Abstract
Burkholderia sp. strain AK-5 utilized 4-aminophenol as the sole carbon, nitrogen, and energy source. A pathway for the metabolism of 4-aminophenol in strain AK-5 was proposed based on the identification of three key metabolites by gas chromatography-mass spectrometry analysis. Strain AK-5 converted 4-aminophenol to 1,2,4-trihydroxybenzene via 1,4-benzenediol. 1,2,4-Trihydroxybenzene 1,2-dioxygenase cleaved the benzene ring of 1,2,4-trihydroxybenzene to form maleylacetic acid. The enzyme showed a high dioxygenase activity only for 1,2,4-trihydroxybenzene, with Km and Vmax values of 9.6 μM and 6.8 μmol min−1 mg of protein−1, respectively.
4-Aminophenol has highly toxic and mutagenic effects and induces DNA cleavage in mouse and human lymphoma cells (12, 22). This compound is an intermediate in the degradation of hydroxyacetanilide (7) and azo dyes (19). However, little is known about the metabolism of 4-aminophenol by bacteria (1). 3-Nitrophenol-grown cells of Ralstonia eutropha JMP 134 convert nitrobenzene to hydroxylaminobenzene, 2-aminophenol, and 4-aminophenol (16). Hydroxylaminobenzene is transformed by 3-nitrophenol-grown cells of Pseudomonas putida 2NP8 to 1,4-benzenediol via 4-aminophenol (25). A number of reports indicate that 4-aminophenol might be a key intermediate in the biodegradation of nitrobenzenes and amines (7, 19, 25). Our aim was to elucidate a biodegradation pathway for 4-aminophenol by analyzing metabolites.
Here we report the isolation of a 4-aminophenol-assimilating bacterium and propose a metabolic pathway for 4-aminophenol. In addition, the characterization of a 1,2,4-trihydroxybenzene 1,2-dioxygenase from strain AK-5 is described.
MATERIALS AND METHODS
Organism and growth conditions.
Strain AK-5 was enriched from rice field soil from the Hyogo Prefecture. The basal medium containing 4-aminophenol was prepared by methods described previously (3). Succinate-glucose medium was a modified basal medium containing 1.0% (wt/vol) sodium succinate, 1.0% (wt/vol) d-glucose, and 0.04% (wt/vol) NH4NO3 as the sole carbon and nitrogen sources instead of 4-aminophenol.
Purification of 1,2,4-trihydroxybenzene 1,2-dioxygenase.
1,2,4-Trihydroxybenzene 1,2-dioxygenase activity was assayed by the method of Latus et al. (10). The molar extinction coefficient of 4.44 × 103 at 243 nm for maleylacetic acid was used (20). Protein concentrations were measured by the method of Lowry et al. (11).
Cells (25 g [wet weight]) of strain AK-5 were suspended in 20 mM Tris-HCl (pH 8.0) (buffer A). Cell extract (fraction 1) was prepared and treated with streptomycin sulfate (fraction 2) as described previously (3). Fraction 2 was fractionated with ammonium sulfate (32 to 50% saturation). After centrifugation (20,000 × g for 10 min), the pelleted precipitate was dissolved in buffer A. The solution was dialyzed against buffer A (fraction 3, 90 ml). Fraction 3 was applied to a DE52 cellulose column (2.1 by 26 cm), and proteins were eluted with a linear gradient (0 to 0.4 M NaCl) at a flow rate of 40 ml h−1. The active fractions were pooled (fraction 4; 60 ml). Fraction 4 was applied to a DEAE-Cellulofine A-800 column (2.0 by 15 cm), and proteins were eluted with a linear gradient (0 to 0.35 M) of NaCl at a flow rate of 30 ml h−1. The active fractions were pooled (fraction 5; 30 ml). Fraction 5 was applied to a phenyl-Cellulofine column (1.6 by 7.5 cm), and proteins were eluted with a linear gradient (0.5 to 0 M) of (NH4)2SO4 at a flow rate of 30 ml h−1. The active fractions were pooled (fractions 6; 28 ml). Fraction 6 was concentrated, and the solution was loaded onto a Cellulofine GCL-1000 sf column (3.2 by 58 cm); proteins were eluted with buffer A containing 0.2 M NaCl at a flow rate of 20 ml h−1. The enzyme purity was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (21).
Substrate specificity.
The benzene ring cleavage of 1,4-benzenediol, catechol, 3-methylcatechol, 4-methylcatechol, 3-chlorocatechol, 4-chlorocatechol, 4-nitrocatechol, protocatechuic acid, and pyrogallol was monitored spectrophotometrically. Inhibition of the enzyme activity by substrate analogues was examined by incubating the enzyme (25 μg ml−1) with each analogue (0.05 mM) in 3 ml of 100 mM sodium-potassium phosphate buffer (pH 7.5) at 24°C for 1 min. The enzyme reaction was started by adding 1,2,4-trihydroxybenzene.
Effect of various compounds on enzyme activity.
The effect of metal salts and chelating and sulfhydryl agents on enzyme activity with 1,2,4-trihydroxybenzene as the substrate was tested by methods described previously (18).
Production and isolation of metabolites.
The reaction mixture contained 84 ml of 100 mM sodium-potassium phosphate buffer (pH 7.5), 3.0 ml of cell suspension (0.57 mg [dry weight] of cells ml−1), and 3 ml of 10 mM 4-aminophenol. After incubation with shaking at 30°C for 5 min, cells were broken by ultrasonic disintegration. The solution was then adjusted to pH 3.0 with 3 N HCl, and precipitated proteins were removed by centrifugation at 20,000 × g for 10 min. The supernatant was concentrated with an evaporator to 20 ml and then extracted with ethyl acetate. The upper layer was recovered and evaporated to dryness. The accumulated products reacted with N,O-bis(trimethylsilyl)-trifluoroacetamide (BSTFA) at 90°C for 1.5 h. The trimethylsilylated products were analyzed by gas chromatography (GC)-mass spectrometry (MS) as described below.
Consumption of molecular oxygen by whole cells.
Possible metabolites were assayed in oxygen uptake experiments with 4-aminophenol-grown cells by using a Clark-type oxygen electrode (Yellow Springs Instruments, Yellow Springs, Colo.). Cells were grown in 4-aminophenol or succinate-glucose medium. The reaction mixture contained 3.0 ml of 50 mM sodium-potassium phosphate buffer (pH 7.0) and 0.1 ml of cell suspension (0.57 mg [dry weight] of cells ml−1). The reaction was started by adding 0.1 ml of 10 mM 4-aminophenol, 1,4-benzenediol, 1,4-benzoquinone, phenol, or catechol, and the reaction mixture was incubated at 24°C.
Identification of the reaction product (compound IV) from 1,2,4-trihydroxybenzene.
The reaction mixture contained 100 ml of 100 mM sodium-potassium phosphate buffer (pH 7.5), 4.0 ml of enzyme solution (25 μg ml−1), and 4 ml of 5 mM 1,2,4-trihydroxybenzene. After incubation at 24°C for 15 min, the mixture was concentrated to 30 ml with a rotary evaporator. The mixture was then extracted and derivatized for GC and GC-MS analysis essentially as described above.
Analytical methods.
UV absorption spectra of reaction products were recorded with a Beckman DU 650 spectrophotometer. The trimethylsilylated compounds were analyzed with a Hitachi M-2500 mass spectrometer under conditions described previously (18). 4-Aminophenol in the growing culture was determined by a diazo coupling reaction (14). A molar extinction coefficient of 3.4 × 103 at 578 nm for the diazotized compound was used. The apparent molecular mass of the native enzyme was determined by gel filtration on Cellulofine GCL-1000 sf. The molecular mass of the enzyme subunits was measured by SDS-PAGE (21).
Chemicals.
4-Aminophenol, 1,2,4-trihydroxybenzene (hydroxyhydroquinone), 1,4-benzenediol (hydroquinone), catechols, and BSTFA were purchased from Wako Pure Chemicals (Osaka, Japan). DE52 cellulose was from Whatman (Madison, Wis.), and DEAE-Cellulofine A-800, phenyl-Cellulofine, and Cellulofine GCL-1000 sf were from Seikagaku (Tokyo, Japan).
RESULTS AND DISCUSSION
Identification of a 4-aminophenol-assimilating bacterium.
Strain AK-5 is a motile rod of 0.8 to 1.2 by 2.4 to 3.6 μm with polar flagella. It is aerobic, gram negative, not spore forming, and catalase and oxidase positive. It produces acid oxidatively from d-glucose, d-fructose, d-sorbitol, d-mannitol, lactose, maltose, and sucrose. It does not produce H2S, indole, or acetoin and does not hydrolyze gelatin. The nucleotide sequence of the 16S rRNA gene of strain AK-5 (1,520 bp; accession no. AB103080) was 97.9 and 98.1% identical to that of Burkholderia sp. strain NF100 (AB025790) and Burkholderia sp. strain S4.9 (AF247496), respectively (6, 8). Thus, strain AK-5 was identified as a species of Burkholderia.
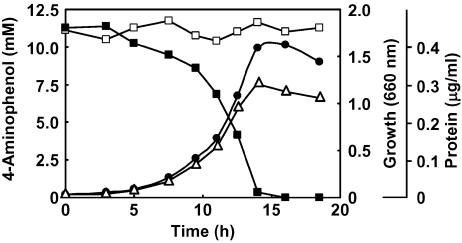
Strain AK-5 grew well on 4-aminophenol as the sole carbon, nitrogen, and energy source (Fig. 1). 4-Aminophenol was rapidly degraded during the exponential phase. The consumption of 4-aminophenol correlated with an increase in cell density and protein content. High concentrations of 4-aminophenol (>18.0 mM) inhibited growth. Strain AK-5 grew well at pHs from 5 to 6.5 and poorly at pHs >6.5.
FIG. 1.
Growth of Burkholderia sp. strain AK-5 on 4-aminophenol. Strain AK-5 was grown in basal medium containing 4-aminophenol (1.2 g · liter−1) without yeast extract in a 500-ml flask at 30°C with shaking. Disappearance of 4-aminophenol (▪; □, control) was measured spectrophotometrically by using a diazo coupling reaction (14). The increase in cell density was determined by measuring the optical density at 660 nm (•) or the protein content (▵) of the culture.
Purification and properties of the purified dioxygenase.
The 1,2,4-trihydroxybenzene 1,2-dioxygenase from strain AK-5 was present in cell extracts of 4-aminophenol-grown cells but not in cell extracts of succinate-glucose-grown cells; therefore, the synthesis of the enzyme was inducible. The enzyme was purified 108-fold, with an overall yield of 0.9% (Table 1). The apparent molecular mass was determined to be 85 kDa by gel filtration, and the molecular mass was determined to be 81 kDa by SDS-PAGE, which indicated that the enzyme is a monomer.
TABLE 1.
Purification of 1,2,4-trihydroxybenzene 1,2-dioxygenase from Burkholderia sp. strain AK-5
| Fractiona | Total activity (U) | Total protein (mg) | Sp act (U · mg−1) | Recovery (%) |
|---|---|---|---|---|
| 1 (cell extract) | 340 | 2,900 | 0.12 | 100 |
| 2 (streptomycin sulfate) | 240 | 2,700 | 0.09 | 71 |
| 3 (ammonium sulfate) | 230 | 1,400 | 0.16 | 68 |
| 4 (DE52) | 80 | 150 | 0.53 | 24 |
| 5 (DEAE-Cellulofine A-800) | 30 | 20 | 1.5 | 8.8 |
| 6 (Phenyl-Cellulofine) | 6.0 | 1.1 | 5.5 | 1.8 |
| 7 (Cellulofine GCL-1000 sf) | 3.0 | 0.24 | 13 | 0.9 |
Fractions 1 to 7 refer to the fractions obtained at the end of steps 1 to 7, respectively, of the purification procedure. See the text for details.
After DE52 chromatography, the dioxygenase from strain AK-5 was stable for several weeks in buffer A containing 250 mM NaCl. The dioxygenase from Burkholderia cepacia AC1100 is also stable at a high salt concentration (5). The enzyme from strain AK-5 maintained more than 100% activity after a 10-min incubation at temperatures up to 50°C and showed maximal activity at pH 7.0. The enzyme had a high activity only for 1,2,4-trihydroxybenzene, with Km and Vmax values of 9.6 μM and 6.8 μmol · min−1 · mg of protein−1, respectively. Such a remarkably narrow substrate specificity is shared with the dioxygenase from Trichosporon cutaneum (17). Among the substrate analogues tested, 1,4-benzenediol, 4-methylcatechol, and 4-chlorocatechol decreased the enzyme activity for 1,2,4-trihydroxybenzene to 58, 66, and 25%, respectively. Among the metal salts tested, the enzyme was completely inhibited by 1 mM HgCl2, 1 mM MgSO4, and 1 mM AgNO3. The addition of 1 mM α,α′-dipyridyl, EDTA, o-phenanthroline, or NaN3 decreased the enzyme activity to 48, 49, 0, and 25%, respectively.
Proposed pathway of 4-aminophenol metabolism.
4-Aminophenol (0.20 mM) was degraded, with elimination of ammonia (0.17 mM), by 4-aminophenol-grown whole cells of strain AK-5. Metabolites were analyzed by GC and GC-MS. Trimethylsilylated 4-aminophenol (M+ = 253) had a GC retention time of 10.7 min. Major peaks at 8.6 and 12.1 min were also observed. The mass spectra (Table 2) and the GC retention times (Rt) of compounds II and III agreed with those of the trimethylsilylated authentic 1,4-benzenediol (Rt = 8.6 min) and 1,2,4-trihydroxybenzene (Rt = 12.1 min), respectively (Fig. 2). The enzymatic reaction product derived from 1,2,4-trihydroxybenzene showed an absorption peak at 243 nm. The mass spectrum of the trimethylsilylated reaction product (compound IV) is in agreement with that of maleylacetic acid (Table 2) (15).
TABLE 2.
Mass spectra of the metabolites from 4-aminophenol and reaction product from 1,2,4-trihydroxybenzene
| Compound | Fragments of the trimethylsilylated product (m/z [assignment, relative intensity {%}]) |
|---|---|
| II (1,4 · benzenediol) | 254 (M+, 92.3), 239 (M+—CH3, 100), 147 ([(CH3)2=O—OSi(CH3)3]+, 2.0), 135 {M+—OSi(CH3)3—CH3×2, 8.4}, 73 {[Si(CH3)3]+, 35.5} |
| III (1,2,4 · trihydroxybenzene) | 342 (M+, 31.4), 327 (M+—CH3, 2.4), 239 {M+—Si(CH3)3—CH3×2, 100}, 147 {[(CH3)2=O—OSi(CH3)3]+, 1.7}, 73 {[Si(CH3)3]+, 22.3} |
| IV (maleylacetic acid) | 374 (M+, 0.8), 359 (M+—CH3, 9.9), 315 (M+—CH3—CO2, 2.1), 257 [M+—COOSi(CH3)3, 100], 241 (1.37), 219 (1.3) 197 (1.0) 147 {[(CH3)2=O—OSi(CH3)3]+, 2.2}, 73 {[Si(CH3)3]+, 43.3} |
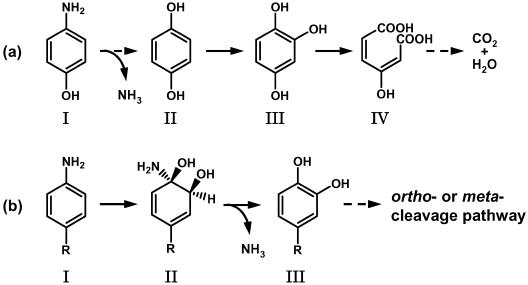
FIG. 2.
Proposed pathway of 4-aminophenol metabolism in Burkholderia sp. strain AK-5 and comparison to the metabolic pathways of aniline and aniline derivatives. (a) Proposed pathway of 4-aminophenol metabolism. I, 4-aminophenol; II, 1,4-benzenediol; III, 1,2,4-trihydroxybenzene; IV, maleylacetic acid. (b) Metabolic pathways of aniline and aniline derivatives in Rhodococcus erythropolis AN-13 (2), Pseudomonas putida mt-2 (12), Moraxella sp. strain G (24), and Hydrogenophaga palleronii S5 (4). I, aniline (R = H), p-toluidine (R = CH3), 4-chloroaniline (R = Cl), 4-aminobenzoic acid (R = COOH), and 4-aminobenzenesulfonic acid (R = SO3H); II, 1-amino-2-hydrodiols; III, catechol (R = H), 4-methylcatechol (R = CH3), 4-chlorocatechol (R = Cl), protocatechuic acid (R = COOH), and 4-sulfocatechol (R = SO3H).
The oxygen uptake rates of 4-aminophenol-grown whole cells with 4-aminophenol, 1,4-benzenediol, and 1,2,4-trihydroxybenzene were 22, 10, and 12 μmol min−1 mg of protein−1, respectively. In contrast, the oxygen uptake rates of succinate-glucose-grown whole cells with these compounds were less than 1 μmol min−1 mg of protein−1. The oxygen uptake rates of 4-aminophenol- and succinate-glucose-grown whole cells with phenol, catechol, or 1,4-benzoquinone were less than 1 μmol min−1 mg of protein−1 These results indicated that the enzymes responsible for 4-aminophenol, 1,4-benzenediol, and 1,2,4-trihydroxybenzene metabolism were induced in 4-aminophenol-grown cells.
Figure 2a shows the proposed metabolic pathway of 4-aminophenol in strain AK-5. 4-Aminophenol was converted to 1,2,4-trihydroxybenzene via 1,4-benzenediol; 1,2,4-trihydroxybenzene 1,2-dioxygenase catalyzed the conversion of 1,2,4-trihydroxybenzene to maleylacetic acid. Presumably, the benzene ring of 4-aminophenol is subjected to two hydroxylation steps to yield 1,2,4-trihydroxybenzene. The proposed pathway differs from previously reported metabolic pathways for aniline and anilines with a methyl-, chloro-, sulfo-, or carboxy-functional-group substituent at the C-4 position (Fig. 2b) (2, 4, 13, 24). The initial reaction in the degradation of anilines is catalyzed by a dioxygenase and yields the corresponding 1-amino-2-hydrodiols as the first metabolites. Subsequent oxidation of 1-amino-2-hydrodiols leads to the formation of the catechols. The dioxygenation and dehydrogenation steps to form catechols in these species are similar irrespective of which functional group at the C-4 position of aniline is the electron donor or electron acceptor.
Hughes et al. (9) have reported that, in Pseudomonas putida strain TW3, 4-hydroxylaminobenzoate lyase converts 4-hydroxylaminobenzoate to protocatechuate, replacing the amino group by a hydroxyl group. Likewise, strain AK-5 could possibly require lyase activity in the initial step of 4-aminophenol metabolism, which we propose to be the direct conversion of 4-aminophenol to hydroquinone.
When the purified enzyme was added to a reaction mixture containing 1,2,4-trihydroxybenzene, the absorption peak at 243 nm, corresponding to maleylacetic acid, and the absorption peak at 260 nm, arising from the auto-oxidation of 1,2,4-trihydroxybenzene (23), increased slowly. In contrast, the auto-oxidation product did not accumulate when cell extract and the (NH4)2SO4 fraction were used in the assay. The cell extract might contain enzymes that inhibit the nonenzymatic reaction or that reduce the product. We are currently investigating enzymes involved in the transformation of the auto-oxidation product.
REFERENCES
- 1.Ahmed, S., M. A. Javed, S. Tanvir, and A. Hameed. 2001. Isolation and characterization of a Pseudomonas strain that degrades 4-acetamidophenol and 4-aminophenol. Biodegradation 12:303-309. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, K., R. Shinike, and H. Nishira. 1983. Metabolism of aniline by Rhodococcus erythropolis AN-13. Agric. Biol. Chem. 47:1611-1616. [Google Scholar]
- 3.Aoki, K., S. Takenaka, S. Murakami, and R. Shinke. 1997. Partial purification and characterization of a bacterial dioxygenase that catalyzes the ring fission of 2-aminophenol. Microbiol. Res. 152:33-38. [Google Scholar]
- 4.Blümel, S., M. Contzen, M. Lutz, A. Stolz, and H. J. Knackmuss. 1998. Isolation of a bacterial strain with the ability to utilize the sulfonated azo compound 4-carboxy-4′-sulfoazobenzene as the sole source of carbon and energy. Appl. Environ. Microbiol. 64:2315-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daubaras, D. L., K. Saido, and A. M. Chakrabarty. 1996. Purification of hydroxyquinol 1, 2-dioxygenase and maleylacetate reductase: the lower pathway of 2,4,5-trichlorophenoxyacetic acid metabolism by Burkholderia cepacia AC1100. Appl. Environ. Microbiol. 62:4276-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedrich, M., R. J. Grosser, E. A. Kern, W. P. Inskeep, and D. M. Ward. 2000. Effect of model sorptive phases on phenanthrene biodegradation: molecular analysis of enrichments and isolates suggests selection based on bioavailability. Appl. Environ. Microbiol. 66:2703-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart, A., and O. L. Orr. 1975. The degradation of paracetamol (4-hydroxyacetanilide) and other substituted acetanilides by a Penicillium species. Antonie Leeuwenhoek. 41:239-247. [DOI] [PubMed] [Google Scholar]
- 8.Hayatsu, M., M. Hirano, and S. Tokuda. 2000. Involvement of two plasmids in fenitrothion degradation by Burkholderia sp. strain NF100. Appl. Environ. Microbiol. 66:1737-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes, M. A., M. J. Baggs, J. al-Dulayymi, M. S. Baird, and P. A. Williams. 2002. Accumulation of 2-aminophenoxazin-3-one-7-carboxylate during growth of Pseudomonas putida TW3 on 4-nitro-substituted substrates requires 4-hydroxylaminobenzoate lyase (PnbB). Appl. Environ. Microbiol. 68:4965-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latus, M., H. Seitz, J. Eberspächer, and F. Lingens. 1995. Purification and characterization of hydroxyquinol 1,2-dioxygenase from Azotobacter sp. strain GP1. Appl. Environ. Microbiol. 61:2453-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 12.Majeska. J. B., and H. E. Holden. 1995. Genotoxic effects of p-aminophenol in Chinese hamster ovary and mouse lymphoma cells: results of a multiple endpoint test. Environ. Mol. Mutagen. 26:163-170. [DOI] [PubMed] [Google Scholar]
- 13.McClure, N. C., and W. A. Venables. 1982. Adaptation of Pseudomonas putida mt-2 to growth on aromatic amines. J. Gen. Microbiol. 132:2209-2218. [DOI] [PubMed] [Google Scholar]
- 14.Norwitz, G., and N. Keliher. 1982. Spectrophotometric determination of aromatic amines by the diazotization-coupling technique with 8-amino-1-hydroxynaphthalene-3,6-disulfonic acid and N-(1-naphthyl)ethylenediamine as the coupling agents. Anal. Chem. 54:807-808. [Google Scholar]
- 15.Rieble, S., D. K. Joshi, and M. H. Gold. 1994. Purification and characterization of a 1,2,4-trihydroxybenzene 1,2-dioxygenase from the basidiomycete Phanerochaete chrysosporium. J. Bacteriol. 176:4838-4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schenzle, A., H. Lenke, P. Fischer, P. A. Williams, and H. J. Knackmuss. 1997. Catabolism of 3-nitrophenol by Ralstonia eutropha JMP134. Appl. Environ. Microbiol. 63:1421-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sze, I. S. Y., and S. Dagley. 1984. Properties of salicylate hydroxylase and hydroxyquinol 1,2-dioxygenase purified from Trichosporon cutaneum. J. Bacteriol. 159:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takenaka, S., S. Murakami, R. Shinke, K. Hatakeyama, H. Yukawa, and K. Aoki. 1997. Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and catalytic properties of the purified enzyme. J. Biol. Chem. 272:14727-14732. [DOI] [PubMed] [Google Scholar]
- 19.Tan, N. C., F. X. Prenafeta-Boldu, J. L. Opsteeg, G. Lettinga, and J. A. Field. 1999. Biodegradation of azo dyes in cocultures of anaerobic granular sludge with aerobic aromatic amine degrading enrichment cultures. Appl. Microbiol. Biotechnol. 51:865-871. [DOI] [PubMed] [Google Scholar]
- 20.Tiedje, J. M., J. M. Duxbury, M. Alexander, and J. E. Dawson. 1969. 2,4-D metabolism: pathway of degradation of chlorocatechols by Arthrobacter sp. J. Agric. Food Chem. 17:1021-1026. [DOI] [PubMed] [Google Scholar]
- 21.Werber, K., and M. Osborn. 1969. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]
- 22.Yoshida. R., S. Oikawa, Y. Ogawa, Y. Miyakoshi, M. Ooida, K. Asanuma, and H. Shimizu. 1998. Mutagenicity of p-aminophenol in E. coli WP2uvrA/pKM101 and its relevance to oxidative DNA damage. Mutat. Res. 415:139-150. [DOI] [PubMed] [Google Scholar]
- 23.Zaborina. O., M. Latus, J. E. Eberspächer, L. A. Golovlea, and F. Lingens. 1995. Purification and characterization of 6-chlorohydroxyquinol 1,2-dioxygenase from Streptomyces rochei 303: comparison with an analogous enzyme from Azotobacter sp. strain GP1. J. Bacteriol. 177:229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeyer, J., A. Wasserfallen, and K. N. Timmis. 1985. Microbial mineralization of ring-substituted anilines through an ortho-cleavage pathway. Appl. Environ. Microbiol. 50:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao, J. S., A. Singh, X. D. Huang, and O. P. Ward. 2000. Biotransformation of hydroxylamine and aminophenol by Pseudomonas putida 2NP8 cells grown in the presence of 3-nitrophenol. Appl. Environ. Microbiol. 66:2336-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]