Abstract
Polyploidy, or whole-genome duplication (WGD), is an important genomic feature for all eukaryotes, especially many plants and some animals. The common occurrence of polyploidy suggests an evolutionary advantage of having multiple sets of genetic material for adaptive evolution. However, increased gene and genome dosages in autopolyploids (duplications of a single genome) and allopolyploids (combinations of two or more divergent genomes) often cause genome instabilities, chromosome imbalances, regulatory incompatibilities, and reproductive failures. Therefore, new allopolyploids must establish a compatible relationship between alien cytoplasm and nuclei and between two divergent genomes, leading to rapid changes in genome structure, gene expression, and developmental traits such as fertility, inbreeding, apomixis, flowering time, and hybrid vigor. Although the underlying mechanisms for these changes are poorly understood, some themes are emerging. There is compelling evidence that changes in DNA sequence, cis- and trans-acting effects, chromatin modifications, RNA-mediated pathways, and regulatory networks modulate differential expression of homoeologous genes and phenotypic variation that may facilitate adaptive evolution in polyploid plants and domestication in crops.
Keywords: polyploidy, nonadditive gene expression, epigenetic regulation, RNA interference, evolution
INTRODUCTION
Polyploids can be classified into allopolyploids and autopolyploids based on the origins and levels of ploidy (25, 49, 135) (Figure 1). An autopolyploid results from doubling a diploid genome (Figure 1a). An allopolyploid is formed by the combination of two or more sets of distinct genomes). Mechanisms include interspecific hybridization followed by chromosome doubling (Figure 1b), fertilization of unreduced gametes between two diploid species (Figure 1c), or interspecific hybridization between two autotetraploids (Figure 1d). The two identical chromosomes (red or blue) within a species are homologous, while the chromosomes derived from different species (red and blue) are orthologous but become homoeologous within an allotetraploid. In an allopolyploid, only bivalents are formed because meiotic pairing occurs between homologous chromosomes (Figure 1b,c,d). If the homoeologous chromosomes have some segments that are homologous (Figure 1e), pairing may occur between the homoeologous chromosomes, resulting in the formation of multivalents and segmental allotetraploids (135). Here we consider allopolyploids and amphidiploids or disomic allopolyploids to be synonyms. Strictly speaking, only bivalents are formed in the amphidiploids and disomic allopolyploids, whereas multivalents may be formed in the allopolyploids or segmental allopolyploids.
Figure 1.

Illustration of auto- and allopolyploids. For simplicity, only one pair of homologous chromosomes (either in red or blue) is shown in a diploid. (a) Formation of an autotetraploid by doubling a basic set of chromosomes. A triploid (not shown) can be formed by hybridization between a diploid and an autotetraploid. (b) Formation of an allotetraploid by interspecific hybridization followed by chromosome doubling. (c) Formation of an allopolyploid by fusion of unreduced gametes in two diploid species. (d) Formation of an allotetraploid by interspecific hybridization between two autotetraploid species. (e) An allotetraploid (left) may become a segmental allotetraploid (right) if homoeologous chromosomes contain some homologous chromosomal segments.
In addition to polyploidy, some plant and animal species exist as intraspecific and inter-specific hybrids (96, 124). Many plants that transmit as diploids are actually paleopolyploids (ancient polyploids), which are derived from at least one event of whole-genome duplication (WGD) followed by massive gene loss and genomic reorganization through a process known as diploidization (152). Arabidopsis (14, 17, 144), rice (154), and maize (45) are good examples of diploidized paleopolyploids. An estimated 30–70% of plant species are of polyploid origin (93, 152). That estimate is as high as 100% if paleopolyploids are included (152).
Polyploidy is a fundamental but relatively underexplored biological process. It is widespread but little is known about how duplicate genes and genomes function in the early stages of hybridization, and how the duplicate genes maintain and diverge functions during plant evolution and crop domestication. Many polyploids are ancient, and their exact progenitors are often unknown. Resynthesized polyploids with known progenitors are excellent materials for dissecting gene expression and genomic changes in early stages and comparisons with older polyploids (28, 79, 132, 149). In addition to the parental phenotypes, polyploids give rise to phenotypes that are intermediate between the two parents and to novel phenotypes that are absent in or exceed features of the contributing parents (77, 82, 119), suggesting nonadditive gene expression. Some traits, such as increasing levels of drought tolerance, apomixis, pest resistance, flowering-time variation, and organ size, may allow polyploids to enter new niches or improve their fitness. Indeed, polyploids may survive better than their diploid progenitors in harsh environments, such as high altitudes and latitudes and cold climates, whereas both diploids and polyploids often thrive and cohabit in mild conditions (49, 135). Moreover, polyploidy is a means of permanent fixation of hybrid vigor and dosage regulation, which may be why many crops (e.g., wheat, cotton, oats, canola, potato, peanuts, sugarcane, coffee, and strawberry) are of polyploid origin (63, 93). Thus, polyploidy has been studied in the context of evolution, genetics, breeding, and molecular biology (25, 29, 50, 76, 79, 86, 95, 111, 132, 149).
Interspecific hybridization and allopolyploidization occur frequently in plant taxa including Brassica (133), Gossypium (150), Senecio (1), Spartina (10), Tragopogon (141), and Triticum (40, 130). Furthermore, hybrids can be formed between different genera including Triticum (wheat)-Secale (rye) (62), Triticum (wheat)-Hordeum (barley) (106), Zea (maize)-Avena (oat) (123), and Zea (maize)-Tripsacum (gamma grass) (58). Some allopolyploids (e.g., Tragopogon miscellus and T. mirus) were produced in natural conditions as recently as ~80 years ago, and new Tragopogon allotetraploids appear to form every year (141). In contrast, polyploids are rarer in animals than in plants (90, 107). Interspecific hybrids occur in vertebrates and mammals (e.g., a mule is a hybrid between a horse and a donkey), but they cannot produce offspring probably because of genomic incompatibility and/or imbalance in imprinting and sex chromosome dosage (107, 112). Polyspermy (fertilization of more than one sperm into one ovum) causes human triploids in 1–3% of conceptions, and the triploid fetuses are aborted (100). An isolated case of a tetraploid South American rodent (Tympanoctomy barrerae) is still debatable (44, 139). Except for endopolyploidy (a diploid individual with cells containing more than 2 C amount of DNA in their nuclei) in some cell types (37), aneuploid and polyploid cells in animals and humans are often associated with malignant cell proliferation or carcinogenesis (136).
MECHANISMS FOR GENE EXPRESSION DIVERGENCE BETWEEN ALLOPOLYPLOIDS AND THEIR PARENTS
Several mechanisms may affect the fate of orthologous and homoeologous genes in polyploids (25, 29, 76, 79, 86, 95, 111, 132, 149) (Figure 2). First, the majority of homoeologous genes are coexpressed. Second, some duplicate genes are lost, mutate, or diverge (due to genetic changes). The half-life of an active paralogous gene that becomes mutated or lost is estimated to be 2–7 million years (87). Third, epigenetic changes may reprogram gene expression and developmental patterns of new allopolyploids. The impact of these mechanisms on various polyploids can be very different. For example, sequence elimination is predominately observed in wheat and Tragopogon allopolyploids (40, 130, 141); chromosomal translocations and transposition (insertion of a DNA fragment into homoeologous chromosomes) are common in Brassica allopolyploids (133); and changes in gene expression appear to be a major consequence in Arabidopsis and cotton allopolyploids (2, 73, 147, 148). Moreover, genetic and epigenetic changes may be interrelated (25); reactivating transposons by chromatin modifications or RNA-mediated pathways may lead to chromosomal breakages and rearrangements. Eliminating DNA sequences (regulatory and/or coding regions) may alter dosage-dependent gene regulation and chromatin structure.
Figure 2.

Hypotheses to explain changes in genome structure and function in polyploids. (a) Genetic and epigenetic changes in polyploid genomes. Genetic changes include sequence mutations and chromosomal instabilities (deletion/insertion, translocation, transposition, etc.). Epigenetic changes may occur at transcriptional and post-transcriptional levels. Transcriptional regulation is associated with the formation of heterochromatin (black segments in red chromosomes) and euchromatin (irregularly-shaped long arms of blue chromosomes), leading to gene silencing or activation. Chromatin modifications [through multiple mechanisms including RNA interference (RNAi)] may transcriptionally silence or activate mobile elements, protein-coding genes, and rRNA genes that are responsible for novel variation in allopolyploids. Post-transcriptional regulation involves RNAi, RNA processing, and stability. AA: diploid genome; AAAA or BBBB: autotetraploid; AABB: allotetraploid. Blue segments in red chromosomes or red segments in blue chromosomes in an allotetraploid indicate nonreciprocal exchanges between homoeologous chromosomes. White segments in the red chromosomes indicate a deletion. (b) A dosage-dependent (12) or “rheostat” (102) model suggests additive effects of gene expression in a diploid (upper panel) and tetraploid (lower panel). The duplicate loci in the tetraploid provide additional levels of controls for gene expression. The levels of gene expression are shown as “off” (0) and by gray arrows (1, 2, and 3), and the maximum level (2 in diploid and 4 in tetraploid) is shown by green arrows. This model may explain dominant effects of hybrid vigor but does not explain overdominant performance or novel variation in allopolyploids. (c) A regulatory compatibility model suggests that regulatory factors (proteins) produced from orthologous genes generate incompatible heterologous products (heterodimers between red squares and blue circles). Alternatively, heterologous proteins may perform better than homologs (homodimers between red squares or blue circles), which may explain overdominant effects and hybrid vigor. (d) Hypotheses for testing additive and nonadditive gene regulation in Arabidopsis allotetraploids. The null hypothesis (Ho) is that gene expression levels in an allotetraploid (Allo) are equal to the sum of two progenitors, A. thaliana autotetraploid (At4) and A. arenosa (Aa). Typical seedling leaves in At4, Aa, and Allo are shown. Ha: alternative hypothesis.
Mutations, Sequence Eliminations, and Chromosomal Rearrangements (Genetic Changes)
Elimination of chromosome- or genome-specific sequences may occur during polyploid formation (Figure 2a). The stochastic changes of duplicate genes may promote polyploid speciation, which is supported by studies in Brassica (133), wheat (40, 130), and Tragopogon(141). In the resynthesized allopolyploids, loss of parental fragments and/or the appearance of novel fragments are commonly observed. Rapid sequence elimination in the resynthesized allopolyploid wheat may account for a relatively high amount (~14%) of genome- or chromosomal-specific DNA sequences (40, 130), suggesting that differential elimination of genome-specific sequences facilitates pairing between homologous chromosomes but not homoeologous chromosomes.
Changes in DNA sequence may contribute to the loss of duplicate gene expression and function. Indeed, many isozyme loci are lost during polyploidization, such as chlorophyll a/b binding protein genes in Polystichum munitum, leucine aminopeptidase loci in tetraploid Chenopodium, and phosphoglucose isomerase loci in homosporous fern and Clarkia (48, 116). Estimates indicate that in the salmonid and cyprinid fish, the loss of duplicate isozyme loci can be as high as 35–65%, suggesting that loss of duplicate gene function is common after polyploidization, which occurred 50 million years ago (Mya) in this lineage (41). In Tragopogon, 9 of 10 genes that display expression differences are associated with changes in allelic DNA sequence (141). However, loss of gene function may also suggest an epigenetic cause (see below).
Epigenetic Regulation of Orthologous Loci
Genetic mutations can explain the cause of gene loss over evolutionary time, but many silencing phenomena may be epigenetically controlled, especially in the early stages of polyploid formation. When two different genomes are combined into a single cell, they must respond to the consequences of genome duplication, especially duplicate copies of genes with similar or redundant functions. Increased gene or genome dosage may induce disease syndromes and abnormal development (7, 38). Thus, the expression of orthologous genes must be reprogrammed through epigenetic mechanisms (Figure 2a) in the early process of polyploidization. This resembles the “genomic shock” phenomenon proposed by McClintock (98). The genomic shock occurs rapidly in interspecific hybrids and allopolyploids, resulting in demethylation of retroelements (92), relaxation of imprinting genes (20, 68, 145), and silencing and activation of homoeologous genes (2, 26, 69, 73, 91, 146-148), including rRNA genes subjected to nucleolar dominance (expression of rRNA genes from only one progenitor in an interspecific hybrid or allopolyploid) (117, 122). Epigenetic changes, which are potentially reversible, provide an effective and flexible means for a polyploid cell to respond to polyploidy or genomic shock. Moreover, gene silencing or activation that is initially epigenetic and reversible could be one step toward a genetically fixed and irreversible state. Epigenetic silencing may also accelerate sequence mutation rates of the affected genes, as observed in repeat-induced point mutations in duplicated genes of Neurospora crassa (128).
Mechanisms for epigenetic regulation of homoeologous genes in the allopolyploids are reminiscent of those for X-chromosome inactivation (75), gametic imprinting (143), paramutation (21, 121, 134), and homology-dependent gene silencing (11, 67, 94). However, ploidy-dependent gene regulation has some unique features. First, epigenetic interactions are established among four alleles of two homoeologous loci in allotetraploids compared with two alleles of one locus in a diploid. Second, homoeologous genes from different parental origins may be up- or downregulated in a chromosomal domain (73, 147), which is different from dosage compensation that often refers to concerted or unidirectional changes in gene expression. Third, at least some epigenetic silencing phenomena in allopolyploids are stochastically established and require multiple generations (24, 104, 148), probably because of the complex process of sorting out chromosome pairing in the allopolyploids. Fourth, pairing occurs mainly between homologous chromosomes, but occasionally between homoeologous chromosomes in allopolyploids (Figure 1e), which may affect gene expression. Finally, because of divergence of regulatory sequences between the progenitors and of heterologous proteins produced in the allopolyploids, cis- and trans- acting effects on homoeologous genes (146, 151) in various biological pathways constitute a major mode of gene regulation in the allopolyploids.
Additive and Nonadditive Expression of Orthologous Loci
Many genes display dosage dependency and are expressed additively in aneuploids and polyploids (53). If the levels of gene expression and phenotypic variation in the progenitors are additive, they would have the midparent values (MPVs) in the polyploids; that is, one plus one is equal to 2 (Figure 2d). If the gene expression is nonadditive (different from the MPV), the values would be larger than 2 or smaller than 2. The former would suggest gene activation including overdominance, whereas the latter would suggest repression and/or silencing. One model to explain additive gene regulation is that there are extra control settings or rheostat potentials (four levels) in a tetraploid compared with two levels in a diploid (Figure 2b). In Arabidopsis and Brassica, the alleles in the FLOWERING LOCUS C (FLC) loci display additive effects on flowering time (102). In Arabidopsis allotetraploids, the expression of A. thaliana and A. arenosa FLC loci is additive, giving rise to a late flowering phenotype (146). Furthermore, up to ~90% of the transcriptome is expressed additively in resynthesized Arabidopsis allotetraploids (147). Although odd and even dosage effects may vary in a ploidy series (see below), coexpression and coevolution of orthologs and paralogs suggest that a selective advantage is obtained from dosage dependency (15, 47).
Similar numbers of genes (5–11%) are differentially expressed between the parents and resynthesized allotetraploids in stable allotetraploids and five selfing generations (148), and a slightly low number of genes (~2.5%) are differentially expressed between a natural allotetraploid, A. suecica, and its assumed progenitors (73). Most gene expression changes observed in early generations are maintained in the late generations and natural allotetraploids, suggesting that rapid and stochastic changes in the resynthesized allotetraploids are responsible for adaptive evolution (148).
Furthermore, allopolyploidy may induce regulatory incompatibilities as well as selective advantage by combining heterologous protein products (Figure 2c). For instance, there is evidence that some protein heterodimers may not function as well as homodimers or vice versa (115, 118). Thus, a silencing strategy could balance regulatory incompatibility and the advantages of having multiple copies of orthologous genes or gene products (e.g., transcriptional factors) spontaneously produced in an allopolyploid cell. Alternatively, novel interactions between heterologous protein products may provide a molecular basis for hybrid vigor and novel adaptation.
INSTANTANEOUS SPECIATION AND A MODEL FOR POLYPLOIDY STUDIES
New species are often gradually formed because of geographical and ecological separations from an ancestral species (49). However, new species are believed to have arisen suddenly via polyploidization in plants and some animals, including vertebrates such as amphibians and lizards (18, 49, 93, 112). For example, Arabidopsis suecica (2n = 4x = 26) is a natural allotetraploid formed 12,000 to 1.5 Mya (64, 71, 125). The two progenitor species, A. thaliana and Arabidopsis arenosa (108, 123), split ~6 Mya (71), similar to the distance between humans and chimpanzees (~6.3 million years) (114). Despite this distance, A. thaliana autotetraploid (2n = 4x = 20) and A. arenosa tetraploid (2n = 4x = 32) can hybridize to produce A. suecica-like plants (2n = 4x = 26) (Figure 3a,b,c). A. arenosa is thought to be an autotetraploid (31), but sequencing analysis sugegsts that it is not a pure autotetraploid (146) (L. Tian, J. Wang & Z.J. Chen, unpublished). The resynthesized allotetraploids are meiotically stable (30, 147) and contain five pairs of A. thaliana chromosomes and eight pairs of A. arenosa chromosomes (28, 30, 147) (Figure 3c). Compared with resynthesized Brassica and wheat allopolyploids that undergo rapid changes in chromosomal structure and DNA sequences (40, 133), the frequency of aneuploids and chromosome abnormalities in Arabidopsis resynthesized allotetraploids is relatively low (30).
Figure 3.
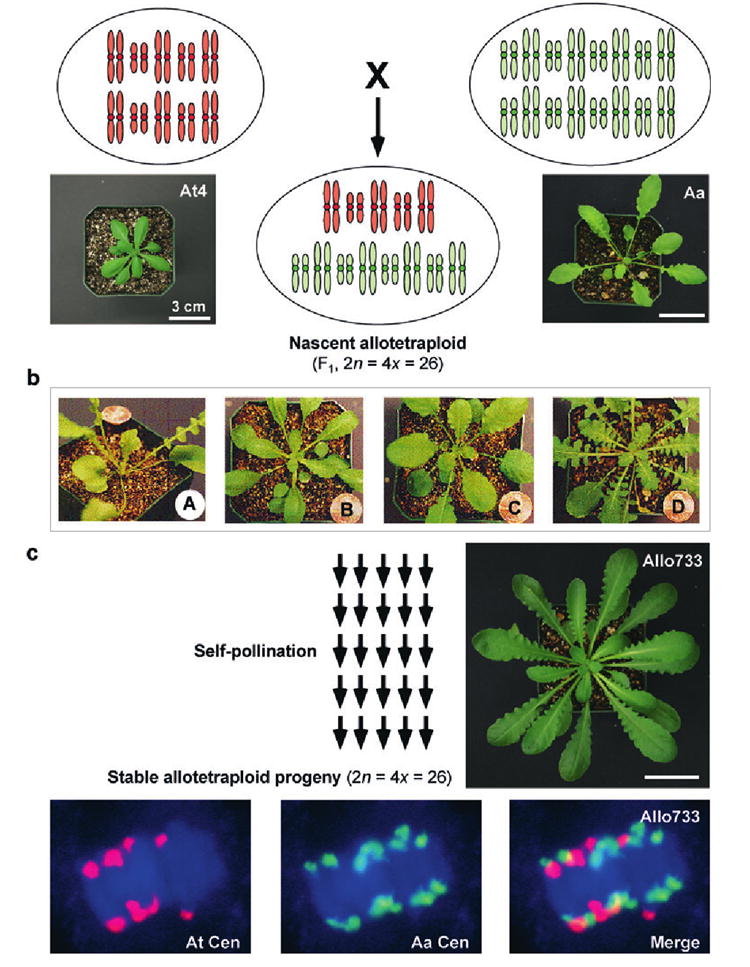
Production of Arabidopsis allotetraploids. (a) Schematic karyotypes of F1 nascent allotetraploids and their progenitors. Seedlings of the two progenitors, A. thaliana autotetraploid (At4) and A. arenosa tetraploid (Aa), are shown. (b) Phenotypic variation was infrequently observed in the F1 allotetraploids but was very common in the segregating populations (F2) (A–D). A and D: A. arenosa-like, B: A. thaliana-like, and C: intermediate. (c) Independent lineages of allotetraploids are produced by selfing multiple F1 lines. Allopolyploids self-pollinate spontaneously (or can be manually self-pollinated), and progressive inbreeding occurs in advanced generations. Karyotypes in a meiotic cell of a resynthesized allotetraploid (Allo733 in the fifth generation) show 5 pairs of A. thaliana centromeres (At Cen, red) and 8 pairs of A. arenosa centromeres (Aa Cen, green) (147) (reproduced with kind copyright permission of the Genetics Society of America). Seedling of Allo733 is shown. All plants were photographed when they were 4–5 weeks old. The size bars indicate 3 cm.
The nascent allotetraploids (F1 individuals) are genetically identical (Figure 3a) and showed subtle phenotypic variation. Some variation among F1 individuls may derive from heterozygosity of the outcrossing tetraploid A. arenosa parent, whereas other variation may result from interactions between A. arenosa and the different genotypes of A. thaliana used in interspecific hybridizations. The degree of variation depends on parental genotypes used in the interspecific hybridization. For example, the seed set is higher in the nascent allotetraploids (F1) between A. arenosa and A. thaliana C24 or Ler ecotype than those between A. arenosa and A. thaliana Columbia (20, 31), indicating genotypic effects on interspecific hybridization. Hybridization was successful only in the crosses using A. thaliana as a maternal parent and A. arenosa as a pollen donor (31) ( J. Wang, L. Tian & Z.J. Chen, unpublished), probably because A. arenosa is outcrossing and self-incompatible. Most F1 individuals and selfing progeny in late generations resemble the A. arenosa parent and A. suecica (31, 91, 147) (Figure 3c), although diverse phenotypes are observed in segregating populations (F2-F3) (Figure 3b). Therefore, A. arenosa appears to be phenotypically dominant over A. thaliana in the allotetraploids (28, 147).
The allotetraploids obtained from selfing the F1s show stable karyoptes in the fifth generation (Figure 3c), but exhibit a wide range of variants, some of which are absent in either parent (transgression) (Figure 4a). Moreover, the allotetraploids display hybrid vigor: larger rosettes, more leaves, longer and wider leaves, and taller plants than the parents. The fertility rate of the plants in the selfing progeny varies from one lineage to another (30, 31). The overall level of fertility improves after each generation of selfing, suggesting that genome incompatibility and gene expression divergence between the progenitors are gradually overcome (148).
Figure 4.

(a) Phenotypic variation of the plants after five generations of selfing. The plants include two parents, A. thaliana autotetraploid (At4) and A. arenosa (Aa), six allotetraploids (S–1 to S–6), and a natural allotetraploid, A. suecica (As). Allotetraploids have the same chromosome number (Figure 3c and data not shown) but display phenotypic and flowering-time variation. Some plants (S5–5 and S5–6) resemble A. suecica, whereas others (S5–1 to S5–4) show novel phenotypes. Differential expression of parental genes may contribute to the phenotypic variation of Arabidopsis allotetraploids. (b) Evidence for epigenetic silencing in resynthesized allopolyploids. A. suecica (As, white flower) is the naturally occurring allotetraploid derived from A. thaliana (not shown) and A. arenosa (Aa), which have white and pink flowers, respectively. The flower colors in the third generation of resynthesized allotetraploids (S3–1, 2, and 3) segregate from all white (S3–1) to all pink (S3–3). Variegated flower colors in the same inflorescence branch (S3–2) indicate epigenetic regulation of genes controlling flower color pathways. The same-size bars are 50 mm in (a), except for At4, and 5 mm in (b), except for As.
The flower colors varied from pink (like A. arenosa) in the early generation (F1) to a mixture of pink and white flowers in the intermediate generations (S2–4) and white in the late generation (S5). During selfing (S3), there is a low frequency of mixed white and pink flowers in the same flower branch (Figure 4b, bottom), which is transient and mosaic (derived from the same zygote). The appearance of varigation within the same flower branch suggests rapid changes in the expression of genes involved in anthocyanin synthesis pathways probably via epigenetic regulation.
NONADDITIVE GENE REGULATION AND TRANSCRIPTOME DOMINANCE IN ALLOPOLYPLOIDS
In 1928, Navashin coined the term “amphiplasty” to describe chromosomal changes in interspecific hybrids of Crepis (110). He defined “differential amphiplasty” as specific changes in a few chromosomes (disappearance of satellites or secondary constrictions) and “general amphiplasty” as the overall changes in chromosomal morphology (shortening, thickening, or lengthening of chromosomes) from one species in the interspecific hybrids or amphidiploids. Changes in chromosomal morphology might also affect gene expression. Indeed, following the pioneering work of Navishin & McClintock (97, 110), several contemporary researchers demonstrated that differential amphiplasty is synonymous to nucleolar dominance (117, 122). The disappearance of the secondary constrictions is caused by silencing of rDNA loci in those chromosomes (117). Nucleolar dominance is observed in Drosophila interspecific hybrids and Xenopus, Arabidopsis, Brassica, and wheat allopolyploids (117, 122). The dominance is reversible and developmentally regulated and is controlled by chromatin modifications involving DNA methylation and histone acetylation (26, 27). Blocking histone acetylation or DNA methylation derepresses the silenced rRNA genes subjected to nucleolar dominance. Both DNA methylation and histone hypoacetylation reinforce the formation of the “inactive” chromatin state, resulting in gene silencing (72). The silencing of rDNA chromatin requires at least one histone deacetylase (AtHDA6) that is localized in nucleoli (35).
General amphiplasty may be similar to the effects of genomic shock (98). Combining two genomes in a “new” polyploid cell may generate the genomic shock and release some constraints imposed on unstable elements locked in a junk yard (e.g., transposable elements in heterochromatin). Little is known about the consequences of general amphiplasty or genomic shock on interspecific hybrids or allopolyploids that have balanced pairs of chromosomes.
To begin to test this, Wang et al. (144) studied transcriptome divergence in Arabidopsis allotetraploids and their progenitors. First, they compared gene expression differences between the two progenitors using the spotted oligo-gene microarrays designed from ~26,000 annotated genes that share a high percentage of sequence identities between A. thaliana and A. arenosa. Most of the oligos can cross-hybridize with both A. thaliana and A. arenosa genes (74). More than 15% of the transcriptome is differentially expressed between A. thaliana and A. arenosa that diverged ~6 Mya. Approximately 2,100 genes (8%) are more abundantly expressed in A. thaliana than in A. arenosa, whereas 1,818 genes (7%) are expressed at higher levels in A. arenosa than in A. thaliana. Second, Wang et al. (144) compared mRNA abundance in an allotetraploid with the mid parental value (MPV: an equal mixture of RNAs from two parents). If the genes from two progenitors are additively expressed (Figure 2d), their cumulative expression levels in the allotetraploid are equal to MPV. Nonadditive expression suggests that at least one of the homoeologous genes is up- or downregulated. There may also be instances in which silencing of a locus is compensated by increased expression of its homoeologous locus, which cannot be detected in this comparison. Wang et al. (146) found that 2,011 genes (~8%) are nonadditively expressed in two independently derived allotetraploids using a common variance analysis, and up to ~38% genes using a pergene variance analysis. Interestingly, ~68% of the genes that are nonadditively expressed in the allotetraploids are differentially expressed between the two parents, suggesting that the genes with species-specific expression patrterns are subjected to expression changes in the allopolyploids. Remarkably, among the nonadditively expressed genes, more than 65% of the genes are downregulated in the allotetraploids. Among them, >94% of the genes that are expressed at higher levels in A. thaliana than in A. arenosa are downregulated in the allotetraploids. These data indicate that the genes with A. thaliana expression patterns tend to be repressed, whereas the genes with A. arenosa expression patterns are transcriptionally dominant in the allotetraploids, coincident with the phenotypic and nucleolar dominance of A. arenosa in the allotetraploids (117, 147) (Figures 3 and 4).
Interestingly, similar levels of transcriptional changes were observed in maize diploid hybrids (9.8%) (140) and polyploid taxa of Senecio (5%) (60), wheat (7.7%) (59), and cotton (5%) (2). The high percentage of gene expression changes in the Tragopogon allopolyploids (~17.5%) (141) is partly associated with a high level of polymorphism (~11%) within populations between the two parents and a moderate amount of variation (>2.5%) among allopolyploid populations. These numbers are also similar to those observed in interspecific hybrids of Drosophila (103, 151), suggesting that the levels of transcriptional changes induced by hybridization may be fairly consistent even across plant and animal kingdoms. Transcriptome dominance is also observed in an analysis of ~210,000 expressed sequence tags (ESTs) derived from an ovular cDNA library of tetraploid cotton (Gossypium hirsutum L.) (153). The upland cotton was formed by ancient interspecific hybridization between AA and DD genome species (150). AA subgenome ESTs of all functional classifications including cell cycle control and transcription factor activity were selectively enriched in G. hirsutum L., a result consistent with the production of long lint fibers in AA genome species. Therefore, transcriptome dominance is likely a general consequence of hybridization effects on gene expression in interspecific hybrids and allopolyploids.
The number of genes displaying expression changes in A. thaliana autopolyploids is much smaller than that in the allotetraploids (147). In yeast, Galitski et al. (43) found that 10 genes are induced and seven genes are reduced in response to an increase in ploidy levels (haploid, diploid, triploid, and tetraploid). The cell size increases with increasing ploidy levels, which is correlated with repression of G1 cyclins (Cln1 and Pcl1). FLO11, a gene important to the invasiveness of the yeast cells, is repressed with increasing ploidy levels. The reduction of FLO11 expression in cells of higher ploidy is correlated with diminished invasion, suggesting a role of ploidy-dependent gene regulation in adaptive evolution. Collectively, the data indicate that genome doubling has smaller effects on gene expression changes than intergenomic hybridization.
What factors affect transcriptome dominance in the allopolyploids? Is the gene repression controlled by widespread chromatin modifications or a few “key” regulatory genes? Over time, the progenitor species may have evolved to possess species-specific gene expression patterns. Modulation of the species-specific expression of these genes may determine the outcome of transcriptional and posttranscriptional competition between the two parental genomes in their offspring. Changes in chromatin landscape on repressed genes may result from concerted modifications of many genes in one species, perhaps by a mechanism similar to that for nucleolar dominance (117). Alternatively, expression changes in a few regulatory genes such as transcription factors and microRNAs may induce trans-acting effects on many downstream pathways (25). For example, a Myb transcription factor gene is responsible for hybrid-induced incompatibilities in Drosophila interspecific hybrids (8). Also, a single miRNA can regulate hundreds of genes involved in the transition from one developmental stage to another (39).
The role of chromatin modifications in silencing or activating protein-coding genes in allopolyploids has been demonstrated in several recent studies (70, 73, 91, 104, 146, 148). Silenced genes can be reactivated by aza-dC (73), a chemical inhibitor of DNA methylation, or by downregulation of the genes encoding DNA methyltransferases using RNA interference (RNAi) (148). Treating allotetraploids with aza-dC generates pleiotropic effects on natural and synthetic allotetraploids including reactivation of mobile elements (91). Reactivation of transposons is also observed in the synthetic allotetraploids (92). The above data suggest that two species may have possessed different levels of chromatin modifications for many genes that display species-specific expression patterns. Perturbation of chromatin structure may have occurred during the formation of interspecific hybrids or allopolyploids, leading to the changes in gene expression.
Factors other than chromatin modifications may also be responsible for genomewide nonadditive gene regulation. Non-additively expressed genes are randomly distributed along the chromosomes (147). Within a small chromosomal region in which TCP3 and RFP genes are located, A. thaliana TCP3 is expressed, whereas A. arenosa TCP3 is silenced (73). For RFP, A. thaliana RFP is repressed, whereas A. arenosa RFP is expressed. Interestingly, the neighboring genes located between TCP3 and RFP loci are coexpressed. The above data are reminiscent of the silenced rRNA genes that are restricted in the rDNA loci (81). Furthermore, in the met1-RNAi A. suecica lines, several silenced genes are not reactivated (148). These data argue that widespread chromatin remodeling does not explain nonadditive regulation for all genes, but support the notion that each gene is regulated through interactions among homoeologous loci such as paramutation-like phenomena observed in A. thaliana tetraploids (104).
ALLOPOLYPLOIDY AND HETEROSIS
Genome-wide nonadditive gene regulation observed in the allotetraploids correlates with expression divergence between the parents. Thus, hybrids derived from distantly related species may induce a high level of gene expression changes in a nonadditive fashion, providing molecular bases of hybrid vigor (13) and phenotypic variation in the allotetraploid progeny (31). Hybrid vigor refers to the performance of an F1 hybrid higher than MPV or the best parent. The genetic basis for heterosis is predicted to be associated with dominant complementation of slightly deleterious recessives (dominance model) (19, 66) or overdominant gene action in which genes have greater expression in heterozygous conditions (overdominance model) (32, 36). According to the dominance model, highest performance should be observed when all dominant favorable genes from both parents are in homozygous conditions. The overdominance model suggests that heterosis should reach its peak at the maximum levels of heterozygosity and dissipate when approaching homozygosity. Moreover, overdominance is accompanied by nonallelic or epistatic interactions, and epistasis is involved in most QTLs associated with inbreeding depression and heterosis in corn (137) and rice (84). Comparing genome-wide gene expression data with phenotypic traits (QTLs) may provide new insights into the role of gene expression changes in various biological pathways that give rise to hybrid vigor.
The gene expression changes observed in maize diploid hybrids (6, 140) and genomewide transcriptome dominance in Arabidopsis allotetraploids (147, 148) support both dominance and overdominance models. Many genes in energy, metabolism, cellular biogenesis, and plant hormonal regulation are upregulated in the allotetraploids (147), which may contribute to the hybrid vigor observed in the allotetraploids. Although the underlying mechanisms are unknown, one possibility is modulation of a few key regulators in the allotetraploids that may control downstream genes in various biological pathways (146, 147) such as photosynthesis and metabolism (Z. Ni & Z.J. Chen, unpublished).
Alternatively, cis- and trans-acting effects involving regulatory sequence changes (see below), chromatin modifications, and RNA-mediated pathways (25) (Figure 2a) may explain dominance, overdominance, and epistasis. The interactions between the diverged orthologous protein products may determine repression or activation of progenitors’ genes in allopolyploids (Figure 2c) of Arabidopsis (148), cotton (2), Senecio (60), and wheat (59, 69), interspecific hybrids (151) in Drosophila, intraspecific diploid hybrids in maize (6, 54, 140), and sex-dependent gene regulation in Drosophila (46, 120). These mechanisms are not mutually exclusive, and the diverged protein-protein and protein-DNA interactions in allopolyploids may trigger repression of the protein-coding genes and rDNA loci derived from one progenitor (e.g., A. thaliana) via chromatin modifications (26, 73, 146, 148) or novel expression patterns leading to hybrid vigor (Figure 2c).
CIS- AND TRANS-ACTING EFFECTS ON TRANSCRIPTIONAL REGULATION IN ALLOPOLYPLOIDS
Stable allopolyploids provide an excellent system for testing cis- and trans-acting effects because a common set of protein factors is present in the same allotetraploid cells. After the unification of the distinct genomes, differences in cis- and trans-regulation contribute to changes in the expression of orthologs that become homoeologous pairs in the allopolyploid or interspecific nucleus (146, 151). Cis-regulatory divergence directly acts on single genes or localized chromatin domains such as promoters or enhancers and may result in asymmetric accumulation of homoeologous transcripts in allpolyploids.
There is evidence for cis- and trans-effects on orthologous or homoeologous genes in the allotetraploids (146) and interspecific hybrids (33, 151). Differential expression of progenitors’ genes in Arabidopsis allopolyploids (73, 148) and interspecific hybrids (33), Drosophila interspecific hybrids (151), and maize diploid hybrids (138) is mainly caused by cis-regulatory changes. Progenitor-specific differences in expression in the same cells are most likely due to allelic or epigenetic differences. In contrast, expression divergence due to alterations in trans-regulatory hierarchies should result in two kinds of expression changes. The first is a difference in the sum of homoeologous mRNAs compared with the mid-value of the two parents or nonadditive gene expression. Indeed, the divergently expressed orthologs comprise ~68% of the genes that were expressed in a nonadditive fashion in two allotetraploids (146, 147), implicating trans-acting effects. The second is a change in the ratio of homoeolog-encoded mRNAs in an allopolyploid compared with the ratio of the two orthologs in a 1:1 mixture of the parental mRNAs (147). Such a difference would demonstrate a regulatory interaction between the parental genomes (for example, the failure of an interspecific heterodimer to activate transcription) (25) (Figure 2c). A regulatory hierarchy (12) model suggests that trans-regulatory differences predominate in allopolyploids (25).
The species-specific expression patterns observed in Arabidopsis allotetraploids (147) may result from sequence divergence at regulatory elements during the ~6 million years that separate the parental species. Cis- and trans-acting regulation and epigenetic modifications of homoeologous genes may change regulatory interactions in a biological pathway (Figure 5a). This has been demonstrated in a subset of genes controlling flowering-time variation. A. arenosa and A. thaliana (Ler) diverged in flowering habits probably because of selective adaptation to cold and warm climates (108, 123), respectively. Natural variation of flowering time is largely controlled by two epistatically acting loci, namely, FRIGIDA (FRI) (65) and FLOWERING LOCUS C (FLC) (102, 131). FRI upregulates FLC expression that represses flowering in A. thaliana. A. thaliana has a nonfunctional AtFRI (65), whereas A. arenosa FRI (AaFRI) is functional (146). Compared with A. thaliana (AtFLC), A. arenosa FLC (AaFLC) loci possess deletions in the promoter and first intron that are important to cis-regulation of FLC expression. In resynthesized allotetraploids, AaFRI complements nonfunctional AtFRI and interacts in trans with AtFLC, making the synthetic allotetraploids winter annual in a dosage-dependent manner. AaFRI acts on AtFLC in trans and on AaFLC in cis because A. thaliana FRI is nonfunctional. The different effects of AaFRI on AtFLC and AaFLC loci are likely dependent on the sequence divergence in their cis-regulatory elements (e.g., deletions in the promoter and first intron). AtFLC and AaFLC upregulation is mediated by H3-K9 acetylation and H3-K4 methylation, suggesting a role of FRI in locus-specific chromatin modifications (146).
Figure 5.

Models for transcriptional and post-transcriptional regulation of nonadditive gene expression in allopolyploids. (a) Cis- and trans-acting effects on transcriptional regulation of nonadditive gene expression in allotetraploids. I. During evolution, changes in cis-regulatory elements may confer “strong/dominant” and “weak/recessive” loci. II. Combination of these loci in the same allotetraploid cells induces cis- and trans-acting effects. Because A. thaliana FRI (AtFRI) is nonfunctional, A. arenosa FRI (AaFRI) trans-activates A. thaliana FLC (AtFLC) and cis-regulates one of the A. arenosa FLC (AaFLC1) loci, leading to overdominance or very late flowering in resynthesized allotetraploids. The cis-regulation and trans-activation are maintained by histone acetylation (Ac in a small purple circle) and methylation (Me in a small orange circle). Functional AaFRI is in a large red circle, and nonfunctional A. thaliana FRI (atflc) is in a white small circle with a slash line. The straight and inclined red arrows indicate cis- and trans-acting effects, respectively, by AaFRI. FLC loci are in oval circles. Blue and green indicates active AtFLC1 and AaFLC1, respectively, and gray indicates silenced AaFLC2. The differences in the expression of three FLC loci may reflect cis-regulatory variation (146). III. Cis- and trans-acting interactions and epigenetic regulation of FRI and FLC loci may facilitate selection for flowering-time variation in resynthesized allotetraploids (indicated by a circular wheel). IV. In A. suecica, additional selection forces may enhance the expression of AaFLC1 (color change from dark green in II to yellow green in IV) and related genes such as MAFs (146), which facilitate adaptation to cold climate. This simple model may be generalized to explain a mechanism for regulatory interactions between divergent loci in other pathways in the allotetraploids (see text for details). (b) Nonadditive accumulation of mRNAs and small RNAs in allotetraploids (null hypothesis Ho: 2 + 1 = 3). There is evidence that 15–40% of the genes are expressed differently between two closely related species of Arabidopsis (147), and a subset of those genes are subjected to nonadditive expression in the allotetraploids. Up- or downregulation of mRNAs may be caused by activation of genes originating in parent A, B, or both. We exclude the situation in which the expression of one locus is compensated by its corresponding homoeologous locus. If the nonadditively accumulated transcripts are small RNAs, such as microRNAs (miRNAs), heterochromatic and transposon-related siRNAs (ht-siRNAs), trans-acting siRNAs (ta-siRNAs), and natural anti-sense (nat-siRNAs), they typically negatively regulate the expression of their target genes. The red and blue wave lines indicate RNA transcripts or small RNAs produced in species A and B, respectively. We hypothetically assign two RNA molecules for species A and one RNA molecule for species B. Note that the number of RNAs in the nascent allotetraploids (F1) is not equal to 3 (2 + 1), suggesting nonadditive regulation (dominance and overdominance if larger than 3 or repression if smaller than 3). Possible changes (dashed arrows) in RNA composition and accumulation (column X) may occur in self-pollinating progeny, which are indicated by orange and green wave lines, respectively. According to the hypothetical outcome, the origins of small RNA upregulation (red in 1–3) and repression (blue in 4–6) are shown in the last column.
Although our model (Figure 5a) simplifies the flowering pathway that involves >80 genes (80), it offers one explanation of the fate of orthologous genes involved in biological pathways during allopolyploidization. Many orthologous genes might have diverged in their cis-regulatory elements that confer strong or weak, dominant or recessive alleles, tissue-specific expression, and/or developmental regulation. The regulatory networks may be reset by cis- and trans-acting effects via chromatin modification immediately after allopolyploidization. Over generations, genetic and epigenetic changes are subject to selection and adaptation, and additional genes (e.g., MAF, a FLC-MAF family member of MADS-box genes, in A. suecica) (146) may be activated for allopolyploids to occupy an environmental niche. A similar mechanism may be responsible for the functional diversification of orthologous genes in developmental regulation of gene expression, a phenomenon known as subfunctionalization of duplicate genes (88). Flowering time directly affects plant reproduction and adaptation. Therefore, sequence evolution and epigenetic regulation play interactive and pervasive roles in mediating the regulatory incompatibilities between divergent genomes, leading to natural variation and selective adaptation during allopolyploid evolution.
POST-TRANSCRIPTIONAL REGULATION IN ALLOPOLYPLOIDS
Some gene expression variation observed in polyploids may be controlled at the level of post-transcriptional regulation (70). Silencing a duplicate copy of homoeologous RNA in polyploids may be part of an RNA-mediated pathway similar to cosuppression (67, 94) or RNAi (42). Silencing of transgenes is correlated with transgene dosage in Drosophila (113) and ploidy levels in Arabidopsis (105). Activation of Wis 2–1A retrotransposon in the newly synthesized wheat allotetraploids drives the readout transcripts from adjacent sequences including the antisense or sense strands of known genes, leading to the silencing or activation of respective corresponding genes (70). RNA-mediated silencing of duplicate genes in polyploids is a developmental strategy. Production of progenitor-dependent RNA transcripts may be associated with mRNA accumulation and stability during growth and development (55). For example, a subset of genes involved in mRNA stability displayed expression variation in allotetraploids (E. Kim & Z.J. Chen, unpublished). CCR4, a gene involved in RNA stability and degradation in yeast and animals (34), is differentially accumulated in leaves and flower buds, suggesting a role of RNA stability in transcript accumulation in allopolyploids.
Over time, species may have adapted to spatial and temporal regulation of RNA transcripts including mRNAs, small RNAs, and additional noncoding RNA transcripts that could accumulate nonadditively in the allotetraploids (Figure 5b). The small RNAs may serve as negative regulators for the expression of target genes originating from two parents. First, the loci encoding miRNAs and siRNAs may diverge during the evolution of the progenitors as observed in the FRI and FLC loci (146). Sequence divergence in promoter regions and cis-acting elements leads to the expression variation when these loci are present in the same cell nuclei. Alternatively, differential expression of trans-acting factors may cause gene expression changes in the allopolyploids, as predicted (25). Second, antiviral RNAi genes involved in the biogenesis of small RNAs such as dicers, Argonaute, and RNA-dependent RNA polymerases (9, 155) generally diverge faster than other proteins during evolution (106). Combining two divergent proteins in the allotetraploids may alter enzymatic activity and specificity. As a result, different pools of small RNAs could accumulate in the allopolyploids. Third, natural sense and anti-sense transcripts and other read-through transcripts may participate in defense mechanisms (16) and may be accumulated differently in the progenitors. These transcripts affect the expression of neighboring loci as well as other loci via trans-acting effects. Fourth, mRNA transcript abundance is different in the progenitors. Although the coding sequences are very similar among Arabidopsis and its related species, sequences at the noncoding regions (5′- and 3′-ends) diverge relatively rapidly (L. Tian, M. Ha & Z.J. Chen, unpublished), which may affect processing and stability of RNA transcripts (55). Finally, each species is differentiated by the presence or absence of species-specific repetitive DNA sequences, including transposons that may affect the chromatin structure and expression of their neighboring genes. Differences in DNA replication and perturbation of chromatin structures among different species may induce the release of transposons and aberrant RNA transcripts that cause “genomic shock” and many downstream effects, as previously predicted (25, 98).
DEVELOPMENTAL REGULATION OF ORTHOLOGOUS AND HOMOEOLOGOUS GENES
During polyploid evolution, both copies of orthologous genes may remain if dosage effects are advantageous (142), or one copy of the gene duplicate may evolve a novel function via neofunctionalization (89). Alternatively, both copies may diverge their functions or expression patterns in different organs or tissues via subfunctionalization (88). Indeed, silenced rRNA genes in vegetative tissues are reactivated during flower development (27). In a survey of 40 genes in cotton, 10 genes (25%) display unequal expression in allotetraploids and exhibit organ-specific expression patterns (2). For 5 genes, the A-subgenome loci are expressed higher than the D-homoeologous loci, whereas for the other 4 genes, the D-subgenome loci are expressed higher than the A-homoeologous loci. For some homoeologous gene pairs, one locus (e.g., AdhA) is silenced in one organ, whereas the other locus is silenced in another organ. This silencing scheme is genotype-independent and occurs in both synthetic and natural cotton allotetraploids (3), suggesting rapid subfunctionalization of duplicate genes and stable maintenance of tissue-specific expression patterns during evolution.
Although the mechanisms for developmental control of the expression of orthologous genes are unclear, developmental regulation of orthologous genes immediately after allopolyploid formation suggests that duplicate genes provide genetic robustness against null mutations (52) and dosage-dependent selective advantage (15, 142). Moreover, immediate divergence in the expression of orthologous genes in allopolyploids provides a virtually inexhaustible reservoir for generating genetic variation and phenotypic diversification, which facilitates natural selection and adaptive evolution.
ODD AND EVEN DOSAGE EFFECTS ON GENE REGULATION IN POLYPLOIDS
Dosage-dependent gene regulation shows odd and even effects, which may affect additive and nonadditive gene regulation in polyploids. Using B-A chromosome translocation lines in maize, Birchler and his colleagues (53) generated a series of lines with different doses of A chromosomes that could be used to measure gene expression in response to changes in chromosome dosage. Gene expression levels are generally positively correlated with the dosage of the genes or chromosomes in these lines. However, the expression levels of ~10% genes are either reduced or negatively correlated with odd chromosome dosages (e.g., one, three, and five). One possibility is that dosage-dependent gene regulation is associated with chromosome pairing because one or more copies of chromosomes in odd dosages cannot pair properly.
The odd and even effects on gene regulation are also observed in the study of transgene expression in diploid and triploid hybrids derived from the crosses of diploid or tetraploid plants with a diploid strain containing a single copy of a transgenic resistance gene in an active state (105). The expression of the transgene is reduced in the triploids compared with the diploid hybrids, leading to the loss of the resistant phenotype at various stages of seedling development in some individuals. The reduction of gene expression was reversible under selective tissue culture conditions. This type of suppression was observed for a single-copy insert in the absence of other trans-acting copies of the transgene and is therefore different from homology-dependent gene silencing. An increase in ploidy or chromosome dosage can give rise to epigenetic gene silencing, generating stochastic variations in gene expression patterns. Although the expression of the transgene in a haploid or a pentaploid was not studied, odd ploidy may result in a new type of epigenetic repression. The expression of the transgene is repressed only in the triploids in which one set of chromosomes is likely not paired or improperly paired.
Ploidy-dependent gene regulation suggests a sensing mechanism for gene dosage and DNA content via chromosome pairing. Although somatic pairing has not been documented in plants, such transient pairing has been observed in humans, Drosophila, and yeast (101). Homologous pairing has been implicated in transvection, position-effect variegation, and transgene gene silencing (5, 61, 113), all of which involve alterations in gene expression.
PARAMUTATION-LIKE EFFECTS IN POLYPLOIDS
Paramutation is the result of heritable changes in gene expression that occur upon interaction between alleles (21). The phenomenon was first discovered in plants and later found in many other organisms including mammals (mouse and human) (21, 121, 134). The paramutagenic allele induces the change in the expression state of the paramutable allele. A paramutation-like phenomenon was also discovered in the tetraploid plants containing active and inactive transgene alleles of hygromycin phosphotransferase (HPT) (104). Active alleles that are trans-inactivated by their silenced counterparts are observed in tetraploid but not in diploid plants, and this occurred only in progeny resulting from self-fertilization of plants heterozygous for the active and inactive HPT allele. The occurrence of transgene paramutation only in tetraploid plants indicates that active and inactive alleles go through meiosis together. This led to the hypothesis of pairing-based trans-inactivation. This predication is consistent with observations in tetraploid tomato, where the frequency of paramutation of a specific paramutagenic allele at the sulfurea locus is different between diploid, triploid, and tetraploid plants and depends on the ratio of paramutagenic to paramutable alleles (57). This suggests a counting mechanism for polyploidy-dependent paramutation, which may be similar to that for X-chromosome inactivation (75).
A paramutation-like phenomenon occurs in the progeny of genetic crosses between heterozygotes and between heterozygotes and wild-type mice independent of gender combination (121). The phenomenon is speculated to be associated with aberrant RNAs resulting from the paramutagenic allele that are packaged in sperm and cause paramutation upon transmission to the next generation. Indeed, paramutation depends on a RNA-dependent RNA polymerase; the rdr101 mutation prevents paramutation in maize (4). However, paramutation in Arabidopsis tetraploids is probably not associated with RNA because trans-activation does not occur in the F1 generation (104). Moreover, crosses of decrease in the DNA methylation (ddm1) mutant with a paramutable tetraploid do not change paramutation phenotypes in the F1 or F2 but do in the F3 family, which is consistent with the gradual loss of DNA methylation by ddm1. The data suggest that methylation occurs later, and is speculated to occur during physical contact of the epialleles during meiosis and after the silencing is established (104). Alternatively, sorting out pairing between homologous and homoeologous chromosomes in polyploids may require a few more rounds of meiosis.
Many paramutation phenomena are associated with repeated sequences (21, 134). Multicopy genes or repetitive intergenic regions are a major trigger for the formation of silenced chromatin. Repeated sequences, whether inverted or tandem, can give rise to the production of dsRNA, an important trigger for RNA silencing as well as heterochromatin formation (85). In addition, repetitive sequences are also able to associate physically with their homologs in nonmeiotic cells (134). It is conceivable that different repeat sequences originating in the progenitors may trigger abnormal siRNA production and heterochromation formation that are responsible for paramutation-like or other epigenetic phenomena in allopolyploids.
FERTILITY, SELF-INCOMPATIBILITY, AND CYTOPLASMIC-NUCLEAR INTERACTIONS IN ALLOPOLYPLOIDS
Incompatibility between alien cytoplasmic and nuclear genomes and between alien nuclear genomes is believed to be a barrier leading to reproductive isolation, speciation, and developmental abnormalities in vertebrates and plants (18, 77, 78, 82, 112). Breaking down this barrier is essential in forming a new polyploid species (78). Seed fertility may be controlled by a few genes or many genetic loci. Three imprinted genes, PHERES1, MEIDOS, and MEDEA, are silenced in allotetraploids in a dosage-dependent manner (68). Disrupting maternal imprinting of AtPHERES1 and paternal imprinting of MEDEA may reduce seed viability in the allopolyploids. Imbalance of paternally and maternally imprinted genes in the endosperm may also cause reproductive failures (20).
Another factor affecting seed fertility is the breeding system. Self-incompatibility is a mechanism for preventing inbreeding in many plant species (22). The sporophytic self-incompatibility system in the family Brassicaceae has been used as a model system to study mating system evolution in plants (99, 108). A. arenosa is an outbreeder (self-incompatible), and A. thaliana is an inbreeding plant (self-compatible). They diverged from the same ancestor ~6 Mya (67, 108). However, the natural allotetraploid A. suecica and the resynthesized allopolyploids are self-compatible (Figure 6), suggesting that the mating system switches immediately following polyploidization. The loss of self-incompatibility in the first generation of allotetraploids is not caused by the segregation of S-alleles in the allotetraploids because all possible alleles are present. The data suggest rapid epigenetic changes in the expression of the genes important to self-incompatibility, probably including the well-characterized loci encoding S-locus receptor kinase (SRK) and S-locus cysteine-rich (SCR) proteins in Brassicaceae (109, 127).
Figure 6.

Switching of self-incompatibility in resynthesized Arabidopsis allotetraploids. (a) Flower morphology of self-compatible A. suecica. (b) Self-incompatible A. arenosa and (c and d) two self-compatible synthetic allotetraploid lines. The altered flower organs (elongated stigma and relatively short stamens) in the synthetic allotetraploids (c and d) may contribute to a low fertility without manual pollination. Size bars (5 mm) are the same in (a) and (b) or in (c) and (d). (e) Diagram of cytoplasmic and nuclear interactions in resynthesized allotetraploids (Allos, 2n = 4x = 26). T or A and tt or aa denote cytoplasm and nuclear genomes, respectively. The cytoplasmic-nuclear genotypes for autotetraploid A. thaliana (At4) are Ttttt, tetraploid A. arenosa (Aa), Aaaaa; and allotetraploids, and either Taatt or Aaatt in reciprocal crosses. Blue and gray indicate A. thaliana and A. arenosa cytoplasms, whereas green, red, and yellow indicate nuclear genomes of A. thaliana, A. arenosa, and allotetraploids, respectively. A. thaliana is self-compatible (SC), whereas A. arenosa is self-incompatible (SI). The resynthesized allotetraploids (Allos) are SC. Fertile seeds are readily obtained from the cross using A. thaliana as a maternal parent and A. arenosa as a pollen donor, and the reciprocal cross is usually unsuccessful (31).
In many cases, polyploidization converts self-incompatible diploids into self-compatible tetraploids in Nicotiana and Solanum with a gametophytic system, and some allopolyploids become self-compatible regardless of the mating types of their parents (18, 49, 82). Selfing in the allopolyploids may have an advantage for adapting new allopolyploids because of increased levels of heterozygosity. Inbreeding depression (1/18 or ~5% of homozygosity in one selfing generation) in allopolyploids is relatively low compared with that in a diploid (50%). An extreme form of reproductive modification is apomixis that is commonly associated with polyploidy (18, 49). Resynthesized allopolyploids may be released from reproductive failure if they are capable of vegetative or seed apomixis.
ADAPTIVE EVOLUTION AND EXPRESSION DIVERGENCE BETWEEN DUPLICATE GENES
Theoretical prediction suggests that one copy of a gene duplicate would become lost by accumulation of deleterious mutations over an evolutionary timescale (87). Evidently, many duplicate genes are retained during evolution, and the redundancy conferred by duplicate genes may facilitate species adaptation (107) and genetic robustness (52) against changes in environmental conditions and developmental programs. Gene expression analyses indicate that duplicate genes offer genetic robustness against null mutations in yeast (52) and tend to experience expression divergence during development and to evolve faster between Drosophila species and within yeast species than single-copy genes (51, 83). Using all duplicate gene pairs derived from a recent WGD (14, 17, 144) and gene expression microarrays in A. thaliana (126), Ha et al. (56) found that expression divergence between gene duplicates is significantly higher in response to external stresses than to internal developmental changes. Rapid divergence between gene duplicates in response to abiotic and biotic stresses may facilitate subfunctionalization (88), neofunctionalization (89), and the evolution of an adaptive mechanism to environmental changes (98, 129). A relatively slow rate of expression divergence between the duplicates may provide dosage-dependent selective advantage (15, 142) and enable organisms to fine-tune complex regulatory networks. Orthologous and homoeologous genes in allopolyploids may have similar evolutionary fates.
The interactions between cytoplasm-nuclear and nuclear-nuclear genomes in the allopolyploids may induce genomic shock (98) and general amphiplasty (110) that are manifested by differential accumulation of transcripts originating from divergent species, leading to transcriptome dominance and activation or silencing of one or both homoeologous loci through genetic and/or epigenetic mechanisms. As a consequence, allopolyploids display hybrid vigor, flowering-time variation, inbreeding, apomixis, and selective advantage. Over time, orthologous or homoeologous loci in the allopolyploids may diverge their functions via neofunctionalization and subfunctionalization (88, 89), as predicted for the paralogous loci (see Sidebar: Adaptive Evolution and Expression Divergence Between Duplicate Genes).
In polyploid populations of separate origin, one population may lose function from one copy of an orthologous gene, while a second population may lose function from a second copy of this ortholog. This “reciprocal silencing” of duplicated genes in polyploid genomes would ultimately lead to hybrid lethality (8), promoting reproductive isolation and the origin of new species. Following this model, the stochastic silencing and subfunctionalization of orthologous genes in different lineages of allopolyploids (148) may play a major role in the origin of new species. Together, these mutually inclusive mechanisms may contribute significantly to the adaptation potential, domestication, and evolution of polyploid plants.
SUMMARY POINTS
Allopolyploidy induces a wider range of gene expression changes than autopolyploidy, suggesting hybridization has greater effects on gene expression and phenotypic variation than genome doubling.
Genetic changes occur immediately after interspecific hybridization or allopolyploidization. These include genome-specific sequence elimination, as observed in Triticum and Tragopogon allopolyploids and chromosomal rearrangements in Brassica allotetraploids. Arabidopsis and cotton (Gossypium) allotetraploids show very low levels of genomic changes.
Approximately 15–43% of the transcriptome diverged in expression between A. thaliana and A. arenosa (split ~6 Mya). The genes that display species-specific expression patterns are preferentially subject to nonadditive regulation in the resynthesized allotetraploids, indicating modulation of transcriptome divergence between the two progenitors.
The genes that are highly expressed in A. thaliana are repressed in the resynthesized allotetraploids, which is correlated with the nucleolar dominance and overall suppression of A. thaliana phenotypes in resynthesized and natural allotetraploids.
Some genes that display nonadditive expression are developmentally regulated, which may lead to subfunctionalization of these homoeologous genes in the allopolyploids.
Establishment of gene expression patterns in the resynthesized allotetraploids is usually rapid and stochastic and in some cases may require several rounds of meiosis (by selfing).
Most nonadditively expressed genes in the resynthesized allotetraploids are present in both parents, suggesting an epigenetic cause and cis- and trans-acting effects on nonadditive gene expression.
Sequence divergence in regulatory elements between the progenitors may cause genetic and epigenetic effects on expression of the genes involved in a biological pathway. Trans-acting differences exert their effects on cis-regulatory elements via epigenetic mechanisms, indicating interrelated regulatory interactions between genetic and epigenetic effects.
Reprogramming gene expression patterns in allopolyploids via chromatin modifications and RNA-mediated pathways may cause changes in adaptive traits such as flowering-time variation, self-incompatibility, and hybrid vigor.
Although the majority of data are documented from studies of resynthesized allotetraploids, similar changes in genomic organization and gene expression may occur in the natural allopolyploids and facilitate polyploid evolution and speciation.
FUTURE ISSUES
Addressing the following questions is essential to illuminate new insights into molecular and evolutionary impact on polyploid formation and speciation.
How and why are genetic dominance and nonadditive expression established rapidly in resynthesized allopolyploids, and how are these molecular processes altered or maintained in natural allopolyploids?
How do changes in gene expression contribute to intricate regulatory hierarchies and complex biological pathways leading to hybrid vigor and adaptive traits in allopolyploids?
Why are ploidy changes more deleterious in animals than in plants, and why are polyploid cells often carcinogenetic in animals?
Acknowledgments
I thank Jianlin Wang, Hyeon-See Lee, Lu Tian, Zhongfu Ni, Misook Ha, Letricia Nogueira, Eun-Deok Kim, Jinsuk Lee, and Suk-Hwan Yang for their contributions to the gene expression data, and Donald Levin, Andrew Woodward, and two anonymous reviewers for critical reading and suggestions to improve the manuscript. I apologize for not citing many enlightening reviews and papers published in this exciting field owing to space limitations. The work was supported by the grants from the National Science Foundation (MCB0608602, DBI0501712 and DBI0624077) and the National Institutes of Health (GM067015).
- Polyploid
an individual or cell that has more than two sets of chromosomes
- Allopolyploid
a polyploid that contains two or more sets of genetically distinct chromosomes, usually by hybridization between different species
- Autopolyploid
a polyploid created by the multiplication of one basic set of chromosomes
- Ploidy
the number of basic chromosome sets
- Homologous
genes or structures that share a common evolutionary ancestor
- Orthologous
chromosomes or genes in different species that have evolved from the same ancestor
- Homoeologous
chromosomes or genes in related species that are derived from the same ancestor and coexist in an allopolyploid
- Segmental allopolyploid
An allopolyploid that contains some homologous segments between homoeologous chromosomes, in which meiotic pairing occurs between homologous chromosomes as well as homoeologous chromosomes
- Paleopolyploid
an ancient polyploid that undergoes chromosomal rearrangements, gene loss, and mutations and eventually becomes diploidized such that it behaves cytogenetically as a diploid
- Nonadditive gene expression
levels of gene expression in an auto- or allopolyploid are not equal to the sum of progenitors, suggesting gene activation including dominance, overdominance, or gene repression
- Apomixis
only one parent (usually female) contributes genes to the offspring
- Aneuploid
an individual in which the chromosome number is not an exact multiple of the typical haploid set for that species
- Paralogous
two or more genes in the same species that share a single ancestral origin
- Epigenetic
heritable changes in gene expression without changes in primary DNA sequences
- Genomic shock
release of genome-wide chromatin constraints of gene expression including activation of transposons in response to genomic, environmental, and physiological changes
- X-chromosome inactivation
during mammalian development, the repression of one of the two X chromosomes in the somatic cells of females as a method of dosage compensation
- Gametic imprinting
the expression of a gene depends on its parental origin in the offspring
- Paramutation
heritable changes in gene expression due to allelic interactions
- Homology-dependent gene silencing
repression of gene expression by homologous DNA sequences such as transgenes or endogenous loci
- Amphiplasty
alterations of chromosomal morphology in interspecific hybrids or amphidiploids
- Neofunctionalization
gain of a novel function (or expression pattern) from one copy of the gene duplicate
- Subfuncationalization
functional (or expression) divergence of gene duplicate from the ancestral gene (e.g., tissue-specific expression of gene duplicate)
LITERATURE CITED
- 1.Abbott RJ, Lowe AJ. Origins, establishment and evolution of new polyploid species: Senecio cambrensis and S. eboracensis in the British Isles. Biol J Linn Soc. 2004;82:467–74. [Google Scholar]
- 2.Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA. 2003;100:4649–54. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]; 2. Examines tissue-specific expression of 40 protein-coding genes in allotetraploid cotton, suggesting subfunctionaliza-tion for 10 homoeologous genes.
- 3.Adams KL, Percifield R, Wendel JF. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics. 2004;168:2217–26. doi: 10.1534/genetics.104.033522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–98. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 5.Aramayo R, Metzenberg RL. Meiotic transvection in fungi. Cell. 1996;86:103–13. doi: 10.1016/s0092-8674(00)80081-1. [DOI] [PubMed] [Google Scholar]
- 6.Auger DL, Gray AD, Ream TS, Kato A, Coe EH, Jr, Birchler JA. Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics. 2005;169:389–97. doi: 10.1534/genetics.104.032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 8.Barbash DA, Siino DF, Tarone AM, Roote J. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci USA. 2003;100:5302–7. doi: 10.1073/pnas.0836927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel B, Bartel DP. MicroRNAs: At the root of plant development? Plant Physiol. 2003;132:709–17. doi: 10.1104/pp.103.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumel A, Ainouche ML, Bayer RJ, Ainouche AK, Misset MT. Molecular phylogeny of hybridizing species from the genus Spartina Schreb. (Poaceae) . Mol Phylogenet Evol. 2002;22:303–14. doi: 10.1006/mpev.2001.1064. [DOI] [PubMed] [Google Scholar]
- 11.Bender J, Fink GR. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83:725–34. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- 12.Birchler JA. Dosage-dependent gene regulation in multicellular eukaryotes: Implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol. 2001;234:275–88. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- 13.Birchler JA, Auger DL, Riddle NC. In search of the molecular basis of heterosis. Plant Cell. 2003;15:2236–39. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–44. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bomblies K, Doebley JF. Pleiotropic effects of the duplicate maize FLORICAULA/ LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics. 2006;172:519–31. doi: 10.1534/genetics.105.048595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–91. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–38. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 18.Briggs D, Walters SM. Plant Variation and Evolution. Cambridge, UK: Cambridge Univ Press; 1997. p. 512. [Google Scholar]
- 19.Bruce AB. The Mendelian theory of heredity and the augmentation of vigor . Science. 1910;32:627–28. doi: 10.1126/science.32.827.627-a. [DOI] [PubMed] [Google Scholar]
- 20.Bushell C, Spielman M, Scott RJ. The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell. 2003;15:1430–42. doi: 10.1105/tpc.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandler VL, Stam M. Chromatin conversations: mechanisms and implications of paramutation. Nat Rev Genet. 2004;5:532–44. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- 22.Charlesworth D, Wright SI. Breeding systems and genome evolution. Curr Opin Genet Dev. 2001;11:685–90. doi: 10.1016/s0959-437x(00)00254-9. [DOI] [PubMed] [Google Scholar]
- 23.Deleted in proof
- 24.Chen ZJ, Comai L, Pikaard CS. Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc Natl Acad Sci USA. 1998;95:14891–96. doi: 10.1073/pnas.95.25.14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen ZJ, Ni Z. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays. 2006;28:240–52. doi: 10.1002/bies.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997;11:2124–36. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]; Discovery of molecular evidence for epigenetic regulation of rRNA genes subjected to nucleolar dominance (silencing of rRNA genes originating from one progenitor) in Brassica allopolyploids, which has provided mechanistic insights into understanding the expression of orthologous genes in allopolyploids
- 27.Chen ZJ, Pikaard CS. Transcriptional analysis of nucleolar dominance in poly-ploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc Natl Acad Sci USA. 1997;94:3442–47. doi: 10.1073/pnas.94.7.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen ZJ, Wang JL, Tian L, Lee HS, Wang JYJ, et al. The development of an Arabidopsis model system for genome-wide analysis of polyploidy effects. Biol J Linn Soc. 2004;82:689–700. doi: 10.1111/j.1095-8312.2004.00351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–46. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 30.Comai L, Tyagi AP, Lysak MA. FISH analysis of meiosis in Arabidopsis allopolyploids. Chromosome Res. 2003;11:217–26. doi: 10.1023/a:1022883709060. [DOI] [PubMed] [Google Scholar]
- 31.Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, et al. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell. 2000;12:1551–68. doi: 10.1105/tpc.12.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first extensive survey of phenotypic and gene expression changes in resynthesized Arabidopsis allotetraploids
- 32.Crow JF. Alternative hypothesis of hybrid vigor. Genetics. 1948;33:477–87. doi: 10.1093/genetics/33.5.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Meaux J, Pop A, Mitchell-Olds T. Cis-regulatory evolution of Chalcone-synthase expression in the genus Arabidopsis. Genetics. 2006;174:2182–202. doi: 10.1534/genetics.106.064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denis CL, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol. 2003;73:221–50. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 35.Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, et al. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006;20:1283–93. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.East EM. Heterosis. Genetics. 1936;21:375–97. doi: 10.1093/genetics/21.4.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 38.Emanuel BS, Shaikh TH. Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nat Rev Genet. 2002;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- 39.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 40.Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM. Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics. 1997;147:1381–87. doi: 10.1093/genetics/147.3.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferris SD, Whitt GS. Loss of duplicate gene expression after polyploidisation. Nature. 1976;265:258–60. doi: 10.1038/265258a0. [DOI] [PubMed] [Google Scholar]
- 42.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 43.Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251–54. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- 44.Gallardo MH, Bickham JW, Honeycutt RL, Ojeda RA, Kohler N. Discovery of tetraploidy in a mammal. Nature. 1999;401:341. doi: 10.1038/43815. [DOI] [PubMed] [Google Scholar]
- 45.Gaut BS. Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res. 2001;11:55–66. doi: 10.1101/gr.160601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson G, Riley-Berger R, Harshman L, Kopp A, Vacha S, et al. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics. 2004;167:1791–99. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottlieb LD. Conservation and duplication of isozymes in plants. Science. 1982;216:373–80. doi: 10.1126/science.216.4544.373. [DOI] [PubMed] [Google Scholar]
- 48.Gottlieb LD, Ford VS. A recently silenced, duplicate PgiC locus in Clarkia. Mol Biol Evol. 1997;14:125–32. doi: 10.1093/oxfordjournals.molbev.a025745. [DOI] [PubMed] [Google Scholar]
- 49.Grant V. Plant Speciation. New York: Columbia Univ Press; 1981. p. 563. [Google Scholar]
- 50.Grant-Downton RT, Dickinson HG. Epigenetics and its implications for plant biology 2. The ‘epigenetic epiphany’: epigenetics, evolution and beyond. Ann Bot. 2006;97:11–27. doi: 10.1093/aob/mcj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu Z, Rifkin SA, White KP, Li WH. Duplicate genes increase gene expression diversity within and between species. Nat Genet. 2004;36:577–79. doi: 10.1038/ng1355. [DOI] [PubMed] [Google Scholar]
- 52.Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li W-H. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 53.Guo M, Davis D, Birchler JA. Dosage effects on gene expression in a maize ploidy series. Genetics. 1996;142:1349–55. doi: 10.1093/genetics/142.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]; An extensive survey of gene expression variation in response to dosage changes in maize
- 54.Guo M, Rupe MA, Zinselmeier C, Habben J, Bowen BA, Smith OS. Allelic variation of gene expression in maize hybrids. Plant Cell. 2004;16:1707–16. doi: 10.1105/tpc.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutierrez RA, Ewing RM, Cherry JM, Green PJ. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA. 2002;99:11513–18. doi: 10.1073/pnas.152204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ha M, Li WH, Chen ZJ. External factors accelerate expression divergence between duplicate genes. Trends Genet. 2006 doi: 10.1016/j.tig.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hagemann R, Berg W. Paramutation at the sulfurea locus of Lycopersicon esculentum Mill. VII. Determination of the time of occurence of paramutation by the quantitative evaluation of the variegation. Theor Appl Genet. 1978;53:709–19. doi: 10.1007/BF00272688. [DOI] [PubMed] [Google Scholar]
- 58.Harlan JR, De Wet JM, Naik SM, Lambert RJ. Chromosome pairing within genomes in maize-Tripsacum hybrids. Science. 1970;167:1247–48. doi: 10.1126/science.167.3922.1247. [DOI] [PubMed] [Google Scholar]
- 59.He P, Friebe BR, Gill BS, Zhou JM. Allopolyploidy alters gene expression in the highly stable hexaploid wheat. Plant Mol Biol. 2003;52:401–14. doi: 10.1023/a:1023965400532. [DOI] [PubMed] [Google Scholar]
- 60.Hegarty MJ, Jones JM, Wilson ID, Barker GL, Coghill JA, et al. Development of anonymous cDNA microarrays to study changes to the Senecio floral transcriptome during hybrid speciation. Mol Ecol. 2005;14:2493–510. doi: 10.1111/j.1365-294x.2005.02608.x. [DOI] [PubMed] [Google Scholar]
- 61.Henikoff S, Comai L. Trans-sensing effects: the ups and downs of being together. Cell. 1998;93:329–32. doi: 10.1016/s0092-8674(00)81161-7. [DOI] [PubMed] [Google Scholar]
- 62.Heslop-Harrison JS. Gene expression and parental dominance in hybrid plants. Development Suppl. 1990;1990:21–28. [PubMed] [Google Scholar]
- 63.Hilu KW. Polyploidy and the evolution of domesticated plants. Am J Bot. 1993;80:2521–28. [Google Scholar]
- 64.Jakobsson M, Hagenblad J, Tavare S, Sall T, Hallden C, et al. A unique recent origin of the allotetraploid species Arabidopsis suecica: Evidence from nuclear DNA markers. Mol Biol Evol. 2006;23:1217–31. doi: 10.1093/molbev/msk006. [DOI] [PubMed] [Google Scholar]
- 65.Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–47. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- 66.Jones DF. Dominance of linked factors as a means of accounting for heterosis. Genetics. 1917;2:466–79. doi: 10.1093/genetics/2.5.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 1990;8:340–44. doi: 10.1016/0167-7799(90)90220-r. [DOI] [PubMed] [Google Scholar]
- 68.Josefsson C, Dilkes B, Comai L. Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol. 2006;16:1322–28. doi: 10.1016/j.cub.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 69.Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–59. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet. 2003;33:102–6. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]; First report of retrotransposon activation and its effects on the expression of adjacent genes in wheat allopolyploids
- 71.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–98. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 72.Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, et al. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 73.Lee HS, Chen ZJ. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc Natl Acad Sci USA. 2001;98:6753–58. doi: 10.1073/pnas.121064698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee HS, Wang JL, Tian L, Jiang HM, Black MA, et al. Sensitivity of 70-mer oligonucleotides and cDNAs for microarray analysis of gene expression in Arabidopsis and its related species. Plant Biotechnol J. 2004;2:45–57. doi: 10.1046/j.1467-7652.2003.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee JT, Jaenisch R. The (epi)genetic control of mammalian X-chromosome inactivation. Curr Opin Genet Dev. 1997;7:274–80. doi: 10.1016/s0959-437x(97)80138-4. [DOI] [PubMed] [Google Scholar]
- 76.Leitch IL, Bennett MD. Polyploidy in angiosperms. Trends Plant Sci. 1997;2:470–76. [Google Scholar]
- 77.Levin DA. Polyploidy and novelity in flowering plants. Am Nat. 1983;122:1–25. [Google Scholar]
- 78.Levin DA. The cytoplasmic factor in plant speciation. Syst Bot. 2003;28:5–11. [Google Scholar]
- 79.Levy AA, Feldman M. The impact of polyploidy on grass genome evolution. Plant Physiol. 2002;130:1587–93. doi: 10.1104/pp.015727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levy YY, Dean C. The transition to flowering. Plant Cell. 1998;10:1973–90. doi: 10.1105/tpc.10.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lewis MS, Pikaard CS. Restricted chromosomal silencing in nucleolar dominance. Proc Natl Acad Sci USA. 2001;98:14536–40. doi: 10.1073/pnas.251424098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewis WH. Polyploidy: Biological Relevance. New York: Plenum; 1980. p. 583. [Google Scholar]
- 83.Li WH, Yang J, Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21:602–7. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 84.Li ZK, Luo LJ, Mei HW, Wang DL, Shu QY, et al. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics. 2001;158:1737–53. doi: 10.1093/genetics/158.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–70. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 86.Liu B, Wendel J. Non-Mendelian phenomenon in allopolyploid genome evolution. Curr Genomics. 2002;3:489–505. [Google Scholar]
- 87.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–55. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 88.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–73. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lynch M, O’Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mable BK. ‘Why polyploidy is rarer in animals than in plants’: Myths and mechanisms. Biol J Linn Soc. 2004;82:453–66. [Google Scholar]
- 91.Madlung A, Masuelli RW, Watson B, Reynolds SH, Davison J, Comai L. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 2002;129:733–46. doi: 10.1104/pp.003095. [DOI] [PMC free article] [PubMed] [Google Scholar]; Documented changes in DNA methylation and transposons in resynthesized Arabidopsis allotetraploids
- 92.Madlung A, Tyagi AP, Watson B, Jiang H, Kagochi T, et al. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005;41:221–30. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- 93.Masterson J. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science. 1994;264:421–24. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- 94.Matzke MA, Matzke AJM. How and why do plants inactivate homologous (trans)genes. Plant Physiol. 1995;107:679–85. doi: 10.1104/pp.107.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matzke MA, Scheid OM, Matzke AJ. Rapid structural and epigenetic changes in polyploid and aneuploid genomes. BioEssays. 1999;21:761–67. doi: 10.1002/(SICI)1521-1878(199909)21:9<761::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 96.Mavarez J, Salazar CA, Bermingham E, Salcedo C, Jiggins CD, Linares M. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–71. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- 97.McClintock B. The relationship of a particular chromosomal element to the development of the nucleoli in Zea mays. Z Zellforsch Mikrosk Anat. 1934;21:294–328. [Google Scholar]
- 98.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 99.McCubbin AG, Kao T. Molecular recognition and response in pollen and pistill interactions. Annu Rev Cell Dev Biol. 2000;16:333–64. doi: 10.1146/annurev.cellbio.16.1.333. [DOI] [PubMed] [Google Scholar]
- 100.McFadden DE, Kwong LC, Yam IY, Langlois S. Parental origin of triploidy in human fetuses: evidence for genomic imprinting. Hum Genet. 1993;92:465–69. doi: 10.1007/BF00216452. [DOI] [PubMed] [Google Scholar]
- 101.McKee BD. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta. 2004;1677:165–80. doi: 10.1016/j.bbaexp.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 102.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–56. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Michalak P, Noor MA. Genome-wide patterns of expression in Drosophila pure species and hybrid males. Mol Biol Evol. 2003;20:1070–76. doi: 10.1093/molbev/msg119. [DOI] [PubMed] [Google Scholar]
- 104.Mittelsten Scheid O, Afsar K, Paszkowski J. Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat Genet. 2003;34:450–54. doi: 10.1038/ng1210. [DOI] [PubMed] [Google Scholar]; Paramutation-like phenomenon was observed in Arabidopsis. tetraploids
- 105.Mittelsten Scheid O, Jakovleva L, Afsar L, Maluszynska J, Paszkowski J. A change of ploidy can modify epigenetic silencing. Proc Natl Acad Sci USA. 1996;93:7114–19. doi: 10.1073/pnas.93.14.7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Molnar-Lang M, Linc G, Logojan A, Sutka J. Production and meiotic pairing behaviour of new hybrids of winter wheat (Triticum aestivum) x winter barley (Hordeum vulgare) Genome. 2000;43:1045–54. [PubMed] [Google Scholar]
- 107.Muller HJ. Why polyploidy is rarer in animals than in plants. Am Nat. 1925;59:346–53. [Google Scholar]
- 108.Nasrallah JB. Cell-cell signalling in the self-incompatibility response. Curr Opin Plant Biol. 2000;3:368–73. doi: 10.1016/s1369-5266(00)00098-4. [DOI] [PubMed] [Google Scholar]
- 109.Nasrallah ME, Liu P, Nasrallah JB. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science. 2002;297:247–49. doi: 10.1126/science.1072205. [DOI] [PubMed] [Google Scholar]
- 110.Navashin M. Chromosomal alterations caused by hybridization and their bearing upon certain general genetic problems. Cytologia. 1934;6:169–203. [Google Scholar]; Discovery of progenitor-dependent chromosome morphological changes in Crepis interspecific hybrids
- 111.Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003;19:141–47. doi: 10.1016/s0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- 112.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–37. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 113.Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–27. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 114.Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–8. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- 115.Phillips JP, Tainer JA, Getzoff ED, Boulianne GL, Kirby K, Hilliker AJ. Subunit-destabilizing mutations in Drosophila copper/zinc superoxide dismutase: neuropathology and a model of dimer dysequilibrium. Proc Natl Acad Sci USA. 1995;92:8574–78. doi: 10.1073/pnas.92.19.8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pichersky E, Soltis D, Soltis P. Defective chlorophyll a/b-binding protein genes in the genome of a homosporous fern. Proc Natl Acad Sci USA. 1990;87:195–99. doi: 10.1073/pnas.87.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pikaard CS. Nucleolar dominance and silencing of transcription. Trends Plant Sci. 1999;4:478–83. doi: 10.1016/s1360-1385(99)01501-0. [DOI] [PubMed] [Google Scholar]
- 118.Prudhomme M, Mejean V, Martin B, Claverys JP. Mismatch repair genes of Streptococcus pneumoniae: HexA confers a mutator phenotype in Escherichia coli by negative complementation. J Bacteriol. 1991;173:7196–203. doi: 10.1128/jb.173.22.7196-7203.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst. 1998;29:467–501. [Google Scholar]
- 120.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–45. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 121.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–74. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 122.Reeder RH. Mechanisms of nucleolar dominance in animals and plants. J Cell Biol. 1985;101:2013–16. doi: 10.1083/jcb.101.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Riera-Lizarazu O, Rines HW, Phillips RL. Cytological and molecular characterization of oat x maize partial hybrids. Theor Appl Genet. 1996;93:123–35. doi: 10.1007/BF00225737. [DOI] [PubMed] [Google Scholar]
- 124.Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–16. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- 125.Sall T, Jakobsson M, Lind-Hallden C, Hallden C. Chloroplast DNA indicates a single origin of the allotetraploid Arabidopsis suecica. J Evol Biol. 2003;16:1019–29. doi: 10.1046/j.1420-9101.2003.00554.x. [DOI] [PubMed] [Google Scholar]
- 126.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–6. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 127.Schopfer CR, Nasrallah ME, Nasrallah JB. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- 128.Selker EU. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- 129.Seoighe C, Gehring C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004;20:461–64. doi: 10.1016/j.tig.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 130.Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell. 2001;13:1749–59. doi: 10.1105/TPC.010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, et al. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–58. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since plant speciation. New Phytol. 2003;161:173–91. [Google Scholar]
- 133.Song K, Lu P, Tang K, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA. 1995;92:7719–23. doi: 10.1073/pnas.92.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]; Discovery of rapid DNA sequence changes in synthetic Brassica allotetraploids, which has renewed the interests in molecular studies on plant polyploids
- 134.Stam M, Mittelsten Scheid O. Paramutation: an encounter leaving a lasting impression. Trends Plant Sci. 2005;10:283–90. doi: 10.1016/j.tplants.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 135.Stebbins GL. Chromosomal Evolution in Higher Plants. London: Edward Arnold; 1971. p. 216. [Google Scholar]
- 136.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 137.Stuber CW, Lincoln SE, Wolff DW, Helentjaris T, Lander ES. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics. 1992;132:823–39. doi: 10.1093/genetics/132.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stupar RM, Springer NM. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics. 2006;173:2199–210. doi: 10.1534/genetics.106.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Svartman M, Stone G, Stanyon R. Molecular cytogenetics discards polyploidy in mammals. Genomics. 2005;85:425–30. doi: 10.1016/j.ygeno.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 140.Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA. 2006;103:6805–10. doi: 10.1073/pnas.0510430103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Microarray analysis of gene expression changes in maize hybrids suggests nonadditive gene regulation, as observed in Arabidopsis allopolyploids
- 141.Tate JA, Ni Z, Scheen AC, Koh J, Gilbert CA, et al. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics. 2006;173:1599–611. doi: 10.1534/genetics.106.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]; An extensive survey of gene expression changes in recent and reciprocally formed Tragopogon allotetraploids in natural populations
- 142.Thomas BC, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–46. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tilghman SM. The sins of the fathers and mothers: Genomic imprinting in mammalian development. Cell. 1999;96:185–93. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 144.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–17. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 145.Vrana PB, Fossella JA, Matteson P, del Rio T, O’Neill MJ, Tilghman SM. Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nat Genet. 2000;25:120–24. doi: 10.1038/75518. [DOI] [PubMed] [Google Scholar]
- 146.Wang J, Tian L, Lee HS, Chen ZJ. Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics. 2006;173:965–74. doi: 10.1534/genetics.106.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence for cis- and trans-acting effects on gene expression of regulatory pathways in allopolyploids
- 147.Wang J, Tian L, Lee HS, Wei NE, Jiang H, et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–17. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first genome-wide analysis of gene expression reveals transcriptome dominance and nonadditive gene regulation in Arabidopsis allotetraploids
- 148.Wang J, Tian L, Madlung A, Lee HS, Chen M, et al. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–73. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–49. [PubMed] [Google Scholar]
- 150.Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–86. [Google Scholar]
- 151.Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- 152.Wolfe KH. Yesterday’s polyploidization and the mystery of diploidization. Nat Rev Genet. 2001;2:333–41. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 153.Yang SS, Cheung F, Lee JJ, Ha M, Wei NE, et al. Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 2006;47:761–75. doi: 10.1111/j.1365-313X.2006.02829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; EST analysis reveals transcription dominance of AA-subgenome in allotetraploid cotton (AADD)
- 154.Yu J, Wang J, Lin W, Li S, Li H, et al. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]


