoxynucleoside triphosphate (dNTP), 0.25 μM each primer, and 2.5 U of Taq DNA polymerase (Promega). The thermal profile was as follows: initial denaturation for 3 min at 94°C followed by 32 cycles of denaturation at 94°C for 40 s, primer annealing at 62°C for 40 s, and elongation at 72°C for 45s. The final extension step was extended to 7 min. Amplification was performed in a total volume of 100 μl in 0.2-ml reaction vessels and a DNA thermal cycler (model 2400; PE Applied Biosystems). Aliquots of the amplicons (10 μl) were checked by electrophoresis on a 1% agarose gel. Amplicons of the expected size were purified and sequenced as described above.
Analysis of sequence data.
Trees were constructed for partial PmoA sequences (151 deduced amino acid positions) by using TreePuzzle and the ARB program package (developed by O. Strunk and W. Ludwig of the Technical University of Munich [http://www.arb-home.de]). TreePuzzle is a quartet maximum-likelihood method for reconstructing tree topologies (32, 33). The amino acid substitution model described by Adachi and Hasegawa (1) was used to construct the trees.
A tree was constructed for the same set of data on the nucleic acid level by the maximum-likelihood method FastDNAml (10).
In order to gain insight into the evolution and expression of the pmoA2 gene, a phylogenetic tree of selected sequences was analyzed with the codeml function of the paml program of Ziheng Yang (http://abacus.gene.ucl.ac.uk/software/paml.html#GetPAML). This program calculates the relative rate of nonsynonomous (amino acid changing) to synonomous (non-amino acid changing) nucleotide substitutions (dN/dS, or ω) in a phylogeny (35). An ω value of <1 indicates that purifying natural selection is acting to prevent changes in a protein, while an ω value of >1 indicates diversifying selection. A ω value = 1 indicates neutral selection and would be expected for a pseudogene.
Nucleotide accession numbers.
The pmoA2 gene sequences obtained in the course of this study from environmental samples and cultured MOB have been deposited in the EMBL, DDBJ, and GenBank databases under the accession no. AJ543418 to AJ543423 and AJ544093 to AJ544102, respectively. The pmoA1 sequences obtained for reference from Methylocaldum strain E10a and Methylomonas strain D1a have been deposited under accession no. AJ5441091 and AJ544092, respectively.
RESULTS
Differential detection of pmoA1 and pmoA2 by T-RFLP analysis.
The first aim of the present study was the development of an appropriate method for detection of multiple, divergent pmoA genes present in a single organism. Using the pmoA2-like environmental clone sequence M84-P3 (19) and the pmoA2 sequence of Methylocystis strain SC2 as the starting point, we designed several forward and reverse primers for specific retrieval of pmoA2 from both environmental samples and cultured MOB. According to the pmoA database available, none of these primers perfectly matched pmoA1 or amoA, suggesting that these primers should be specific for pmoA2. Environmental DNA from rice paddy soil is composed of genomic DNA from a complex community of phylogenetically diverse microorganisms, including various type I and II methanotrophs (19). Consequently, to assess the target specificity and the range of pmoA2-like sequence diversity detectable, we tested various primer combinations in first- and second-round PCRs using environmental DNA extracted from flooded rice microcosms in a previous study (19). This led to the retrieval of various pmoA2-like sequences from bulk soil and rice roots of flooded rice microcosms. Six representatives of these environmental clone sequences have been deposited in the EMBL, DDBJ and GenBank databases (see Materials and Methods) and are shown in Fig. 1. The best performance with regard to both target specificity and range of pmoA2-like sequence diversity detectable was exhibited by the primer combination pmoA206f-pmoA703b (Table 2).
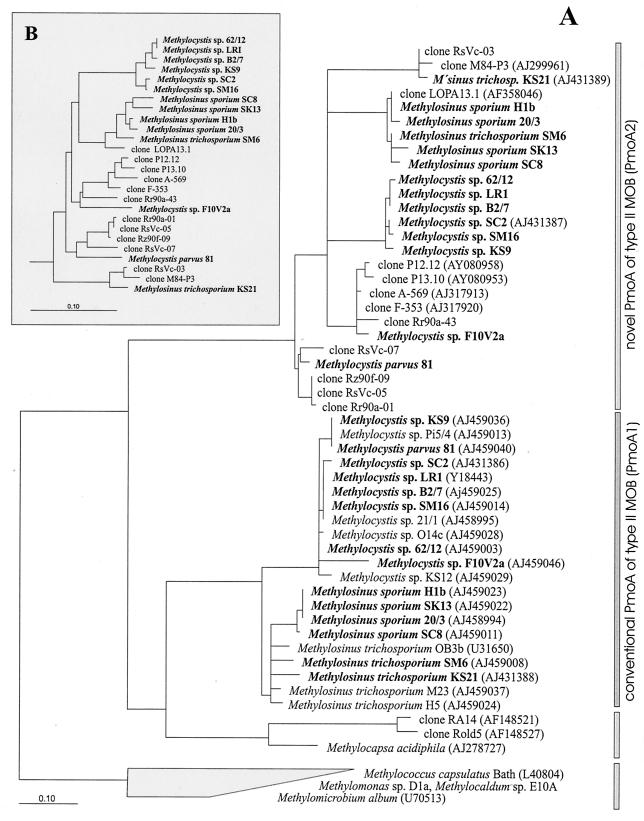
FIG. 1.
Phylogenetic trees constructed for partial pmoA sequences and deduced amino acid sequences (PmoA1 and PmoA2). (A) PmoA tree showing the relationship of PmoA1 and PmoA2 of type II MOB of the Methylosinus-Methylocystis group to environmental clones as well as to Methylocapsa acidiphila and selected type I MOB. The environmental sequences were retrieved by cultivation-independent methods from the following habitats: forest soil (clones RA14, Rold5 [18], and P12.12 and P13.10 [27]), peat soil (clone LOPA13.1 [25]), natural and manipulated organic soils (clones A-569 and F-353), as well as bulk soil (clones RsVc-03, RsVc-05, and RsVc-07) and rice roots (clones M84-P3 [19], Rr90a-01,Rr90a-43, and Rz90f-09) of flooded rice microcosms. In the present study, the retrieval of environmental pmoA2-like sequences from the bulk soil and rice roots of flooded rice microcosms was based on total DNA extracted in a previous study (19). Type II MOB for which both PmoA1 and PmoA2 sequences could be detected are shown in boldface. GenBank accession numbers are given in parentheses after the species or clone names for sequences that were not obtained in this study. The tree was constructed by using TreePuzzle, a quartet maximum-likelihood method (32, 33). Lineages for which, based on 25,000 puzzling steps, the exact branching order could not be unambiguously determined are shown by multifurcation. The scale bar represents 0.1 change per amino acid position. (B) pmoA tree showing the pmoA2 intracluster relationship at the nucleic acid level. The tree was constructed with the same data set as that on the PmoA level and by the maximum-likelihood method (FastDNAml [10]); however, the presentation of the tree is restricted to the pmoA2 cluster. The distance bar represents 0.1 substitution per nucleotide position.
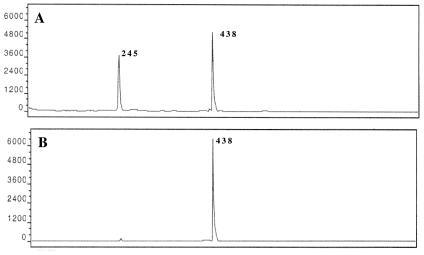
Using Methylocystis strain SC2 as the model organism, this primer combination was therefore chosen to establish for cultured type II MOB a tool for differential detection of pmoA1 and pmoA2 by T-RFLP analysis. The T-RF size distribution of pmoA1 and pmoA2 was assessed by computer simulation. The T-RF sizes (5′ or 3′ termini) were predicted for as many as 10 different restriction endonucleases. For MspI, we identified two distinct 5′-T-RFs, which corresponded to pmoA1 (245 bp) and pmoA2 (438 bp), respectively. Based on the in silico analysis, we concluded that PCR with primer pair pmoA206f-pmoA703b followed by digestion with MspI was the simplest way to classify the two different pmoA sequence types present in Methylocystis strain SC2. To test our predictions experimentally, PCR amplification of pmoA was conducted for subsequent T-RFLP analysis. The effect of annealing temperature used in PCR on the pmoA sequence type detectable via T-RFLP analysis was investigated by performing the amplifications at various annealing temperatures. Two different T-RFs with sizes of 245 and 438 bp were observed at an annealing temperature of 60°C, whereas only one distinct T-RF was observed at an annealing temperature of 66°C (Fig. 2). Thus, no discrepancy was observed between the results expected by in silico analyses and the empirical data obtained. However, to verify that only pmoA2 was amplified at 66°C, another PCR was carried out with the annealing temperature of 66°C, and the resulting PCR product was sequenced. Comparative sequence analysis confirmed that the product was derived from pmoA2. This finding indicated that T-RFLP analysis in combination with comparative sequence analysis could be a rapid and reliable method for screening methanotrophic pure cultures for the presence of multiple, divergent pmoA gene copies.
FIG. 2.
pmoA-based T-RFLP profiles of Methylocystis strain SC2. PCRs were carried out with primers pmoA206f and pmoA703b at two different annealing temperatures: 60°C (A) and 66°C (B). On the y axis, the intensities of fragments are given in arbitrary units. Numbers indicate the lengths of T-RFs corresponding to pmoA1 (245 bp) and pmoA2 (438 bp).
Screening for pmoA2 in single MOB strains by T-RFLP analysis.
In addition to Methylocystis strain SC2 and M. trichosporium strain KS21, 30 pure cultures, including 25 type II MOB and 5 type I MOB, were screened for the presence of pmoA2 by differential T-RFLP analysis. Prior to pmoA-based analysis, the identity of each strain was confirmed by comparative sequence analysis of its 16S rRNA gene (16).
Except for M. trichosporium strains OB3b and H5, PCR products were obtained for all type II MOB at both annealing temperatures (60 and 66°C). The five type I MOB tested failed to give a positive PCR with the primer combination pmoA206f-pmoA703b (Table 1). The T-RFLP patterns obtained for the type II MOB species revealed multiple, distinct T-RFs within single strains, suggesting that some degree of pmoA sequence diversity was present. For all of the strains that tested positive at an annealing temperature of 66°C, a single T-RF was observed. Depending upon the strain, these T-RFs were 115, 159, 209, 245, 279, 350, or 438 bp in size (Table 1). Representatives of each of these different T-RFs were further investigated by direct sequencing of the pmoA amplicon generated at an annealing temperature of 66°C. Translated sequences of PCR products were aligned and compared with a set of representative PmoA sequences. It was confirmed that the 209- and 245-bp T-RFs were derived from typical pmoA1 sequences of type II MOB (Methylosinus-Methylocystis group) that could still be amplified with the presumed pmoA2-specific primer set at 66°C. However, T-RFs with sizes of 115, 159, 279, 350, and 438 bp were indicative of pmoA2 (Table 1). The treeing analysis showed that all PmoA2 sequences obtained from type II MOB of the Methylosinus-Methylocystis group (Table 1) formed a coherent cluster clearly distinct from that of PmoA1 (Fig. 1). In addition, cloned sequences retrieved by cultivation-independent approaches from various environments, including forest soil, peat soil, natural and manipulated organic soils, and flooded rice microcosms grouped within the PmoA2 cluster.
The intercluster PmoA identity values between sequence types of PmoA1 and PmoA2 ranged from 64 to 74.5%. The lowest pmoA2 and PmoA2 intracluster identity values were 18.2 and 21.5%, respectively. For comparison, the corresponding values for pmoA1 and PmoA1 were 14.7 and 12.4%, respectively. The PmoA2 cluster was divided into five sublineages. These sublineages were characterized by Methylocystis parvus strain 81, Methylocystis strain F10V2a, M. trichosporium strain KS21, or by a set of either Methylocystis or Methylosinus spp.
Screening for pmoA2 by Southern hybridization.
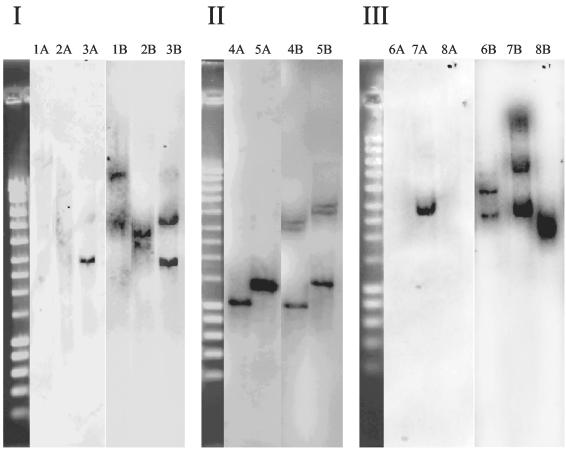
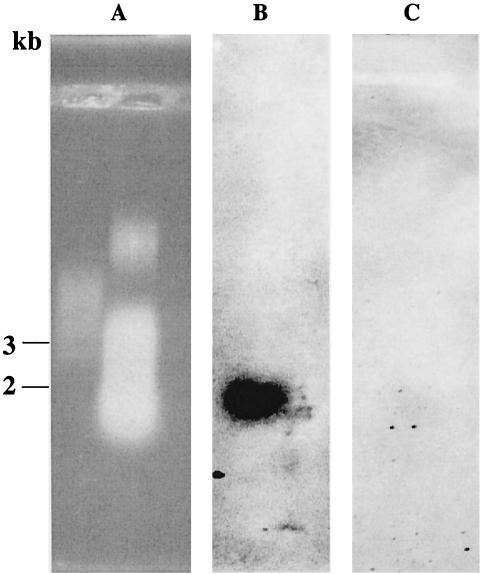
In order to provide further evidence for the absence of pmoA2 in M. trichosporium strain OB3b and three type I MOB by a second, PCR-independent approach, a 26-bp oligonucleotide probe was formulated for the specific detection of pmoA2 (termed pmoA636b) (Table 2) by Southern hybridization. Based on the matching target region of pmoA2-like sequences initially available for the design of this probe, including environmental clone sequences directly retrieved from flooded rice microcosms, Methylocystis strain SC2 (one weak G:T mismatch) and M. trichosporium KS21 (Fig. 3), we concluded that pmoA636b should target a wide range of diverse pmoA2 sequence types (compare Fig. 1 and 3). The probe exhibited various mismatches to the corresponding target sites of pmoA1 of the MOB investigated by Southern hybridization. Southern blots of digested chromosomal DNA were hybridized, respectively, with the oligonucleotide probe pmoA636b and a pmoA gene probe (generated as a mixture of DIG-labeled pmoA amplicons of the different strains tested). All hybridization experiments were carried out with Methylocystis strain SC2 as a positive control. Under the hybridization and washing conditions used, probe pmoA636b did not detect pmoA1 for any of the MOB tested. As expected, no pmoA2 signal was observed for the type I MOB M. capsulatus strain Bath, Methylocaldum strain E10a, or Methylomonas strain D1a (Fig. 4). A pmoA2 signal was also not obtained for M. trichosporium strain OB3b, but the presence of pmoA2 was confirmed in another M. trichosporium strain (SM6) and in Methylocystis strain SC2. These results agreed well with those obtained by PCR. Full-length gene probes of pmoA detected multiple pmoA copies in several of the strains. Three gene copies, including pmoA2, were detected in the genomic DNA of M. trichosporium strain SM6 and Methylocystis strain SC2, while two pmoA copies were observed for M. trichosporium strain OB3b and the three type I MOB (Fig. 4).
FIG. 3.
Alignment showing the targeting region of probe pmoA636b among pmoA2 and corresponding pmoA1 sequences of those MOB investigated by Southern hybridization. The clone M84-P3 is shown as a representative of a set of diverse environmental pmoA2 sequences that perfectly match in the target region probe pmoA636b, including clones RsVc-03, Rr90a-43, RsVc-07, Rz90f-09, RsVc-05, and Rr90a-01 (Fig. 1). Letters shown in boldface indicate weak G:T mismatching nucleotide positions, while those shown in boldface and underlined indicate stronger mismatches (e.g., C:T, C:C, or G:G). Strains shown in the alignment are Methylocystis strain SC2, M. trichosporium strains OB3b and SM6, Methylomonas strain D1a, Methylocaldum strain E10a, and M. capsulatus strain Bath.
FIG. 4.
Southern hybridization of genomic DNA to oligonucleotide probe pmoA636b (A lanes) or PCR-generated universal pmoA gene probe (B lanes). Under the hybridization conditions used in this study, probe pmoA636b is specific for pmoA2 (Fig. 3), while the universal pmoA gene probe detects both pmoA1 and pmoA2. (I) EcoRI digests. Lanes: 1, M. trichosporium strain OB3b; 2, Methylocaldum strain E10A; 3, Methylocystis strain SC2. (II) XhoI digests. Lanes: 4, Methylosinus trichosporium strain SM6; 5, Methylocystis strain SC2. (III) PstI digests. Lanes: 6, Methylococcus capsulatus strain Bath; 7, Methylocystis strain SC2; 8, Methylomonas strain D1a. Methylocystis strain SC2 was used as positive control (9). The PstI digest of DNA of phage λ was used as a size marker.
Expression of pmoA2 in Methylocystis strain SC2 (theoretical considerations).
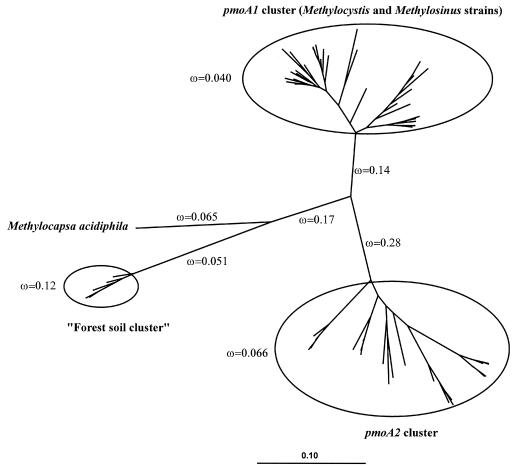
The paml model (35) was applied to a phylogenetic tree incorporating selected pmoA1 and pmoA2 gene sequences as shown in Fig. 5. The phylogeny was fit to a model incorporating eight separate ω values. All estimated ω values were <1. The ω values within the pmoA1 and pmoA2 clusters were <0.1, indicating that in recent evolutionary history, strong purifying selection has been acting on both genes. This is strong evidence that both genes are expressed and have important cellular functions. Interestingly, the branch connecting the pmoA2 cluster to the other branches of the phylogeny has an elevated ω value of 0.28. While still <1, this increase may indicate a relaxation of purifying selection or even an increase in diversifying selection forces acting at particular times or on particular codons. Although purely speculative, a functional differentiation of the pmoA1 and pmoA2 genes may have occurred. Two different experimental strategies (i.e., Northern hybridization and RT-PCR) were applied to assess whether pmoA2 was expressed when strain SC2 was grown under standard laboratory growth conditions (see Materials and Methods section “MOB strains”).
FIG. 5.
Analysis of ω values within an unrooted phylogenetic tree incorporating 61 selected pmoA nucleotide sequences. The branching topology was calculated by using a neighbor-joining algorithm (29) with a Jukes-Cantor correction (20) (2,000 data resamplings), and branch lengths were adjusted by the paml program. A maximum-likelihood algorithm produced nearly the same tree topology. The distance bar indicates 0.1 nucleotide substitution per position. The paml model (model 2 in codeml) was fit with eight separate ω values as indicated: one for each of the five longest branches and one each for the pmoA1 cluster, the pmoA2 cluster, and the “forest soil cluster,” respectively, which are enclosed by circles. The pmoA1 cluster included sequences from various Methylocystis and Methylosinus strains (accession no. U31569, U31651, AJ431386, AJ431388, AJ458994, AJ458998, AJ459000-AJ459003, AJ459008, AJ459011, AJ459013, AJ459014, AJ459019, AJ459021-AJ459025, AJ459027-AJ459029, AJ459034, AJ459036, AJ459038, AJ459046-AJ459048, and AJ459052). The pmoA2 cluster included sequences from pure cultures of Methylocystis and Methylosinus and sequences retrieved from soils by cultivation-independent methods (accession no. AJ431389, AJ299961, AY080950, and AJ459013 and sequences from the present study). The “forest soil cluster” contains sequences retrieved by Holmes et al. (18) using cultivation-independent methods (accession no. AF148525, AF148521, AF148527, AF148528, and AF148523).
Expression of pmoA2 in Methylocystis strain SC2 (experimental assessment).
Total RNA of Methylocystis strain SC2 was extracted and separated on a 1% formaldehyde agarose gel and blotted onto a nylon membrane for Northern hybridization. Probing was carried out with oligonucleotide A593b or pmoA636b specific for either pmoA1 or pmoA2. In comparison to a strong signal observed for pmoA1, no conclusive signal was obtained for pmoA2 (Fig. 6).
FIG. 6.
Northern hybridization of total RNA from Methylocystis strain SC2 to pmoA-targeted oligonucleotide probes. (A) Agarose gel electrophoresis of total RNA in presence of an RNA marker (RNA ladder; New England Biolabs). (B) Hybridization with probe A593b to specifically detect mRNA expressed from pmoA1. (C) Hybridization with probe pmoA636b to specifically detect mRNA expressed from pmoA2. Lanes contain approximately 60 μg of total RNA from strain SC2.
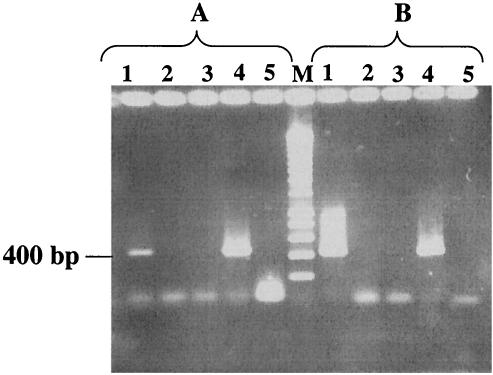
Consequently, we applied RT-PCR as a second, methodologically more sensitive, approach. Initially, we used the primers A593b and pmoA636b (Table 2) for RT. The primer combinations A189f-A593b and pmoA206f-pmoA636b were used to specifically amplify cDNA from either pmoA1 or pmoA2. Sequencing of RT-PCR products confirmed strong expression of pmoA1. PCR of the cDNA generated with probe pmoA636b gave a product of nearly the size we expected. Sequencing of that product determined that it was not pmoA; it corresponded to a portion of 23S rRNA. We therefore assumed that nonspecific cDNA was formed, perhaps because the primer may have bound to a nonspecific target at the lower temperature of RT. A similar problem occurred in our earlier attempts to perform RT-PCR (9) and may have contributed to our initial failure to detect pmoA2 expression in strain SC2. In an attempt to overcome the problem, we used the universal pmoA primer A682b to generate the cDNA. Using the cDNA as template, we carried out specific PCR assays in which we applied either A593b or pmoA636b as the reverse primer. In both cases, products of the expected size were obtained (Fig. 7), and comparative sequence analysis of the RT-PCR product showed that sequences corresponded either to pmoA1 or to pmoA2.
FIG. 7.
Detection of pmoA-like mRNA in Methylocystis strain SC2 by RT-PCR. RT was carried out with primer A682b. (A) Use of the primer combination pmoA206f-pmoA636b for specific detection of functional activity of pmoA2. (B) Use of the primer combination A189f-A593b for specific detection of functional activity of pmoA1. Lanes: 1, total RNA plus RT-PCR (positive detection); 2, total RNA plus PCR without the RT step (negative control); 3, RNase-treated RNA sample plus PCR (negative control); 4, genomic DNA plus PCR (positive control); 5, no DNA plus PCR (negative control).
DISCUSSION
The newly developed PCR assay enabled the differential detection of pmoA1 and pmoA2 in single MOB strains by T-RFLP analysis. The pmoA-based T-RFLP fingerprinting and comparative sequence analysis of 27 different type II MOB strains revealed that pmoA2 is widely distributed among type II MOB, including strains of Methylosinus sporium, M. trichosporium, Methylocystis echinoides, and M. parvus, as well as a set of other Methylocystis strains (16). The MOB strains that tested positive for pmoA2 can be considered to reflect the full phylogenetic diversity known within the Methylosinus-Methylocystis group. The treeing analysis clearly showed that pmoA2 and deduced PmoA2 sequences form a coherent cluster clearly distinct from that of pmoA1 and deduced PmoA1, respectively. It is noteworthy that PmoA1 sequences of the Methylosinus-Methylocystis group clustered in the maximum-likelihood tree (Fig. 1) more closely to the PmoA sequence from Methylocapsa acidiphila than to their own corresponding PmoA2 sequences (Fig. 1).
The data obtained also suggested that pmoA2 is not present in all type II MOB investigated. M. trichosporium strains OB3b and SM6 exhibit an overall 16S rRNA gene sequence identity of 98.2%. PCR-based screening led to the identification of pmoA2 in strain SM6, but pmoA2 could not be detected in strain OB3b. Further evidence that, in contrast to strain SM6, M. trichosporium strain OB3b does not harbor a pmoA2 gene copy was provided by Southern hybridization using probe pmoA636b (Fig. 3 and 4). It should also be noted that the gene probe of PCR-amplified pmoA (containing both the conventional pmoA1 and the novel pmoA2) led to the detection of two pmoA gene copies in M. trichosporium strain OB3b (Fig. 4). This is consistent with previous reports that strain OB3b harbors two nearly identical pmoCAB operons that correspond to pmoA1 (11). Thus, the two different experimental approaches (PCR-based screening and Southern blot analysis) taken together provide strong evidence that closely related type II MOB genospecies may or may not harbor pmoA2. Interestingly, a similar observation has been made for the distribution of mmoX genes among type II MOB. Nearly identical species appear to differ in whether they possess or do not possess the mmoX gene (13, 16).
However, we cannot completely rule out the possibility that variations in the target site of the primer combination pmoA206f-pmoA703b may be the reason why pmoA2 was not detected in all type II MOB tested. Thus, the distribution of pmoA2 among type II MOB might still have to be addressed in further studies. This may also be true for type I MOB, although in all five strains tested, the detection of a pmoA2-like gene copy by PCR (Table 1)—and in some cases by Southern hybridization (Fig. 4)—failed.
The second major aim of the present study was to determine whether pmoA2 is expressed and encodes a functionally active protein or is instead an unexpressed pseudogene. Proteins with important basic cellular functions are often subject to strong purifying selection. Under such selection, a synonomous (non-amino acid changing) nucleotide mutation in the encoding gene has a much greater chance of becoming fixed than does a nonsynonomous (amino acid changing) mutation. Nonsynonomous/synonomous substitution rate ratios (ω) can typically be 0.1 or less for genes under strong purifying selection (36). Using the paml model (35), our phylogenetic analysis indicated that the pmoA1 and pmoA2 genes have both displayed similarly low ω values in recent evolutionary history. This is strong evidence that both genes are expressed and have important cellular functions. In contrast to our previous study (12), experimental evidence that pmoA2 of Methylocystis strain SC2 is expressed could be obtained by RT-PCR (Fig. 7). However, as concluded from Northern hybridization experiments (Fig. 6), the level of pmoA2 expression under the laboratory standard growth conditions applied was clearly lower than that of pmoA1.
Further studies will need to focus on the functional role of the enzyme expressed by the pmo operon that corresponds to pmoA2. Multiple pMMOs with different substrate specificities would provide MOB with a means of adapting to changing concentrations of methane and competitive substrates such as ammonia (13). There is some evidence for such a mechanism. Increasing Cu concentration has been shown to allow M. capsulatus to form a pMMO with a higher affinity for CH4 (23). Methylocystis strain LR1, which possesses a pmoA2, displays a variable affinity for methane depending on the growth conditions (7), although this may be due to physiological conditions rather than to genetic changes (8). Ammonia does not appear to affect all soil MOB communities in the same way, suggesting that different MOB have different sensitivities to this cosubstrate (12). Thus, further investigation will require various cultivation conditions and the use of knockout mutations.
Acknowledgments
This work was supported partly by grants from the Deutsche Forschungsgemeinschaft (SFB 395 to W. Liesack and grant DU377/1-1 to P. Dunfield) and Bundesministerium für Bildung und Forschung (bmb+f, GenoMik Network, Göttingen, Germany). M. Tchawa Yimga was supported by the German Academic Exchange Service (DAAD). J. Heyer was supported in part by the bmb+f BIOLOG project.
REFERENCES
- 1.Adachi, J., and M. Hasegawa. 1996. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J. Mol. Evol. 42:459-468. [DOI] [PubMed] [Google Scholar]
- 2.Bowman, J. P., L. I. Sly, P. D. Nichols, and A. C. Hayward. 1993. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int. J. Syst. Bacteriol. 43:735-753. [Google Scholar]
- 3.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. R. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dedysh, S. N., W. Liesack, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, A. M. Bares, N. S. Panikov, and J. M. Tiedje. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50:955-969. [DOI] [PubMed] [Google Scholar]
- 6.Dedysh, S. N., V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, W. Liesack, and J. M. Tiedje. 2002. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 52:251-261. [DOI] [PubMed] [Google Scholar]
- 7.Dunfield, P. F., W. Liesack, T. Henckel, R. Knowles, and R. Conrad. 1999. High-affinity methane oxidation by a soil enrichment culture containing a type II methanotroph. Appl. Environ. Microbiol. 65:1009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunfield, P. F., and R. Conrad. 2000. Starvation alters the apparent half-saturation constant for methane in the type II methanotroph Methylocystis strain LR1. Appl. Environ. Microbiol. 66:4136-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunfield, P. F., M. Tchawa Yimga, S. N. Dedysh, U. Berger, W. Liesack, and J. Heyer. 2002. Isolation of a Methylocystis strain containing a novel pmoA-like gene. FEMS Microbiol. Ecol. 41:17-26. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 11.Gilbert, B., I. R. McDonald, R. Finch, G. P. Stafford, A. K. Nielsen, and J. C. Murrell. 2000. Molecular analysis of the pmo (particulate methane monooxygenase) operons from two type II methanotrophs. Appl. Environ. Microbiol. 66:966-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulledge, J., A. P. Doyle, and J. P. Schimel. 1997. Different NH4+-inhibition patterns of soil CH4 consumption: a result of distinct CH4-oxidizer populations across sites? Soil Biol. Biochem. 29:13-21. [Google Scholar]
- 13.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyer, J., Y. Malashenko, U. Berger, and E. Budkova. 1984. Verbreitung methanotropher Bakterien. Z. Allg. Mikrobiol. 24:725-744. [Google Scholar]
- 16.Heyer, J., V. F. Galchenko, and P. F. Dunfield. 2002. Molecular phylogeny of type II methane-oxidizing bacteria isolated from various environments. Microbiology 148:2831-2846. [DOI] [PubMed] [Google Scholar]
- 17.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, A. J., P. Roslev, I. R. McDonald, N. Iversen, K. Henriksen, and J. C. Murrell. 1999. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl. Environ. Microbiol. 65:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horz, H.-P., M. Tchawa Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, N.Y.
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J. Wiley & Sons, Ltd., Chichester, United Kingdom.
- 22.Leadbetter, E. R., and J. W. Forster. 1958. Studies on some methane-utilising bacteria. Arch. Microbiol. 30:91-118. [DOI] [PubMed] [Google Scholar]
- 23.Lontoh, S., and J. D. Semrau. 1998. Methane and trichloroethylene degradation by Methylosinus trichosporium OB3b expressing particulate methane monooxygenase. Appl. Environ. Microbiol. 64:1106-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukow, T., P. F. Dunfield, and W. Liesack. 2000. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32:241-247. [DOI] [PubMed] [Google Scholar]
- 25.Morris, S. A., S. Radajewski, T. W. Willison, and J. C. Murrell. 2002. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murrell, J. C., B. Gilbert, and I. R. McDonald. 2000. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 173:325-332. [DOI] [PubMed] [Google Scholar]
- 27.Radajewski, S., G. Webster, D. S. Reay, S. A. Morris, P. Ineson, D. B. Nedwell, J. I. Prosser, and J. C. Murrell. 2002. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331-2342. [DOI] [PubMed] [Google Scholar]
- 28.Reeburgh, W. S., S. C. Whalen, and M. J. Alperin. 1993. The role of methylotrophy in the global methane budget, p. 1-14. In J. C. Murrell and D. P. Kelly (ed.), Microbial growth on C1 compounds. Intercept Ltd., Andover, United Kingdom.
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Semrau, J. D., A. Chistoserdov, J. Lebron, A. Costello, J. Davagnino, E. Kenna, A. J. Holmes, R. Finch, J. C. Murrell, and M. E. Lidstrom. 1995. Particulate methane monooxygenase genes in methanotrophs. J. Bacteriol. 177:3071-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolyar, S., A. M. Costello, T. L. Peeples, and M. E. Lidstrom. 1999. Role of multiple gene copies in particulate methane monooxygenase activity in the methane-oxidizing bacterium Methylococcus capsulatus Bath. Microbiology 145:1235-1244. [DOI] [PubMed] [Google Scholar]
- 32.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 33.Strimmer, K., N. Goldman, and A. von Haeseler. 1997. Bayesian probabilities and quartet puzzling. Mol. Biol. Evol. 14:210-211. [Google Scholar]
- 34.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, Z., and J. P. Bielawski. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, Z., R. Nielsen, N. Goldman, and A. M. K. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]