Abstract
Culture-independent techniques, denaturing gradient gel electrophoresis (DGGE) analysis, and random cloning of 16S rRNA gene sequences amplified from community DNA were used to determine the diversity of microbial communities in gas industry pipelines. Samples obtained from natural gas pipelines were used directly for DNA extraction, inoculated into sulfate-reducing bacterium medium, or used to inoculate a reactor that simulated a natural gas pipeline environment. The variable V2-V3 (average size, 384 bp) and V3-V6 (average size, 648 bp) regions of bacterial and archaeal 16S rRNA genes, respectively, were amplified from genomic DNA isolated from nine natural gas pipeline samples and analyzed. A total of 106 bacterial 16S rDNA sequences were derived from DGGE bands, and these formed three major clusters: beta and gamma subdivisions of Proteobacteria and gram-positive bacteria. The most frequently encountered bacterial species was Comamonas denitrificans, which was not previously reported to be associated with microbial communities found in gas pipelines or with microbially influenced corrosion. The 31 archaeal 16S rDNA sequences obtained in this study were all related to those of methanogens and phylogenetically fall into three clusters: order I, Methanobacteriales; order III, Methanomicrobiales; and order IV, Methanosarcinales. Further microbial ecology studies are needed to better understand the relationship among bacterial and archaeal groups and the involvement of these groups in the process of microbially influenced corrosion in order to develop improved ways of monitoring and controlling microbially influenced corrosion.
Corrosion is a leading cause of pipe failure and is a main component of the operating and maintenance costs of gas industry pipelines (3, 10, 18, 23, 30, 31, 42-44, 54). Quantifying the cost of corrosion generally, and more specifically the cost associated with microbial corrosion, in the gas industry is not easily done and is controversial. Pipeline corrosion was estimated in 1996 to cost the gas industry about $840 million/year (10), and in 2001 it was estimated that the annual cost of all forms of corrosion to the oil and gas industries was $13.4 billion, of which microbially influenced corrosion accounted for about $2 billion (31). While it is well recognized that chemical and microbial mechanisms both contribute to corrosion, it is uncertain what the relative contribution of microbial activity to overall pipe corrosion is. It has been estimated that 40% of all internal pipeline corrosion in the gas industry can be attributed to microbial corrosion (23, 44), but data are needed to confirm or revise this estimate. Basic research to increase our understanding of the microbial species involved in microbial corrosion and their interactions with metal surfaces and with other microorganisms will be the basis for the development of new approaches for the detection, monitoring, and control of microbial corrosion. A thorough knowledge of the causes of microbially influenced corrosion and an efficient and effective means of detecting and preventing corrosion are lacking. It is well recognized that microorganisms are a major cause of corrosion of metal pipes, but despite decades of study it is still not known with certainty how many species of microorganisms contribute to corrosion, how to reliably detect their presence prior to corrosion events, or how to rapidly assess the efficacy of biocides and mitigation procedures (2, 5, 17, 18, 23, 30, 42, 43, 54).
Investigations of microbial species present in gas industry pipelines have traditionally relied upon the use of samples obtained from pipelines to grow bacterial cultures in the laboratory (42). Laboratory growth media cannot accurately reflect the true conditions within pipelines, and microbiologists have recognized that the vast majority of microbial species cannot currently be grown in the laboratory (35, 61); thus, culture-dependent approaches underestimate the biocomplexity of microbial communities. The purpose of this study was to apply molecular techniques to investigate the microbial species found in gas pipeline liquids or in biofilms attached to metal surfaces that exhibited corrosion. These data allow a better appreciation of the compositions and variability of gas pipeline microbial communities and may contribute to new and improved ways to detect, monitor, and control microbial corrosion of gas industry pipelines.
MATERIALS AND METHODS
Origin of gas pipeline samples.
Eight samples were received from three gas companies. Six liquid samples (A1, A2, B1, B2, S3, and S4) were centrifuged for collection of the biomass. Two samples (S2 and S5) were inoculated into sulfate-reducing bacterium (SRB) medium (American Type Culture Collection culture medium 292) containing a submerged metal coupon (1/16-in.-thick C1018 mild steel, 7.87 g/cm3; Metal Samples Co., Munford, Ala.) (25). After incubation for 50 days at room temperature in an anaerobic chamber, the biomass on the coupon surface was scraped off with a sterile razor blade. Biomass derived from SRB medium inoculated with sample S2 was recovered by two-step centrifugation and designated sample S2L. Iron sulfide was first removed by centrifugation at 1,000 × g for 5 min, and then biomass was recovered from the supernatant by centrifugation at 10,000 × g for 15 min. This sample was used only for characterization of archaea. Sample B2 was also used in a laboratory reactor to mimic the gas pipeline environment. Six metal coupons were placed in a 2-in.-diameter pipe above the liquid sample and exposed to a low flow rate (1/10 vol/min) of humidified technical grade natural gas at an ambient temperature and pressure. After a 1-month exposure, the biomass was scraped off from the coupons with a razor blade and the sample was designated B2R. All collected biomass was washed three times with PBS (10 mM phosphate [pH 7.4], 2.7 mM KCl, and 137 mM NaCl) and stored at −80°C until DNA extraction (59, 61). The metal coupons were also examined by scanning electron microscopy to visualize biofilm growth and detect corrosion of the metal surface beneath biofilms (21).
PCR and DGGE analysis of bacterial 16S rRNA genes.
For samples S2, S5, and A2, V2-V3 variable regions of eubacterial 16S rDNA corresponding to positions 101 to 518 in Escherichia coli (9) were directly amplified with primer pair BA101F-GC-BA518R (52). The primer BA101F-GC included a GC clamp at the 5′ end (37). The sequences of primers used in this study (Table 1) were synthesized by MWG Biotech. The amplifications were performed with a Mastercycler gradient thermocycler (Eppendorf AG, Hamburg, Germany). Fifty microliters of PCR mixture contained 1 μl of template DNA, 400 nM (each) primers, 1× high fidelity buffer with Mg2+, 200 μM (each) deoxynucleoside triphosphates, and 1 U of TripleMaster enzyme mix (Brinkman Instruments Inc., Westbury, N.Y.). After 5 min of initial denaturation at 94°C, TripleMaster enzyme mix was added to the reaction mixture, and then a “touchdown” PCR was performed (13a, 61). The annealing temperature was decreased by 0.5°C per cycle from 57 to 47°C, at which temperature 10 additional cycles were carried out. Amplification was performed with 1 min of denaturation at 94°C, 1 min of primer annealing, and 2 min of primer extension at 72°C, followed by 7 min of final primer extension.
TABLE 1.
Primers used for amplification of 16S rRNA genes
| Primera | Sequence (5′ to 3′) | Positionb | Specificity | Reference(s) |
|---|---|---|---|---|
| BA8F | AGTTTGATCCTGGCTCAG | 8-25 | Bacteria | 49 |
| UN1492R | GGYTACCTTGTTACGACTT | 1474-1492 | Universal | 49 |
| BA101F-GC | CGCCCGCCGCGCCCCGCGCCCGTCCCGCC | |||
| GCCCCCGCCCGTGGCGGACGGGTGAGTAA | 101-118 | Bacteria, V2-V3 region | 37, 52 | |
| BA518R | CGTATTACCGCGGCTGCTGG | 499-518 | Bacteria, V2-V3 region | 52 |
| AR21F | TCCGGTTGATCCYGCCGG | 21-38 | Archaea | 48 |
| AR340F | CCCTACGGGGYGCASCAG | 340-358 | Archaea, V3-V6 region | 39 |
| AR1100R | YGGGTCTCGCTCGTTRCC | 1083-1100 | Archaea, V3-V6 region | 39 |
F, forward primer; R, reverse primer.
Position in the 16S rRNA gene of E. coli (9).
For samples A1, B1, B2, B2R, S3, and S4, no PCR products were obtained from direct amplification with primers BA101F-GC and BA518R; therefore, nested PCR was applied to these samples. The templates were first amplified with primer pair BA8F-UN1492R (49) with the following program: 94°C for 5 min; 30 cycles of denaturation at 94°C for 45 s, annealing at 46°C for 30 s, and extension at 72°C for 1 min; and a single final extension at 72°C for 7 min. Then 0.5 μl of these first PCR products was used as a template to amplify the V2-V3 regions of 16S rDNA with primer pair BA101F-GC and BA518R.
Four hundred nanograms of purified PCR products was loaded onto an 8% (wt/vol) polyacrylamide gel with denaturing gradients ranging from 35 to 60% (100% denaturant contains 7 M urea and 40% formamide). Denaturing gradient gel electrophoresis (DGGE) was performed in 1× TAE buffer (40 mM Tris, 20 mM acetate, 1 mM EDTA [pH 8.0]) on a Dcode universal mutation detection system (Bio-Rad Laboratories, Hercules, Calif.) at 100 V at 60°C overnight. DNA bands were visualized by silver staining, and DNA from individual bands was eluted into 30 μl of 0.1× TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0) at 4°C overnight. Three microliters of eluate was reamplified with primers and under conditions described above, but 0.1% bovine serum albumin was added to the PCR mixture. The amplified products were run on DGGE gels to ascertain their mobility and then sequenced (SeqWright, Inc., Houston, Tex.) by using primer BA518R. The sequence data were inspected for the presence of ambiguous base assignments and subjected to the Check Chimera program from the Ribosomal Database Project (35) before the sequences were submitted for BLAST searches (1). The sequences were aligned by using Clustal_X version 1.81 (56), and phylogenetic trees were constructed by using the neighbor-joining method (51) with 1,000 bootstrap replicates and viewed by using TreeView (40). For classification into phylogenetic groups, sequences were entered into the Sequence Match program (version 2.7) from the Ribosomal Database Project (35).
PCR and clone library construction of archaeal 16S rRNA genes.
Nested PCR products of archaeal 16S rRNA genes from primer pairs AR46F-AR1100R and AR340F-UN519R (39) were subjected to DGGE analysis.
In addition, nested PCR products of the V3-V6 variable regions of 16S rRNA genes (9) from primer pairs AR21F-UN1492R (48, 49) and AR340F-AR1100R (24, 39) were cloned into pGEM-T Easy cloning vector (Promega Corp., Madison, Wis.). The PCR conditions were the same as those for amplification of bacterial sequences, except that 5% (wt/vol) acetamide was added in the first PCR mixture to minimize nonspecific amplification from bacterial templates present in the samples (39). Fifty white colonies were randomly selected, and DNA inserts were amplified with M13 forward and M13 reverse primers. The amplified insert was digested with restriction enzyme HaeIII, and the resulting restriction enzyme fragment patterns were compared visually on 3% NuSieve agarose gel (BioWhittaker Molecular Applications, Rockland, Maine). DNA sequence data was generated from unique clones by using M13 forward primer and analyzed as described above.
Nucleotide sequence accession numbers.
Bacterial and archaeal 16S rDNA sequences obtained in this study were deposited with GenBank and are available under accession numbers AY256577 to AY256647.
RESULTS AND DISCUSSION
Characterization of bacterial communities in gas pipelines by DGGE and 16S rRNA gene sequencing.
The DGGE gel in Fig. 1 illustrates the variety of 16S rRNA gene fragments amplified from six gas pipeline liquids (A1, A2, B1, B2, S3, and S4) and three metal coupon biofilm samples (S2, S5, and B2R) analyzed in this study. Informative comparisons are of samples S3 and S4 versus S2 and S5, B2 versus B2R, and A1 and A2 versus B1 and B2 versus S3 and S4.
FIG. 1.
DGGE profiles of PCR-amplified V2-V3 variable regions of eubacterial 16S rRNA genes from gas industry pipeline samples.
The S samples are derived from the same gas pipeline company. The DGGE patterns produced by biofilm samples S2 and S5 appear to be similar to each other but are somewhat different from the patterns produced by pipeline liquid samples S3 and S4 and also appear to be less complex (fewer dominant bands are apparent). There is some similarity between the DGGE patterns of pipeline liquid sample B2 and biofilm sample B2R from a laboratory reactor, but each appears to have several unique bands. The analysis of the metal coupon from sample B2R by scanning electron microscopy indicated that the growth of biofilm was associated with corrosion of the metal surface (Fig. 2). The DGGE band patterns of samples B1 and B2 show significant differences although the samples came from the same pipeline company. The DGGE patterns of the A samples are likewise very different from each other and from those of the samples derived from the other two pipeline companies. The appearance of the DGGE band patterns suggests that the microbial communities in different pipelines are rather different and that perhaps the greatest biodiversity is found in sample A2.
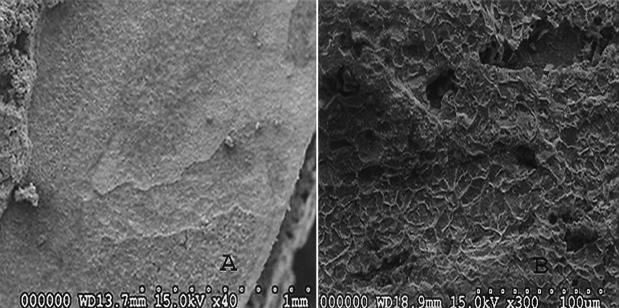
FIG. 2.
Scanning electron microscopy indicated that the bacterial cultures obtained from gas industry pipelines were capable of forming biofilms on metal coupons, and corrosion of the metal coupons occurred beneath these biofilms. Panel A illustrates the intact biofilm at the top left corner and the metal surface beneath. Panel B illustrates the corrosion of the metal coupon under higher magnification.
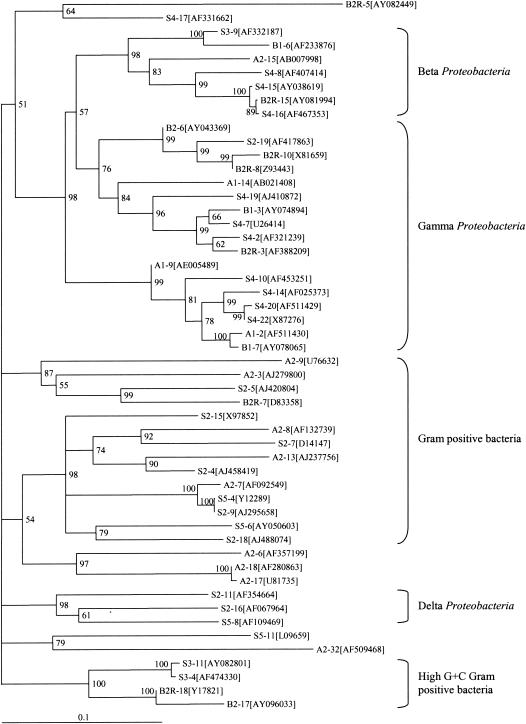
A total of 106 bacterial 16S rDNA sequences derived from DGGE bands of nine pipeline samples were determined and summarized in Table 2. Eighty-six sequences were more than 98% identical to sequences already deposited in the GenBank database, while 20 sequences showed 97% or less identity with sequences in the database. It is generally assumed that an accurate identification of a 16S rRNA gene to the species level requires a 98% or better match to a sequence in the database, while identification to the genus level requires a 97% or better match (27, 35, 53). DNA sequences that show lower similarity to those in the database may be derived from previously uncultivated or unknown bacterial species. Of the 106 bacterial 16S rDNA sequences analyzed in this study, some were detected multiple times. Fifty-two unique sequences were used as representatives to construct a phylogenetic tree by using the maximum likelihood method (Fig. 3). The phylogenetic analysis showed that the sequences retrieved in this study formed three major phylogenetic clusters: beta and gamma subdivisions of Proteobacteria and gram-positive bacteria. Two smaller clusters of high-G+C-content gram-positive bacteria and the delta subdivision of Proteobacteria were also observed. Forty-three of 106 sequences determined in this study were closely related to those of the gamma subdivision of Proteobacteria (mainly pseudomonads and enteric organisms, with 95 to 100% similarity to the sequences in GenBank), 20 sequences belonged to beta Proteobacteria (Acidovorax and Bordetella groups, with more than 96% similarity), and 20 sequences were related to those of gram-positive bacteria (mainly Eubacterium and Clostridium).
TABLE 2.
Bacterial 16S rDNA sequences obtained from natural gas pipeline samples
| Closest relative in GenBank (accession no.) | No. of sequences derived from DGGE bands for sample:
|
% Identity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S3 | S4 | S2 | S5 | B1 | B2 | B2R | A1 | A2 | ||
| Acetobacterium psammolithicum (AF132739) | 1 | 96 | ||||||||
| Acidovorax delafieldii isolate N7-28 (AF332187) | 1 | 100 | ||||||||
| Acinetobacter junii (AF417863) | 2 | 1 | 1 | 1 | 97-100 | |||||
| Acinetobacter sp. (DSM590) (X81659) | 1 | 99 | ||||||||
| Acinetobacter sp. (ATCC17924) (Z93443) | 1 | 98 | ||||||||
| Acinetobacter sp. strain AU1523 (AY043369) | 1 | 99 | ||||||||
| Anaerofilum pentosovorans (X97852) | 1 | 1 | 1 | 98 | ||||||
| Bacteroides sp. strain 253c (AY082449) | 1 | 2 | 99 | |||||||
| Citrobacter werkmanii (AF025373) | 1 | 99 | ||||||||
| Clostridium acidisoli strain CK74 (AJ237756) | 1 | 92 | ||||||||
| Clostridium algidixylanolyticum (AF092549) | 1 | 98 | ||||||||
| Clostridium butyricum strain E5 (AJ458419) | 1 | 2 | 98-100 | |||||||
| Clostridium sp. (Y12289) | 1 | 99 | ||||||||
| Clostridium sp. strain IrT-JG1-67 (AJ295658) | 1 | 99 | ||||||||
| Comamonas denitrificans strain 110 (AF233876) | 2 | 3 | 1 | 2 | 1 | 2 | 96-100 | |||
| Desulfovibrio aminophilus (AF067964) | 2 | 1 | 95-98 | |||||||
| Desulfovibrio desulfuricans (AF354664) | 1 | 99 | ||||||||
| Desulfovibrio sp. strain zt10e (AF109469) | 1 | 94 | ||||||||
| Enterococcus casseliflavus strain CECT969T (AJ420804) | 1 | 100 | ||||||||
| E. coli (AF511430) | 1 | 1 | 6 | 3 | 6 | 1 | 96-99 | |||
| E. coli O157:H7 (AE005489) | 1 | 99 | ||||||||
| E. coli O39:NM (AY078065) | 2 | 99-100 | ||||||||
| Geotoga aestuarianus strain T3B (AF509468) | 2 | 99-100 | ||||||||
| Halanaerobium congolense (U76632) | 1 | 99 | ||||||||
| Klebsiella pneumoniae (AF453251) | 1 | 95 | ||||||||
| Klebsiella pneumoniae (AF511429) | 1 | 2 | 99-100 | |||||||
| Klebsiella pneumoniae (X87276) | 1 | 99 | ||||||||
| Microbacterium sp. strain 44574 (AY082801) | 1 | 99 | ||||||||
| Microbacterium testaceum strain CE648 (AF474330) | 1 | 98 | ||||||||
| Peptostreptococcus indolicus (D14147) | 1 | 99 | ||||||||
| Propionibacterium sp. strain V07/12348 (Y17821) | 2 | 97 | ||||||||
| Propionibacterium sp. strain WJ6 (AY096033) | 1 | 1 | 100 | |||||||
| Pseudomonas aeruginosa strain H13 (AY074894) | 1 | 2 | 99-100 | |||||||
| Pseudomonas iners (AB021408) | 1 | 99 | ||||||||
| Pseudomonas sp. strain NZ066 (AF388209) | 1 | 99 | ||||||||
| Pseudomonas sp. strain PM-2001 (AF321239) | 1 | 100 | ||||||||
| Pseudomonas stutzeri strain DNSP21 (U26414) | 1 | 98 | ||||||||
| Pseudomonas stutzeri strain YPF-41 (AJ410872) | 1 | 99 | ||||||||
| Ralstonia sp. strain MBIC3838 (AB007998) | 1 | 99 | ||||||||
| Rhizobium sp. strain RM1-2001 (AF331662) | 1 | 99 | ||||||||
| Sporomusa ovata (DSM2662) (AJ279800) | 2 | 98-99 | ||||||||
| Staphylococcus auricularis (ATCC33753T) (D83358) | 1 | 99 | ||||||||
| Sulfurospirillum sp. strain 18.1 (AF357199) | 3 | 91-92 | ||||||||
| Thermus thermophilus HB27 (L09659) | 1 | 100 | ||||||||
| Uncultured bacterium clone GOUTA13 (AY050603) | 1 | 93 | ||||||||
| Uncultured bacterium clone IA-23 (AJ488074) | 2 | 94-95 | ||||||||
| Uncultured bacterium clone KRA30-58 (AY081994) | 1 | 99 | ||||||||
| Uncultured bacterium clone RB13C11 (AF407414) | 1 | 1 | 98-99 | |||||||
| Uncultured bacterium mle1-42 (AF280863) | 1 | 89 | ||||||||
| Uncultured beta proteobacterium clone 1518 (AF467353) | 1 | 100 | ||||||||
| Uncultured eubacterium clone GL182.6 (AY038619) | 1 | 1 | 1 | 98-99 | ||||||
| Unidentified eubacterium clone vadinHA73 (U81735) | 1 | 89 | ||||||||
| Total | 7 | 15 | 15 | 9 | 7 | 12 | 17 | 9 | 15 | |
FIG. 3.
Neighbor-joining phylogenetic tree constructed by using bacterial 16S rRNA gene sequences retrieved from DGGE bands of gas industry pipeline samples and amplified with primer pair BA101F-GC and BA518R. Numbers at nodes represent the percentages of occurrence of nodes in 1,000 bootstrap trials. Only bootstrap values greater than 50% are indicated. GenBank accession numbers of sequences of the most closely related bacteria are shown in brackets. The scale bar represents the expected number of substitutions per nucleotide position.
The appearance of the DGGE gel shown in Fig. 1 suggested that the biocomplexity of sample S2 may be relatively low, but the results of DNA sequencing experiments demonstrate otherwise, as shown in Table 2. Samples S2, S4, and B2R appear to have the greatest biocomplexity, with 12 different species each, as determined by sequencing of DNA bands derived from DGGE gels (Table 2). Samples A1 and B1 appear to have the least biocomplexity as determined by DNA sequencing, as only four unique species were detected in each sample. This contrasts sharply with the appearance of the DGGE gel, which shows multiple bands for each sample. Since individual bands eluted from DGGE gels were used to obtain DNA sequence data, the apparent discrepancy in sample diversity as indicated by DGGE versus DNA sequence data cannot be explained by cloning bias. These results are most easily explained by the fact that most bacterial species contain multiple copies of the 16S rRNA gene in their chromosomes (7) so that pure bacterial cultures can give rise to several bands with different mobilities in DGGE analyses. It is also frequently observed that the most abundant bacterial species present in a given environment may be a group of closely related bacterial species or strains (20, 36) so that while a large number of bands are observed in DGGE analysis the actual number of unique species detected by DNA sequence analysis is much smaller. This indicates that the appearance of DGGE gels may have some use as a quick diagnostic tool to determine whether a given sample has a “fingerprint” matching that of a known environmental sample but that the results of DGGE gels are not useful in predicting the biocomplexity of environmental samples in terms of the bacterial species represented by unique 16S rDNA sequences.
Of particular interest are those bacterial species that were detected in multiple environmental samples and may therefore represent bacterial species commonly found in gas pipelines. Comamonas denitrificans and E. coli were the most frequently detected species, occurring in six of the nine samples and often with more than one example of these sequences within individual environmental samples. The frequent occurrence of Comamonas denitrificans in these samples is unexpected as this species has not previously been reported to be associated with microbially influenced corrosion or found in gas pipelines. This finding suggests that nitrogen metabolism may have a major impact on the composition of bacterial communities within pipeline environments and warrants further investigation, particularly as recent research indicates that the presence of nitrate (6 to 10 mM) can result in increased metal corrosion rates (38). The frequent occurrence of E. coli sequences could simply be an artifact derived from trace amounts of E. coli DNA present in reagents and the laboratory environment. However, the care taken in sample preparation and the isolation of three distinct E. coli sequences suggests that E. coli is actually present in some gas pipeline samples.
Pseudomonads and Acinetobacter species were found in five and four of the nine samples, respectively. Pseudomonads and Acinetobacter have minimal nutritional requirements and are often present in aquatic environments that are rich in organic pollutants such as gasoline and solvents, etc. (33, 57, 58). In addition, pseudomonads contribute to biofilm formation by producing exopolysaccharides and facilitating the attachment of other microorganisms (34, 41) and hence accelerate the corrosion process (5). Pseudomonads are also capable of both complete and incomplete denitrification (6, 12, 14). Propionibacterium, Clostridium, and Anaerofilum were each detected in three of nine samples, and Bacteroides, Desulfovibrio, and Klebsiella were each found twice. Clostridium butyricum, Clostridium algidixylanolyticum, Anaerofilum pentosovorans, Bacteroides sp., Acinetobacter sp., and Propionibacterium sp. produce organic acids such as acetic, butyric, formatic, lactic, succinic, and propionic acid that may contribute to corrosion (8, 22, 28, 58, 60). Desulfovibrio desulfuricans and Desulfovibrio aminophilus are two important SRB that play an important role in metal corrosion (15, 16, 60) by reducing sulfate and/or sulfur to hydrogen sulfide (H2S) (4). Hydrogen sulfide reacts with iron to form black ferrous sulfide (FeS); it also reacts with water to produce an acid condition, accelerating the corrosion process.
The most pronounced differences observed in bacterial species when S2 and S5 (biofilms from metal coupons submerged in SRB medium) and S3 and S4 (pipeline liquids) are compared is the presence of Anaerofilum pentosovorans, Clostridium species, and Desulfovibrio species in samples S2 and S5 but not in S3 and S4. It is important to note that SRB, such as Desulfovibrio species, were detected only in samples that had been enriched in bacterial growth medium appropriate for SRB. Previous microbiological studies have suggested that SRB play a key role in microbially influenced corrosion (16, 23, 25, 42), yet SRB were not detected in the genetic analysis of liquids directly obtained from gas pipelines in this study. These results do not demonstrate that SRB were not present in these pipeline liquids. Rather, these results demonstrate that SRB are present and can be detected after enrichment for their growth but that they were not present among the most abundant bacterial species in these gas pipeline liquids. It is also worth noting that no SRB were detected in sample B2R, which is a sample of biofilm obtained from a metal coupon that exhibited corrosion. This demonstrates that while SRB are indeed contributors to microbially influenced corrosion, they need not be present in abundance in all microbial communities responsible for microbially influenced corrosion. The presence of the acid-forming bacterial species Clostridium butyricum and Anaerofilum pentosovorans in samples S2 and S5, but not in S3 and S4, suggests that acid-forming bacteria may play a key role in corrosion, and these species are more abundant in biofilms formed on metal coupons submerged in SRB medium than in the planktonic bacterial population in gas pipelines. Klebsiella pneumoniae, which was detected only in samples S3 and S4, is a facultative anaerobe, often present in soil and water, that can fix nitrogen under anaerobic or microaerobic conditions (13). It produces nitrates and/or nitric acid that may contribute to the corrosion of metal. Clostridium acidisoli detected in sample A2 has a function similar to that of Klebsiella pneumoniae (32). Those bacterial species found in biofilm sample B2R, but not in the planktonic sample B2, include Anaerofilum pentosovorans, Bacteroides, Pseudomonas sp., and Staphylococcus auricularis. These results lend further evidence to the hypothesis that acid-forming bacteria play a key role in microbially influenced corrosion.
In addition to the denitrifying bacteria, SRB, and acid-forming bacteria, we also retrieved rDNA sequences from bacterial species capable of various biodegradation processes. We retrieved sequences closely related to those of thiosulfate- or sulfur-reducing anaerobes (Geotoga aestuarianus, Halanaerobium congolense, and Sulfurospirillum sp.) (11, 47), tetrachloroethene-degrading anaerobes (Sporomusa ovata) (55), triethanolamine-degrading bacteria (Acetobacterium sp.) (19), poly(3-hydroxybutyrate-co-3-hydroxyvalerate)-degrading denitrifiers (Acidovorax sp., Pseudomonas sp., and Comamonas sp.) (29), and xylan-degrading bacteria (Clostridium algidixylanolyticum) (8). The nature of the pipeline environment justified the presence of biodegrading bacteria. Since it is not clear whether the pipelines were undergoing corrosion when the samples were taken, it is premature to conclude which types of bacteria detected in this study play a role in microbial corrosion. The data in Table 2 also show that the traditional analysis of gas pipeline samples by using cultivation in growth medium for specific types of bacteria may yield misleading results, as our retrieved sequences showed that the dominant species in the gas pipeline environment and those from SRB growth medium are often different.
Characterization of archaeal communities in gas pipelines by cloning and sequencing of 16S rRNA genes.
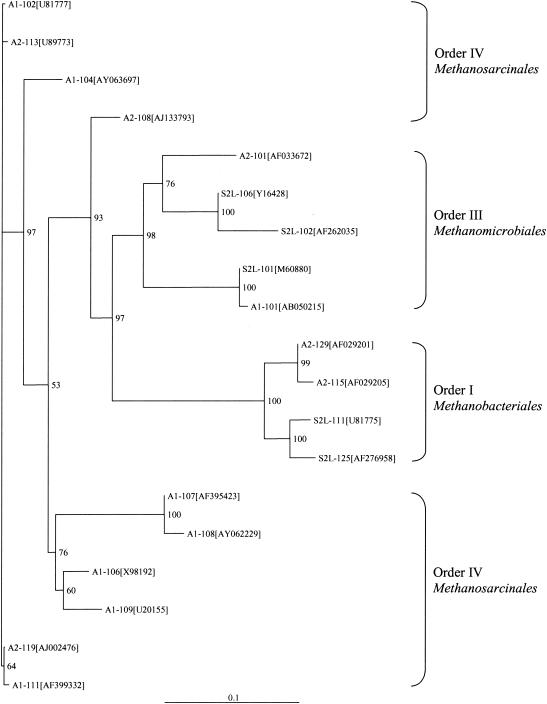
The PCR-DGGE approach to characterizing archaeal populations in gas pipeline samples failed to generate clear and sharp DGGE bands (data not shown). Therefore, cloning of nested PCR products with restriction enzyme screening was used in this study. Archaeal sequences were amplified in only three out of 10 samples (A1, A2, and S2L), and the sequence data were summarized in Table 3. Of a total of 31 archaeal 16S rDNA sequences (11 from A1, 10 from A2, and 10 from S2L) retrieved, 22 were more than 97% identical to the sequences in GenBank while nine sequences showed 90 to 96% identity to those in GenBank, indicating that they may be from currently unknown species (27, 53). Some sequences were observed more than once, so the 19 unique sequences from these three samples were used for the construction of a phylogenetic tree by using the maximum likelihood method (Fig. 4). The result indicated that all the sequences in archaeal libraries from gas pipeline samples obtained in this study are related to those of methanogenic archaea. They formed three phylogenetic clusters, corresponding to three orders: order I, Methanobacteriales; order III, Methanomicrobiales; and order IV, Methanosarcinales (35). Most of the sequences (13 of 17) in order IV belong to the family Methanosarcinaceae, the members of which can form methane from a variety of substrates, such as acetate, H2 and CO2, methanol, and methylamines (45, 46); four sequences belong to the family Methanosaetaceae, whose members can use only acetate as a substrate to produce methane and CO2. The families Methanomicrobiaceae and Methanospirillaceae account for five and three sequences in order III, respectively. Methanomicrobiaceae are limited to the use of H2 and CO2 and formate as substrates to form methane and water. All six sequences retrieved in order I belong to the family Methanobacteriaceae, the members of which use H2 and CO2 and/or formate as substrates for methanogenesis (45, 46, 50).
TABLE 3.
Archaeal 16S rDNA sequences obtained from natural gas pipeline samples
| Closest relative in GenBank (accession no.) | Retrieved from sample:
|
% Identity | ||
|---|---|---|---|---|
| A1 | A2 | S2L | ||
| Methanobacterium curvum (AF276958) | + | 99 | ||
| Methanocalculus halotolerans (AF033672) | + | 99 | ||
| Methanoculleus sp. clone A3 (AJ133793) | + | 90 | ||
| Methanofollis liminatans (DSM4140) (Y16428) | + | + | + | 95-98 |
| Methanofollis sp. strain N2F9704 (AF262035) | + | 94 | ||
| Methanohalophilus euhalobius (X98192) | + | + | 96 | |
| Methanolobus vulcani (U20155) | + | 97 | ||
| Methanosarcina barkeri strain CM1 (AJ002476) | + | + | + | 99 |
| Methanosarcina siciliae (U89773) | + | + | 99 | |
| Methanospirillum hungatei (M60880) | + | + | 96 | |
| Uncultured archaeon Arc No. 5 (AF395423) | + | + | + | 96-99 |
| Uncultured archaeon clone GW70-10-2 (AY062229) | + | 98 | ||
| Uncultured archaeon clone RS500-12 (AY063697) | + | 96 | ||
| Uncultured archaeon SAGMA-J2 (AB050215) | + | 99 | ||
| Uncultured Methanosarcinaceae archaeon Gap-A19 (AF399332) | + | 99 | ||
| Unidentified archaeon clone vadinCA25 (U81777) | + | + | 99 | |
| Unidentified archaeon clone vadinDC06 (U81775) | + | 98 | ||
| Unidentified methanogen ARC43 (AF029201) | + | 97 | ||
| Unidentified methanogen ARC62 (AF029205) | + | + | + | 97 |
FIG. 4.
Neighbor-joining phylogenetic tree constructed by using archaeal 16S rRNA gene sequences amplified with primer pair AR340F and AR1100R from clone libraries of gas industry pipeline samples. Numbers at nodes represent the percentages of occurrence of nodes in 1,000 bootstrap trials. Only bootstrap values greater than 50% are indicated. GenBank accession numbers of sequences of the most closely related archaea are shown in brackets. The scale bar represents the expected number of substitutions per nucleotide position.
Hydrogen-consuming organisms, such as methanogens, are capable of accelerating corrosion by cathodic depolarization (a process which pulls the cathodic reduction of protons by removal of the product and thereby accelerates anodic metal dissolution) (2, 26); therefore, more information regarding the abundance of methanogens in gas pipeline biofilms may help us to better understand microbially influenced corrosion. It was unexpected that the only archaeal sequences detected in natural gas pipeline samples were those of methanogens, since methane is the main component of natural gas. The difficulty in amplifying archaeal 16S rDNA sequences from natural gas pipeline samples suggests that they are probably present in low numbers, but further research is required before the archaeal community in this environment can be fully described.
The results reported here demonstrate that molecular genetic analyses are capable of providing more in-depth analyses of the composition of microbial communities in gas pipelines than was previously possible, but further study is needed to determine which organisms play a key role in microbial corrosion. Research is currently under way to establish correlations between the presence of various types of microorganisms in complex biofilms and metal corrosion rates. It is likely that with an improved understanding of the compositions and variability of microbial communities present in gas pipelines we will be able to develop better means of monitoring and preventing microbially influenced corrosion.
Acknowledgments
We thank Kevin J. Kayser for helpful discussions.
This work was supported by the Gas Research Institute under contract no. 8473.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Angell, P. 1999. Understanding microbially influenced corrosion as biofilm-mediated changes in surface chemistry. Curr. Opin. Biotechnol. 10:269-272. [DOI] [PubMed] [Google Scholar]
- 3.Angostini, R. A., and R. D. Young. 1990. A case history: investigations of microbially influenced corrosion in a west Texas waterflood, p. 1-14. In Proceedings of the NACE Annual Conference, Corrosion/90, paper 119. NACE International, Houston, Tex.
- 4.Baena, S., M. L. Fardeau, M. Labat, B. Ollivier, J. L. Garcia, and B. K. Patel. 1998. Desulfovibrio aminophilus sp. nov., a novel amino acid degrading and sulfate reducing bacterium from an anaerobic dairy wastewater lagoon. Syst. Appl. Microbiol. 21:498-504. [DOI] [PubMed] [Google Scholar]
- 5.Batista, J. F., R. F. Pereira, J. M. Lopes, M. F. Carvalho, M. J. Feio, and M. A. Reis. 2000. In situ corrosion control in industrial water systems. Biodegradation 11:441-448. [DOI] [PubMed] [Google Scholar]
- 6.Bothe, H., G. Jost, M. Schloter, B. B. Ward, and K. Witzel. 2000. Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol. Rev. 24:673-690. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, S. L., V. R. Flechtner, and J. R. Johansen. 2001. Is the 16S-23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol. Biol. Evol. 18:1057-1069. [DOI] [PubMed] [Google Scholar]
- 8.Broda, D. M., D. J. Saul, R. G. Bell, and D. R. Musgrave. 2000. Clostridium algidixylanolyticum sp. nov., a psychrotolerant, xylan-degrading, spore-forming bacterium. Int. J. Syst. Evol. Microbiol. 50(Pt. 2):623-631. [DOI] [PubMed] [Google Scholar]
- 9.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 10.Buck, E., G. C. Maddux, and R. L. Sullivan. 1996. Internal corrosion cost impact study—United States natural gas exploration and production industry. GRI-96/0056 document no. 96-1466. Gas Research Institute, Des Plaines, Ill.
- 11.Campbell, B. J., C. Jeanthon, J. E. Kostka, G. W. Luther III, and S. C. Cary. 2001. Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 67:4566-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuypers, H., and W. G. Zumft. 1993. Anaerobic control of denitrification in Pseudomonas stutzeri escapes mutagenesis of an fnr-like gene. J. Bacteriol. 175:7236-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon, R., R. R. Eady, G. Espin, S. Hill, M. Iaccarino, D. Kahn, and M. Merrick. 1980. Analysis of regulation of Klebsiella pneumoniae nitrogen fixation (nif) gene cluster with gene fusions. Nature 286:128-132. [DOI] [PubMed] [Google Scholar]
- 13a.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. [DOI] [PMC free article] [PubMed]
- 14.Drysdale, G., H. Kasan, and F. Bux. 1999. Denitrification by heterotrophic bacteria during activated sludge treatment. Water SA 25:357-362. [Google Scholar]
- 15.Dzierzewicz, Z., B. Cwalina, L. Weglarz, and S. Glab. 1992. Isolation and evaluation of corrosive aggressivity of wild strains of sulfate reducing bacteria. Acta Microbiol. Pol. 41:211-221. [Google Scholar]
- 16.Dzierzewicz, Z., B. Cwalina, E. Chodurek, and T. Wilczok. 1997. The relationship between microbial metabolic activity and biocorrosion of carbon steel. Res. Microbiol. 148:785-793. [DOI] [PubMed] [Google Scholar]
- 17.Emde, K. M. E., D. W. Smith, and R. Facey. 1992. Initial investigation of microbially influenced corrosion (MIC) in a low temperature water distribution system. Water Res. 26:169-175. [Google Scholar]
- 18.Farthing, S. 1997. Company combats MIC with aggressive control program. Pipe Line Gas Ind. 80(10):43-47. [Google Scholar]
- 19.Frings, J., C. Wondrak, and B. Schink. 1994. Fermentative degradation of triethanolamine by a homoacetogenic bacterium. Arch. Microbiol. 162:103-107. [DOI] [PubMed] [Google Scholar]
- 20.Fuhrman, J., and L. Campbell. 1998. Microbial microdiversity. Nature 393:410-411. [Google Scholar]
- 21.Geiger, S. L., T. J. Ross, and L. L. Barton. 1993. Environmental scanning electron microscope (ESEM) evaluation of crystal and plaque formation associated with biocorrosion. Microsc. Res. Tech. 25:429-433. [DOI] [PubMed] [Google Scholar]
- 22.Gibson, G. R., and X. Wang. 1994. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J. Appl. Bacteriol. 77:412-420. [DOI] [PubMed] [Google Scholar]
- 23.Graves, J. W., and E. H. Sullivan. 1996. Internal corrosion in gas gathering systems and transmission lines. Mater. Prot. 5:33-37. [Google Scholar]
- 24.Grosskopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton, W. A. 1985. Sulphate-reducing bacteria and anaerobic corrosion. Annu. Rev. Microbiol. 39:195-217. [DOI] [PubMed] [Google Scholar]
- 26.Horn, J., and D. Jones. 2002. Microbiologically influenced corrosion: perspectives and approaches, p. 1072-1083. In C. Hurst, R. Crawford, G. Knudsen, M. McInerney, and L. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 27.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 28.Kanauchi, O., Y. Fujiyama, K. Mitsuyama, Y. Araki, T. Ishii, T. Nakamura, Y. Hitomi, K. Agata, T. Saiki, A. Andoh, A. Toyonaga, and T. Bamba. 1999. Increased growth of Bifidobacterium and Eubacterium by germinated barley foodstuff, accompanied by enhanced butyrate production in healthy volunteers. Int. J. Mol. Med. 3:175-179. [DOI] [PubMed] [Google Scholar]
- 29.Khan, S. T., Y. Horiba, M. Yamamoto, and A. Hiraishi. 2002. Members of the family Comamonadaceae as primary poly(3-hydroxybutyrate-co-3-hydroxyvalerate)-degrading denitrifiers in activated sludge as revealed by a polyphasic approach. Appl. Environ. Microbiol. 68:3206-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kholodenko, V. P., S. K. Jigletsova, V. A. Chugnov, V. B. Rodin, V. S. Kobelev, and S. V. Karpov. 2000. Chemicomicrobiological diagnostics of stress corrosion cracking of trunk pipelines. Appl. Biochem. Microbiol. 36:594-601. [Google Scholar]
- 31.Koch, G. H., M. P. H. Brongers, N. G. Thompson, Y. P. Virmani, and J. H. Payer. 2001. Corrosion costs and preventive strategies in the United States. FHWA-RD-01-156. [Online.] Federal Highway Administration, Washington, D.C. http://www.corrosioncost.com/.
- 32.Kuhner, C. H., C. Matthies, G. Acker, M. Schmittroth, A. S. Gossner, and H. L. Drake. 2000. Clostridium akagii sp. nov. and Clostridium acidisoli sp. nov.: acid-tolerant, N2-fixing clostridia isolated from acidic forest soil and litter. Int. J. Syst. Evol. Microbiol. 50:873-881. [DOI] [PubMed] [Google Scholar]
- 33.Leahy, J., A. Byrne, and R. Olsen. 1996. Comparison of factors influencing trichloroethylene degradation by toluene-oxidizing bacteria. Appl. Environ. Microbiol. 62:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeChevallier, M. W., C. D. Cawthon, and R. G. Lee. 1988. Factors promoting survival of bacteria in chlorinated water supplies. Appl. Environ. Microbiol. 54:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. L. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 37.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 38.Nemati, M., G. E. Jenneman, and G. Voordouw. 2001. Impact of nitrate-mediated microbial control of souring in oil reservoirs on the extent of corrosion. Biotechnol. Prog. 17:852-859. [DOI] [PubMed] [Google Scholar]
- 39.Ovreas, L., L. Forney, F. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 41.Page, S., and C. C. Gaylarde. 1990. Biocide activity against Legionella and Pseudomonas. Int. Biodeterior. Bull. 26:139-148. [Google Scholar]
- 42.Pope, D. H., and R. M. Pope. 1998. Guide for the monitoring and treatment of microbiologically influenced corrosion in the natural gas industry. GRI report GRI-96/0488. Gas Research Institute, Des Plaines, Ill.
- 43.Pope, D. H., T. P. Zintel, B. A. Cookingham, R. G. Morris, D. Howard, R. A. Day, J. R. Frank, and G. E. Pogemiller. 1989. Mitigation strategies for microbially influenced corrosion in gas industry facilities, p. 1-15. In Proceedings of the NACE Annual Conference, Corrosion/89, paper 192. NACE International, Houston, Tex.
- 44.Pound, B. G. 1998. Gap analysis of the Pipeline Research Committee International (PRCI)/Gas Research Institute (GRI) research program on internal corrosion. GRI contract 6008. Topical report SF26363.000/AOTO/1198/BP02. Gas Research Institute, Des Plaines, Ill.
- 45.Raskin, L., L. Poulsen, D. Noguera, B. Rittmann, and D. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raskin, L., J. Stromley, B. Rittmann, and D. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravot, G., M. Magot, B. Ollivier, B. K. Patel, E. Ageron, P. A. Grimont, P. Thomas, and J. L. Garcia. 1997. Haloanaerobium congolense sp. nov., an anaerobic, moderately halophilic, thiosulfate- and sulfur-reducing bacterium from an African oil field. FEMS Microbiol. Lett. 147:81-88. [DOI] [PubMed] [Google Scholar]
- 48.Reysenbach, A.-L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a mid-Atlantic ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reysenbach, A.-L., G. S. Wickham, and N. R. Pace. 1994. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 60:2113-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rocheleau, S., C. W. Greer, J. R. Lawrence, C. Cantin, L. Laramee, and S. R. Guiot. 1999. Differentiation of Methanosaeta concilii and Methanosarcina barkeri in anaerobic mesophilic granular sludge by fluorescent in situ hybridization and confocal scanning laser microscopy. Appl. Environ. Microbiol. 65:2222-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 52.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 54.Strickland, L. N., R. T. Fortnum, and B. W. DuBose. 1996. A case history of microbiologically influenced corrosion in the Lost Hills oilfield, Kern County, California, p. 1-11. In Proceedings of the NACE Annual Conference, Corrosion/96, paper 297. NACE International, Houston, Tex.
- 55.Terzenbach, D. P., and M. Blaut. 1994. Transformation of tetrachloroethylene to trichloroethylene by homoacetogenic bacteria. FEMS Microbiol. Lett. 123:213-218. [DOI] [PubMed] [Google Scholar]
- 56.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsoi, T. V., E. G. Plotnikova, J. R. Cole, W. F. Guerin, M. Bagdasarian, and J. M. Tiedje. 1999. Cloning, expression, and nucleotide sequence of the Pseudomonas aeruginosa 142 ohb genes coding for oxygenolytic ortho dehalogenation of halobenzoates. Appl. Environ. Microbiol. 65:2151-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vetting, M. W., D. A. D'Argenio, L. N. Ornston, and D. H. Ohlendorf. 2000. Structure of Acinetobacter strain ADP1 protocatechuate 3,4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase. Biochemistry 39:7943-7955. [DOI] [PubMed] [Google Scholar]
- 59.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., Brooklyn, N.Y. [DOI] [PubMed]
- 60.Zellner, G., E. Stackebrandt, D. Nagel, P. Messner, N. Weiss, and J. Winter. 1996. Anaerofilum pentosovorans gen. nov., sp. nov., and Anaerofilum agile sp. nov., two new, strictly anaerobic, mesophilic, acidogenic bacteria from anaerobic bioreactors. Int. J. Syst. Bacteriol. 46:871-875. [DOI] [PubMed] [Google Scholar]
- 61.Zhu, X. Y., T. Zhong, Y. Pandya, and R. D. Joerger. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124-137. [DOI] [PMC free article] [PubMed] [Google Scholar]