Abstract
Mycobacterium tuberculosis and Mycobacterium avium are pathogenic slow-growing mycobacteria that cause distinct human diseases. In contrast to recent advances in M. tuberculosis genetics and pathogenesis investigation, M. avium has remained genetically intractable and, consequently, its pathogenic strategies remain poorly understood. Here we report the successful development of efficient allelic exchange and transposon mutagenesis in an opaque clinical strain of M. avium by specialized transduction. Efforts to disrupt the leuD gene of M. avium by specialized transduction were successful but were complicated by inefficient isolation of recombinants secondary to high spontaneous antibiotic resistance. However, by using this leucine auxotroph as a genetic host and the Streptomyces coelicolor leuD gene as a selectable marker, we achieved efficient allelic exchange at the M. avium pcaA locus. A leuD-marked transposon delivered by specialized transduction mutagenized M. avium with efficiencies similar to M. tuberculosis. These results establish a system for random and directed mutagenesis of M. avium. In combination with the forthcoming M. avium genome sequence, these tools will allow the distinct physiologic and pathogenic properties of M. avium to be dissected in molecular detail.
Mycobacterium avium subsp. avium is a slow growing pathogenic Mycobacterium that causes a variety of clinical infections in both humans and animals. M. avium is often grouped with its close mycobacterial relative Mycobacterium intracellulare and designated M. avium complex (MAC). These organisms cause disseminated infections in advanced AIDS, pulmonary disease in immunocompetent adults, and localized lymphadenitis in children. Serious M. avium infections are often difficult to treat because many first line antituberculosis agents are not active against MAC. In addition, as a pathogenic slow-growing Mycobacterium sp. that can be handled in BSL2 conditions, M. avium can serve as an initial model for certain aspects of mycobacterial pathogenesis (28) that can then be confirmed in Mycobacterium tuberculosis.
In recent years, advances in the genetic manipulation of pathogenic mycobacteria have resulted in an explosion of knowledge about the molecular basis for mycobacterial pathogenicity. In M. tuberculosis, allelic exchange, transposon mutagenesis, and specialized transduction have been achieved (5, 6, 8, 21, 23, 24) and revealed important pathogenesis determinants. However, M. avium has remained somewhat intractable to genetic manipulation. Allelic exchange has been successful in M. intracellulare by using a two-step suicide vector strategy, but these efforts either required an avirulent laboratory strain of M. intracellulare or a chemically mutagenized clinical strain or required extensive laborious screening to isolate rare recombinants (16-18). In one of these studies, deletion of katG in M. intracellulare required screening 448 clones to isolate one mutant strain. Transposon mutagenesis has been achieved in M. avium subsp. paratuberculosis by using phage delivery of an antibiotic-marked transposon (12) but not in M. avium subsp. avium. Thus, new genetic tools for the creation of random and directed mutants of M. avium would expand our understanding of M. avium pathogenicity and the molecular basis for its environmental persistence.
We attempted to use specialized transduction to mediate allelic exchange and transposon mutagenesis in a clinical strain of M. avium, as has been done recently for M. tuberculosis (3). Our initial attempts to disrupt the leuD gene were successful but replicated the difficulties of prior published attempts, with inefficient recombination and a high level of spontaneous antibiotic resistance. However, when this M. avium leucine auxotroph was used as a host for genetic manipulation with leuD as a selectable marker, allelic exchange and transposon mutagenesis were achieved with efficiencies similar to those achieved with M. tuberculosis. These studies establish an efficient random and directed mutagenesis system for M. avium and make genetic studies with this organism feasible with the relative ease now possible for M. tuberculosis. Genetic studies in M. avium will reveal the aspects of mycobacterial pathogenesis unique to M. avium in the immunocompromised host and may allow M. avium to serve as a BSL2 model organism for certain aspects of M. tuberculosis physiology and pathogenesis.
MATERIALS AND METHODS
Bacterial strains, phages, and culture conditions.
All chemicals were purchased from Sigma unless otherwise specified. Major plasmids used in the present study are listed in Table 1. The bacterial strains and phages used in the present study are summarized in Table 2. The M. avium strain mc22500 is a clinical isolate from a patient with AIDS and was obtained from the Montefiore Medical Center clinical microbiology laboratory. This strain displays a smooth transparent (SmT) colony morphology. M. avium strain mc22501 is an opaque variant (SmD) of strain mc22500 that arose spontaneously after in vitro passage. M. avium was grown in Middlebrook 7H9 or 7H10 media (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC; Becton Dickinson), 0.5% glycerol, and 0.05% Tween 80 (liquid). When appropriate, hygromycin (Roche) was used at 50 μg/ml and leucine was used at 50 μg/ml.
TABLE 1.
Major plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pMSG110 | M. avium leuC; leuD::hyg; hupC; leuD knockout construct | This work |
| pMSG206 | S. coelicolor leuD fused to constitutive mycobacterial hsp60 promoter | This study |
| pMSG207 | leuD complementation cassette from pMSG206 cloned into pmv306kan; single-copy vector | This study |
| pMSG208 | leuD transposon vector derived from Tn5370 | This study |
| pMSG210 | S. coelicolor leuD expression cassette from pMSG206 with flanking multiple cloning site (MCS), lambda COS, and PacI sites for phage packaging | This study |
| pMSG212 | M. avium pcaA::hsp60-leuD knockout construct | This study |
TABLE 2.
Bacterial strains and phages used in this study
| Strain or phage | Description | Source or reference |
|---|---|---|
| Strain | ||
| mc22500 | M. avium clinical isolate; SmT colony type | Montefiore Medical Center |
| mc22501 | mc22500 SmD colony type after in vitro passage | This study |
| mgm93 | mc22501 (leuD::hyg) | This study |
| mgm100 | mgm93 (pcaA::hsp60-Streptomyces leuD) | This study |
| JCM34 | mc2155 (leuD::Tn5370); leucine auxotroph | Jeffery Cox |
| Phage | ||
| phAE85 | Temperature-sensitive derivative of TM4 expressing firefly luciferase | 7 |
| phAE87 | Temperature-sensitive derivative of TM4 | 7 |
| phMSG110 | pMSG110 packaged into phAE87.leuD knockout specialized transducing phage | This study |
| phMSG208 | Streptomyces leuD transposon delivery phage | This study |
| phMSG212 | phAE87 (pMSG212); M. avium pcaA knockout phage | This study |
Luciferase phage infections.
Luciferase phage infections of M. avium and M. bovis BCG (BCG) were performed on log-phase cultures at an optical density at 600 nm of 0.6 to 0.8, essentially as described previously (27). The luciferase reporter mycobacteriophage phAE85 is a temperature-sensitive derivative of TM4 expressing firefly luciferase and has been previously described (7).
Disruption of leuD in M. avium.
Preliminary sequence data from M. avium was obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org). The M. avium leuC-leuD region was identified by basic local alignment search tool (BLAST) searching of the unfinished M. avium genome sequence with the M. tuberculosis leuC-leuD operon as the query nucleotide sequence. To construct an M. avium leuD-null allele, the flanking regions of leuD were amplified by PCR and inserted on either side of a hygromycin resistance gene. The 5′ flanking region was amplified from M. avium genomic DNA with the primer pair omsg47 and omsg48 (5′-CTTAAGCACCGAGGTCTATCTG-3′ and 5′-TCTAGACTCATCCCTTCACGG-3′), which amplify a 616-bp product 5′ of leuD that includes the first six nucleotides of the predicted leuD open reading frame and introduce XbaI and AflII sites at the 3′ and 5′ ends of the PCR product. The 3′ flanking region was amplified with omsg49 and omsg50 (5′-AAGCTTGACGACCACACCGC-3′ and 5′-CCTCGAGACAACCGCTTTGAA-3′), which amplify a 622-bp fragment that includes the last 138 nucleotides of leuD and introduce the HindIII and XhoI sites at the 5′ and 3′ ends of the PCR product. Both of these PCR products were cloned, sequenced, and subcloned on either side of a hygromycin resistance gene to produce pMSG110. pMSG110 was packaged into the mycobacteriophage phAE87 exactly as previously described (10, 11). Strains mc22500 and mc22501 were infected at a multiplicity of infection of 10 at 39°C for 4 h, washed once with medium, and cultured on 7H9-OADC medium supplemented with 50 μg of leucine and 50 μg of hygromycin B/ml. Genomic DNA from hygromycin-resistant, leucine auxotrophic transductants was digested with SmaI and NotI and analyzed by Southern hybridization with the omsg49-omsg50 PCR product as a probe. The leuD mutant of M. avium 2501 (mc22501 leuD::hyg) was designated mgm93 and used in further studies.
Heterologous complementation of mycobacterial leuD mutant with Streptomyces coelicolor leuD.
To construct a complementation cassette for the M. avium leuD mutation, we identified the predicted leuD operon in the unfinished S. coelicolor genome sequence. At the time this work was performed, the S. coelicolor genomic sequence was available in unannotated form from the Sanger Center BLAST server. This sequence has subsequently been annotated and published (4). The native Streptomyces leuCD operon was amplified from S. coelicolor genomic DNA (a kind gift of Apoorva Bhatt, John Innes Center) in two pieces with the oligonucleotides omsg115 and omsg116 (GATATCCAGAACGCCGACGCCG and GGTCAGACCGAGCTGCGGGC) and omsg117 and omsg118 (GCGGTCCTGGGCCACCTGGC and GATCCGGAAACGGCCGTACCC). The omsg115-116 PCR product contains putative promoter of leuCD and the first 353 nucleotides of leuC. The omsg117-118 PCR product contains the entire leuD open reading frame and the last 72 nucleotides of leuC. These PCR products were cloned and sequenced. The leuCD operon was reconstructed with an in-frame deletion of leuC formed by the fusion of two AatII sites at the 5′ and 3′ ends of leuC, creating a truncated leuC and an intact leuD, both expressed from the native promoter. This ΔleuC-leuD expression construct in pmv306kan, an integrating mycobacterial vector, is pmsg205. For constitutive heterologous expression of Streptomyces leuD, the omsg117-118 PCR product was ligated as a 947-bp EcoRI fragment from pMSG196 into pMV261, placing the last 72 nucleotides of leuC and the entire leuD open reading frame under the constitutive mycobacterial hsp60 promoter.
Deletion of M. avium pcaA by using leu complementation.
An M. avium pcaA-null allele was constructed similarly to the M. avium leuD null allele. The pcaA locus was identified from preliminary sequence data from TIGR as described for leuD. The predicted PcaA protein in M. avium is 82% identical to M. tuberculosis PcaA. In addition, M. avium pcaA is adjacent to a homologue of umaA1, and therefore the genomic organization of these two genes in M. avium replicates the organization found in the M. tuberculosis chromosome. The flanking regions of the M. avium pcaA gene were amplified by PCR with the primer pairs omsg102 and omsg103 (AAGCTTTCCAAAATGCGGCGTG and ACTAGTGATGATGCGGTTCACC) and omsg104 and omsg105 (TCTAGAGAAGAAGTAGCCGTCC and GGTACCCTGGAGAAGTATGACG). These PCR products were cloned, sequenced, and inserted flanking the Streptomyces leuD expression cassette to create pMSG212. This plasmid was packaged into phAE87 to create phMSG212, which was used to infect mgm93 as described for the leuD disruption of mc22501. This transduction was selected on solid medium lacking leucine, and leucine prototrophs that arose after ca. 10 days incubation were screened for allelic exchange at pcaA by Southern blotting.
Construction of leuD transposon for use in mgm93.
To construct a leuD transposon, the previously characterized Tn5370 was modified (8). This transposon has been used successfully for transposon mutagenesis of M. tuberculosis (19) and signature-tagged mutagenesis (8). The hygromycin cassette in Tn5370 was replaced by the constitutive Streptomyces leuD cassette and a kanamycin marker inserted outside of the inverted repeats to facilitate cloning. This transposon, contained in pMSG208, was packaged into phAE87 and delivered to mgm93 as described above. Transposition events were selected by growth on minimal medium, and individual leucine prototrophs were analyzed by Southern hybridization for the presence of the transposable element.
To determine the site of transposon insertion, we used an inverse PCR strategy. Genomic DNA from individual transposon mutants was digested to completion with BssHII, which does not cut inside of the inverted repeats of the transposon. After inactivation of BssHII, an aliquot of DNA was self-ligated to form genomic circles and amplified by PCR with primers (omsg131 and omsg132, CGGTTCTAGCCGGTAGCGTC and ACACCCCAAGCCAACCAGACC) complementary to the leuD transposon cassette ends. This PCR primer pair amplifies around the genomic circle to create PCR products of diverse sizes, depending on the size of the circle. The site of transposon insertion was determined by direct sequencing of these PCR products with omsg131 and omsg132 and BLAST searching against the M. tuberculosis and M. avium genome sequences.
RESULTS
Quantitative assessment of TM4 infection of clinical M. avium strains.
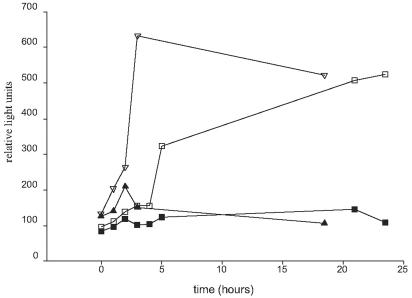
Recently, specialized transduction has been used to mediate allelic exchange in M. tuberculosis. The phages used in these prior studies were temperature-sensitive derivatives of the mycobacteriophage TM4 (1, 3). To assess the utility of these phages in the genetic manipulation of M. avium, we compared the efficiency of infection of BCG and M. avium by using a TM4 derivative expressing luciferase. A luciferase expressing TM4 derivative was used to infect BCG and two clinical strains of M. avium: one opaque colony morphotype (strain mc22501) and one SmT colony morphotype (strain mc22500). Compared to BCG, the efficiency of infection of M. avium was lower by more the 2 orders of magnitude (data not shown). Nevertheless, infection of M. avium cells was discernible in both M. avium colony morphotypes compared to uninfected control cells (Fig. 1). The relative light output of M. avium strains was 5- to 10-fold higher than with control cells, suggesting some phage DNA entry and consequent light production. Based on these results, we attempted to develop a specialized transduction system for M. avium by using the TM4 derivative phAE87 that has been used previously in M. tuberculosis. Although the relative efficiency of phage infection of M. avium strains is substantially lower than in M. tuberculosis and BCG, allelic exchange may still be feasible with this system.
FIG. 1.
Quantitative assessment of TM4 infection of clinical M. avium strains by using luciferase reporter mycobacteriophages. The clinical M. avium strains mc22500 and mc22501 were infected with phAE85, a temperature-sensitive derivative of TM4 expressing firefly luciferase. Symbols: ▪, mc22501, no phage; □, mc22501, phAE85; ▴, mc22500, no phage; ▵, mc22500, phAE85.
Successful deletion of leuD in M. avium by specialized transduction.
To test specialized transduction in M. avium, we constructed a null allele of the leucine biosynthetic gene leuD. leuD encodes the small subunit of 3-isopropylmalate dehydratase, the second step in leucine biosynthesis. leuD has been successfully disrupted in other mycobacteria, resulting in leucine auxotrophy (20), a phenotype that can be easily scored if the efficiency of allelic exchange is low. Using M. avium genomic sequence information available through the M. avium genome sequencing project at TIGR, we constructed a specialized transducing phage carrying a null allele of the avium leuD gene in phAE87. Both transparent (strain mc22500) and opaque (mc22501) colony morphotypes were infected with this phage. After ca. 200 hygromycin-resistant transductants were screened for leucine auxotrophy, we determined that none of the SmT transductants had undergone leuD allelic exchange. However, one transductant out of ca. 200 of the opaque strain displayed leucine auxotrophy. Analysis of this strain by Southern blotting revealed allelic exchange at leuD (Fig. 2). Thus, allelic exchange through specialized transduction is feasible in the opaque colony morphotype of M. avium. However, the efficiency of allelic exchange in comparison to spontaneous hygromycin resistance or illegitimate recombination was very low. Without an easily scoreable phenotype, such as auxotrophy, this system would not be practical for routine allelic exchange. The M. avium leuD deletion mutant was named mgm93 and was used in further studies described below.
FIG. 2.
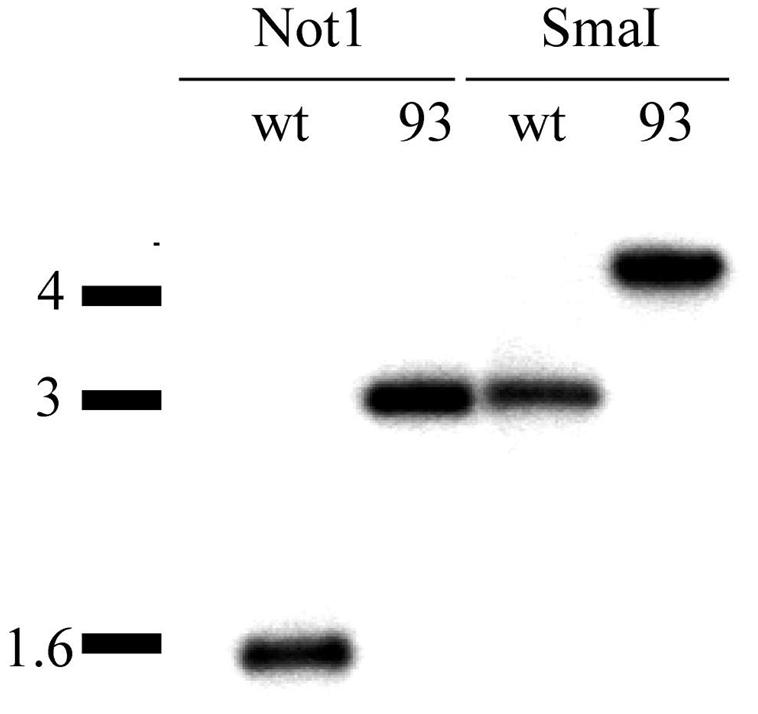
Disruption of leuD in M. avium strain mc22501 by using specialized transduction. Predicted sizes (in base pairs) are as follows: NotI, WT-1544, mgm93-2999; SmaI, WT-2991, mgm93-4446. The leuD-null mutant is designated mgm93.
The S. coelicolor leuD gene can complement a mycobacterial leuD mutation.
To improve the efficiency of allelic exchange in M. avium, we developed a leuD complementation cassette that could be used as a selectable marker when introduced into the mgm93 auxotrophic strain. This strategy is designed to utilize the extremely low (<10−11) reversion frequency of leuD-null mutants (13) and thereby avoid the recurrent problem of spontaneous antibiotic resistance in M. avium that complicates genetic analysis. Under this system, allelic exchange could be selected on minimal medium (leucine prototrophy). To avoid the possibility of allelic exchange of the complementing leuD cassette with the chromosomal leuD deletion, thereby regenerating a wild-type leuD locus, we developed a heterologous leuD complementation strategy. We chose the leuD gene from S. coelicolor because it has a GC content similar to that of M. avium but is 65% identical on the nucleotide level. We hypothesized that this degree of nucleotide sequence divergence would reduce allelic exchange between the complementing cassette and the chromosomal leuD locus. However, the S. coelicolor LeuD protein is 59% identical and 73% similar to its M. avium counterpart, suggesting that the S. coelicolor structural gene could function in mycobacteria. This idea is plausible since the E. coli leuCD cassette can complement a BCG leuD mutant (2).
The Streptomyces leuD gene was cloned from S. coelicolor genomic DNA by PCR with sequence information from the S. coelicolor genome sequencing project. To test the hypothesis that the Streptomyces leuD gene could complement a mycobacterial leuD-null mutant, we transformed an M. smegmatis leuD-null mutant with Streptomyces leuD expression constructs. Somewhat surprisingly, Streptomyces leuD expressed from its native promoter (pmsg205) could not complement an M. smegmatis leuD-null mutation (Fig. 3). However, when expressed from a constitutive mycobacterial promoter, the Streptomyces gene did complement the leuD mutation (pmsg207, Fig. 3). These results suggest that leuD from Streptomyces can form the appropriate dimer with the M. avium leuC to form a functional 3-isopropylmalate dehydratase. This complementation system for mycobacterial leuD can be used as a selectable marker for genetic manipulations in the mgm93 strain background. Since the reversion frequency of the leucine auxotrophy in mgm93 is <10−9 (data not shown), this strategy may surmount the recurrent problem of spontaneous antibiotic resistance in M. avium that has hampered the present study and previous similar published attempts.
FIG. 3.
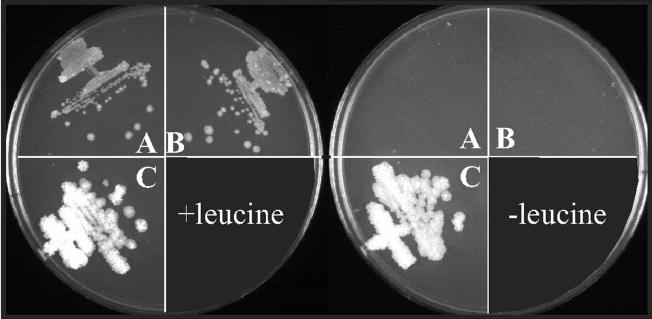
Heterologous complementation of an M. smegmatis leuD mutation by the S. coelicolor leuD structural gene. A leuD transposon mutant of M. smegmatis (JCM34) was transformed with vector DNA (A), a plasmid carrying the S. coelicolor leuCD operon (pmsg205) (B), and Streptomyces leuD expressed from a constitutive M. tuberculosis promoter (pmsg207) (C). The left panel shows the transformants growing on kanamycin and leucine, and the right panel shows the same strains on kanamycin without leucine. See Table 1 for detailed strain descriptions.
Efficient allelic exchange in M. avium with leucine prototrophy as a selectable marker.
To test the feasibility of allelic exchange in mgm93 with leuD-marked null alleles, we targeted the M. avium homologue of pcaA, the mycolic acid cyclopropane synthase previously shown to be important for M. tuberculosis pathogenesis (11). We constructed a deletion allele for M. avium pcaA by using the constitutively expressed Streptomyces leuD cassette described above. This pcaA-leuD construct (pMSG212) was packaged into phAE87 and used to infect mgm93. Transductants were selected on minimal medium, and leucine prototrophs were screened for allelic exchange at pcaA by Southern hybridization. Of seven leucine prototrophs examined, four contained the pcaA disruption (Fig. 4). Three leucine prototrophs appeared to be illegitimate recombinants, as evidenced by the presence of the wild-type pcaA locus and random integration of the targeting construct into the chromosome (Fig. 4). These results indicate that allelic exchange in M. avium is efficient when not limited by spontaneous antibiotic resistance. In addition, these results confirm our hypothesis that the Streptomyces cassette can function in M. avium, but that it does not recombine with the chromosomal M. avium leuD locus to regenerate prototrophic cells.
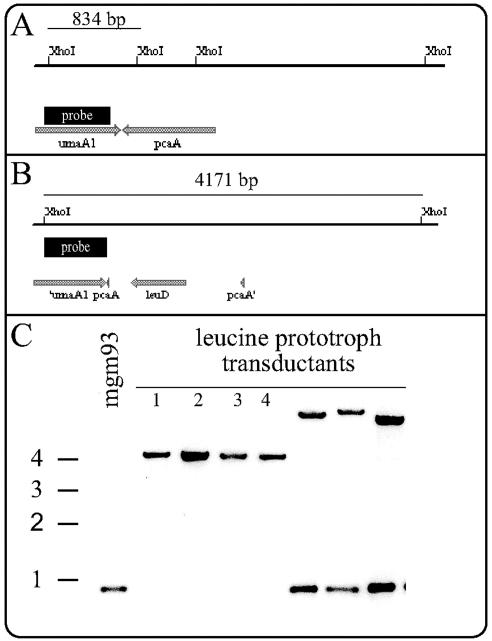
FIG. 4.
Disruption of pcaA by specialized transduction of leuD-marked construct into mgm93. (A and B) Maps of pcaA chromosomal loci in wild-type (A) and pcaA knockout (B) strains with restriction sites, probe locations, and predicted sizes indicated. (C) Southern hybridization of genomic DNA digested with XhoI and probed with the fragments indicated in panels A and B.
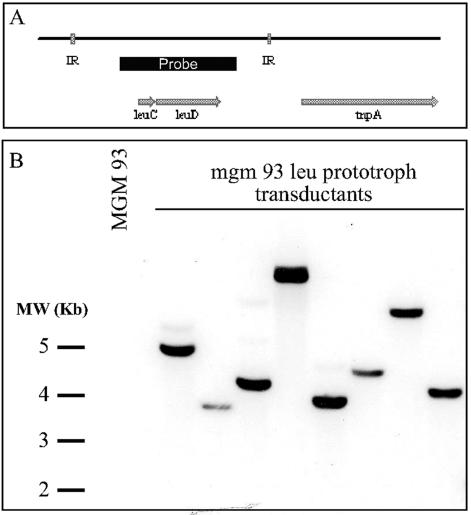
Efficient transposon mutagenesis in M. avium with leuD as a selectable marker.
Given our success with allelic exchange with mgm93 as a genetic host, we attempted transposon mutagenesis by the same approach. We modified Tn5370, previously used to mutagenize M. tuberculosis, by replacing the hygromycin resistance gene with the leuD expression construct. This transposon was introduced into phAE87 and used to infect mgm 93. After selection on minimal medium, leucine prototrophs were examined by Southern blotting to confirm the presence of the transposable element. The frequency of transposition in two independent experiments was 5.4 × 10−6 and 1.5 × 10−6 at a multiplicity of infection of 10. All of the leucine prototrophs examined contained the transposable element (Fig. 5). To confirm that the transposition had occurred in diverse locations in the circular chromosome and not in the plasmids that have been reported in some M. avium clinical strains, we determined the site of transposon insertion in eight leucine prototrophs by inverse PCR as described in Materials and Methods. All eight transposition events had occurred in distinct genetic loci in genes that are within the circular chromosome. Although this small number of transposon insertions does not establish the degree of randomness of this transposon, it does suggest that this transposon does not have strong preferential insertional loci that would reduce the efficiency of random mutagenesis.
FIG. 5.
Transposon mutagenesis in M. avium. (B) Southern blot of genomic DNA from individual leucine prototrophs that arose from transduction of mgm93 with a leuD transposon. (The probe location is given in the diagram in panel A.)
DISCUSSION
This study establishes an efficient specialized transduction system for allelic exchange and transposon mutagenesis in a clinical strain of M. avium. When combined with the recently completed M. avium genome sequence, these tools will allow new insights into M. avium physiology. Several aspects of M. avium physiology differ substantially from M. tuberculosis, including colony morphotype switching (22), environmental persistence (9), plasmid maintenance (15), and antibiotic resistance. These distinct properties of M. avium can now be investigated genetically with the tools presented here.
Unfortunately, we were unsuccessful in generating a leuD mutant in the SmT colony morphotype (mc22500). The high rate of spontaneous hygromycin resistance in both SmD and SmT colony types clearly impaired the efficient selection of recombinants and suggests that this antibiotic marker is not useful for M. avium. Although mc22501 is derived from an original clinical isolate, it is a spontaneous opaque colony variant. Colony morphology in M. avium is associated with virulence, with SmT types being more virulent for mice than SmD variants (14, 25, 26). In addition, although opaque types are generally less virulent than SmT types, some opaque strains are isolated from patients and retain the ability to replicate within mice (26). Thus, it is possible that mgm93 could be used to examine the genetic basis for virulence in M. avium, depending upon its in vivo characteristics in mice. We plan to test the in vivo growth characteristics of mc22501 in immunocompetent mice to assess whether mgm93 derivatives will be useful for pathogenesis studies. In addition, it may be possible to isolate a SmT revertant of mgm93 after in vivo passage that would then be suitable for genetic manipulation.
A minimal set of genetic tools for M. avium would include efficient transformation, which has not been routinely achieved. The isolation of new mycobacterial plasmid types from M. avium (15) may allow efficient transformation vectors to be created, thereby further expanding the set of genetic tools for M. avium. The efficiency of transposon mutagenesis in the present study is comparable to rates reported previously for M. tuberculosis by using specialized transduction of a related transposon. This high rate of transposition in M. avium was achieved despite poor phage entry into M. avium cells, as measured by luciferase reporter phages. This result suggests that entry of transposon encoding DNA into the host cell is not the limiting factor in efficient transposon mutagenesis in mycobacteria. The leuD transposon constructed here may insert more efficiently in the chromosome due to its small size, thereby compensating for poor phage entry.
In summary, the present study extends the utility of specialized transduction to include allelic exchange and transposon mutagenesis to a clinical strain of M. avium. Our use of a leucine auxotrophic strain of M. avium as a genetic host eliminated the spontaneous antibiotic resistance that had limited prior genetic studies in M. avium. In conjunction with the recently completed M. avium genome sequence, these genetic tools will allow direct study of M. avium physiology and may allow investigation into M. avium pathogenesis.
Acknowledgments
We thank Jeff Cox and Apoorva Bhatt for strains and reagents.
This work was supported by NIH grants AI01534 (M.S.G.) and AI27160 (W.R.J.).
REFERENCES
- 1.Banaiee, N., M. Bobadilla-Del-Valle, S. Bardarov, Jr., P. F. Riska, P. M. Small, A. Ponce-De-Leon, W. R. Jacobs, Jr., G. F. Hatfull, and J. Sifuentes-Osornio. 2001. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. J. Clin. Microbiol. 39:3883-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bange, F. C., A. M. Brown, and W. R. Jacobs, Jr. 1996. Leucine auxotrophy restricts growth of Mycobacterium bovis BCG in macrophages. Infect. Immun. 64:1794-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardarov, S., Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG, and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Berthet, F. X., M. Lagranderie, P. Gounon, C. Laurent-Winter, D. Ensergueix, P. Chavarot, F. Thouron, E. Maranghi, V. Pelicic, D. Portnoi, G. Marchal, and B. Gicquel. 1998. Attenuation of virulence by disruption of the Mycobacterium tuberculosis erp gene. Science 282:759-762. [DOI] [PubMed] [Google Scholar]
- 6.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 7.Carriere, C., P. F. Riska, O. Zimhony, J. Kriakov, S. Bardarov, J. Burns, J. Chan, and W. R. Jacobs, Jr. 1997. Conditionally replicating luciferase reporter phages: improved sensitivity for rapid detection and assessment of drug susceptibility of Mycobacterium tuberculosis. J. Clin. Microbiol. 35:3232-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs, Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 9.Falkinham, J. O., III. 2002. Nontuberculous mycobacteria in the environment. Clin. Chest Med. 23:529-551. [DOI] [PubMed] [Google Scholar]
- 10.Glickman, M. S., S. M. Cahill, and W. R. Jacobs, Jr. 2001. The Mycobacterium tuberculosis cmaA2 gene encodes a mycolic acid trans cyclopropane synthetase. J. Biol. Chem. 276:2228-2233. [DOI] [PubMed] [Google Scholar]
- 11.Glickman, M. S., J. S. Cox, and W. R. Jacobs, Jr. 2000. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5:717-727. [DOI] [PubMed] [Google Scholar]
- 12.Harris, N. B., Z. Feng, X. Liu, S. L. Cirillo, J. D. Cirillo, and R. G. Barletta. 1999. Development of a transposon mutagenesis system for Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 175:21-26. [DOI] [PubMed] [Google Scholar]
- 13.Hondalus, M. K., S. Bardarov, R. Russell, J. Chan, W. R. Jacobs, Jr., and B. R. Bloom. 2000. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infect. Immun. 68:2888-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kansal, R. G., R. Gomez-Flores, and R. T. Mehta. 1998. Change in colony morphology influences the virulence as well as the biochemical properties of the Mycobacterium avium complex. Microb. Pathog. 25:203-214. [DOI] [PubMed] [Google Scholar]
- 15.Kirby, C., A. Waring, T. J. Griffin, J. O. Falkinham III, N. D. Grindley, and K. M. Derbyshire. 2002. Cryptic plasmids of Mycobacterium avium: Tn552 to the rescue. Mol. Microbiol. 43:173-186. [DOI] [PubMed] [Google Scholar]
- 16.Mahenthiralingam, E., B. I. Marklund, L. A. Brooks, D. A. Smith, G. J. Bancroft, and R. W. Stokes. 1998. Site-directed mutagenesis of the 19-kilodalton lipoprotein antigen reveals no essential role for the protein in the growth and virulence of Mycobacterium intracellulare. Infect. Immun. 66:3626-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marklund, B. I., E. Mahenthiralingam, and R. W. Stokes. 1998. Site-directed mutagenesis and virulence assessment of the katG gene of Mycobacterium intracellulare. Mol. Microbiol. 29:999-1008. [DOI] [PubMed] [Google Scholar]
- 18.Marklund, B. I., and R. W. Stokes. 1998. Gene replacement in Mycobacterium intracellulare. Methods Mol. Biol. 101:217-224. [DOI] [PubMed] [Google Scholar]
- 19.McAdam, R. A., S. Quan, D. A. Smith, S. Bardarov, J. C. Betts, F. C. Cook, E. U. Hooker, A. P. Lewis, P. Woollard, M. J. Everett, P. T. Lukey, G. J. Bancroft, W. R. Jacobs, Jr., and K. Duncan. 2002. Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology 148:2975-2986. [DOI] [PubMed] [Google Scholar]
- 20.McAdam, R. A., T. R. Weisbrod, J. Martin, J. D. Scuderi, A. M. Brown, J. D. Cirillo, B. R. Bloom, and W. R. Jacobs, Jr. 1995. In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect. Immun. 63:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavelka, M. S., Jr., and W. R. Jacobs, Jr. 1999. Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guerin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J. Bacteriol. 181:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedrosa, J., M. Florido, Z. M. Kunze, A. G. Castro, F. Portaels, J. McFadden, M. T. Silva, and R. Appelberg. 1994. Characterization of the virulence of Mycobacterium avium complex (MAC) isolates in mice. Clin. Exp. Immunol. 98:210-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919-925. [DOI] [PubMed] [Google Scholar]
- 25.Reddy, V. M., J. Luna-Herrera, and P. R. Gangadharam. 1996. Pathobiological significance of colony morphology in Mycobacterium avium complex. Microb. Pathog. 21:97-109. [DOI] [PubMed] [Google Scholar]
- 26.Reddy, V. M., K. Parikh, J. Luna-Herrera, J. O. Falkinham III, S. Brown, and P. R. Gangadharam. 1994. Comparison of virulence of Mycobacterium avium complex (MAC) strains isolated from AIDS and non-AIDS patients. Microb. Pathog. 16:121-130. [DOI] [PubMed] [Google Scholar]
- 27.Riska, P. F., and W. R. Jacobs, Jr. 1998. The use of luciferase-reporter phage for antibiotic-susceptibility testing of mycobacteria. Methods Mol. Biol. 101:431-455. [DOI] [PubMed] [Google Scholar]
- 28.Sturgill-Koszycki, S., P. H. Schlesinger, P. Chakraborty, P. L. Haddix, H. L. Collins, A. K. Fok, R. D. Allen, S. L. Gluck, J. Heuser, and D. G. Russell. 1994. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263:678-681. [DOI] [PubMed] [Google Scholar]