Abstract
DNA and peptide nucleic acid (PNA) molecular beacons were successfully used to detect rRNA in solution. In addition, PNA molecular beacon hybridizations were found to be useful for the quantification of rRNA: hybridization signals increased in a linear fashion with the 16S rRNA concentrations used in this experiment (between 0.39 and 25 nM) in the presence of 50 nM PNA MB. DNA and PNA molecular beacons were successfully used to detect whole cells in fluorescence in situ hybridization (FISH) experiments without a wash step. The FISH results with the PNA molecular beacons were superior to those with the DNA molecular beacons: the hybridization kinetics were much faster, the signal-to-noise ratio was much higher, and the specificity was much better for the PNA molecular beacons. Finally, it was demonstrated that the combination of the use of PNA molecular beacons in FISH and flow cytometry makes it possible to rapidly collect quantitative FISH data. Thus, PNA molecular beacons might provide a solution for limitations of traditional FISH methods, such as variable target site accessibility, poor sensitivity for target cells with low rRNA content, background fluorescence, and applications of FISH in microfluidic devices.
Rapid monitoring of changes in microbial population abundance is important in several fields of microbiology. To this end, rRNA molecules have been used widely as target molecules for nucleic acid-based monitoring techniques. In particular, rRNA-targeted hybridization assays have been very useful for quantifying population levels (23, 30). Most hybridization assays rely on the immobilization of target nucleic acids, intact cells, or probes on membranes or other solid support materials, require the removal of unbound probe or target (through one or several wash steps) before hybridization signals can be detected, and use oligonucleotide probes labeled with radioactive isotopes, enzymes, or fluorescent dyes. The use of fluorescently labeled oligonucleotide probes to detect and quantify whole cells by using fluorescence in situ hybridization (FISH) (2, 5) has been particularly popular during the last decade. Despite the success of FISH in many applications, some challenges remain, such as nonuniform cell permeability, variable target site accessibility, poor sensitivity if the rRNA content of target cells is low (2, 31), and background fluorescence derived from autofluorescence of samples, insufficient washing, or unspecific binding of probes to compounds present in samples that cannot be removed by washing. Here we address several of these challenges by using DNA and peptide nucleic acid (PNA) molecular beacons (MBs).
An MB is a single-stranded oligonucleotide that fluoresces only upon hybridization to its target nucleic acid (32, 33). A DNA MB consists of a probe sequence flanked by two complementary sequences, which allow formation of a stem-loop structure when the MB is free in solution. A reporter fluorophore is attached to one end, and a quencher at the other end quenches the fluorescence when both ends are in close proximity in the absence of target nucleic acids. The fluorophore provides a signal when the MB hybridizes to a target sequence and opens its stem. Therefore, they can be used to detect specific DNA or RNA sequences in an aqueous solution without the prerequisite of immobilizing either the target nucleic acid or the probe as in traditional hybridization assays or in DNA microarrays. In addition, the wash steps that typically are needed posthybridization to remove unbound probes can be eliminated.
DNA MBs have been used since 1996 primarily for the detection of DNA, e.g., in real-time PCR monitoring and detection of single nucleotide polymorphisms and for the monitoring of mRNA (http://www.molecular-beacons.org). Attempts have been made to use DNA MBs to detect rRNA (15, 25; K. R. Hristova, M. Balberg, D. Frigon, M. Mau, D. Brady, D. Beebe, and L. Raskin, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. N-26, 2000).
PNA is a DNA analogue with a polyamide backbone instead of a sugar phosphate backbone (6). Due to the neutral backbone of PNA, PNA probes have hybridization characteristics, such as faster hybridization kinetics, that are superior to those of DNA probes. In addition, PNA probes can hybridize to targets at a very low salt concentration (20, 27). These properties should make PNA probes ideal for detection of rRNA, since rRNA molecules exhibit extensive secondary structures and since the secondary structures of rRNA targets and oligonucleotide probes greatly impact hybridization kinetics (16). Most of the secondary structure disappears when rRNA is denatured and immobilized on solid supports. However, when rRNA is used as the target in solution-based hybridizations, the secondary structure is retained. Since very low salt concentrations promote denaturation of rRNA, PNA MBs should help overcome some of the difficulties associated with solution-based hybridizations targeting rRNA (35). Furthermore, using PNA MBs in whole-cell hybridization assays should help address some of the challenges associated with FISH discussed above. Recently, fluorescently labeled PNA probes but not PNA MBs have been used for the detection of bacterial cells in a number of applications (21, 26, 28, 29). The use of PNA MBs to detect DNA in solution (PCR amplicons) was introduced in 1998 (19).
In this study, we used DNA and PNA MBs to detect and quantify rRNA in solution and in whole cells and demonstrated the applicability of PNA MBs in quantitative FISH for studying rRNA degradation.
Quantification of rRNA extracted from Escherichia coli by using DNA and PNA MBs.
One of the DNA MBs used in this study, S-D-Bact-0338-a-A-(6+)18(+1) (Hristova et al., Abstr. 100th Gen. Meet. Am. Soc. Microbiol.) (referred to as DNA MB Bact0338 hereafter), is a modification of a commonly used oligonucleotide probe specific for the bacterial domain, S-D-Bact-0338-a-A-18 (1) (5′-GCTGCCTCCCGTAGGAGT-3′). The modification consists in the addition of a sequence of nucleotides to the 5′ end of the original probe, so that the added sequence is complementary to the 3′ end of the probe and so that a stem-loop structure can be formed. To further stabilize the stem structure, one additional nucleotide was added to each end of the MB (Hristova et al., Abstr. 100th Gen. Meet. Am. Soc. Microbiol.). This MB was synthesized by Megabases, Inc. (Evanston, Ill.) with fluorescein as the fluorescent marker at the 5′ end and DABCYL [4-(4-dimethylaminophenylazo)-benzoic acid] as the quencher at the 3′ end (fluores-cein-5′-CATCCGCTGCCTCCCGTAGGAGTG-3′-DABCYL; bacterial domain probe sequence is in bold).
The PNA MB used in this study (fluorescein-E-GCTGCCTCCCGTAGGA-K-K-DABCYL; hereafter referred to as PNA MB Bact0338) was designed following guidelines for PNA probes provided by the synthesizer (Applied Biosystems, Mountain View, Calif.). These rules include limitation of probe length to 18 nucleotides, limitation of purine content to 60%, and restriction of self-complementarity to 3 bp (http://www.appliedbiosystems.com/support/seqguide.cfm). This PNA MB, specific to Bacteria, was designed by simply eliminating two nucleotides at the 3′ end of probe S-D-Bact-0338-a-A-18. Since PNA is very hydrophobic and folds on itself when in solution, there is no need for additional nucleotides to form a stem-loop structure, as required for DNA MBs. In addition to the hydrophobic properties of PNA, the opposite charges of glutamic acid (E) and lysine (K) at the ends of the PNA MB help assure that the fluorophore and quencher are in close proximity when PNA is in solution.
In a previous study, Hristova et al. (Hristova et al., Abstr. 100th Gen. Meet. Am. Soc. Microbiol.) optimized hybridization conditions for the detection of rRNA by using DNA MB Bact0338. Their optimized hybridization buffer consisted of a 10-mM phosphate buffer (pH 7.8), containing 900 mM NaCl, 1.3% polyethylene glycol, and 30% formamide (referred to as NaCl-polyethylene glycol hybridization buffer). This buffer was used in all subsequent experiments with the DNA MBs for the detection and quantification of rRNA in solution and in whole cells.
A hybridization buffer containing 25 mM Tris-HCl and 100 mM NaCl (modified from reference 20) was used for experiments with PNA MBs (referred to as Tris-HCl-NaCl hybridization buffer). In order to determine the optimal formamide concentration in the hybridization buffer for detecting rRNA by using PNA MB Bact0338, total RNA extracted from a pure culture of E. coli containing around 25 nM 16S rRNA was incubated in triplicate for 1 h at room temperature in a microtiter plate with 50 nM PNA MB Bact0338 in the Tris-HCl-NaCl hybridization buffer containing concentrations of formamide ranging from 0 to 50%. The fluorescence intensity of each reaction was measured with a SPECTRAmax GEMINI XS Microplate spectrofluorometer (Molecular Devices, Sunnyvale, Calif.). The highest fluorescence intensity was obtained at 40% formamide (Fig. 1). Thus, for subsequent experiments with the PNA MB, the Tris-HCl-NaCl hybridization buffer amended with 40% formamide was used.
FIG. 1.
Optimization of formamide concentration in Tris-HCl-NaCl hybridization buffer for PNA MB Bact0338 to detect rRNA in solution. The hybridization buffer contained 25 mM Tris-HCl and 100 mM NaCl and different concentrations of formamide. Each value shown is the mean plus or minus standard deviation of the mean of three replicates. PNA MB background represents fluorescence intensity of 50 nM PNA MB Bact0338 (control); PNA MB + E. coli 16S rRNA indicates fluorescence intensity of 50 nM PNA MB Bact0338 incubated with total RNA extracted from a pure culture of E. coli containing around 25 nM 16S rRNA; Difference refers to the absolute value of fluorescence intensity of hybridization signals obtained by subtracting results obtained with the PNA MB alone from hybridization signals. a.u. indicates arbitrary units.
To detect and quantify rRNA in solution, different concentrations of total RNA extracted from a pure culture of E. coli were incubated in triplicate for 2 h at room temperature in a microtiter plate with 50 nM PNA and DNA Bact0338 MBs in their respective optimized hybridization buffers. The fluorescence intensity of each reaction was measured by using the microplate spectrofluorometer (Fig. 2). The hybridization signal obtained with the DNA MB was above the background level only for the two highest E. coli 16S rRNA concentrations evaluated (12.5 and 25 nM). For the lower concentrations, the difference between hybridization and background intensities for different 16S rRNA concentrations was not obvious. In addition, the use of the DNA MB resulted in a higher background signal than the use of the PNA MB (data not shown). In contrast, substantial differences between hybridization and background intensities for all 16S rRNA concentrations were observed for the PNA MB and hybridization signals increased in a linear fashion with 16S rRNA concentrations ranging from 0.39 to 25 nM (R2 = 0.9991).
FIG. 2.
Quantification of E. coli 16S rRNA with 50 nM PNA and DNA Bact0338 MBs. Concentrations of 16S rRNA in E. coli total RNA extraction were approximately 0 (control), 0.39, 0.78, 1.56, 3.125, 6.25, 12.5, and 25 nM. The MB background fluorescence was subtracted from each sample. a.u. indicates arbitrary units. Each value shown is the mean plus or minus standard deviation of the mean of three replicates.
These results demonstrated that both DNA and PNA MBs can be used to detect rRNA in solution. However, for the hybridization conditions evaluated here, PNA MBs were much more useful for quantifying rRNA than were DNA MBs.
Detection of whole cells using DNA and PNA MBs.
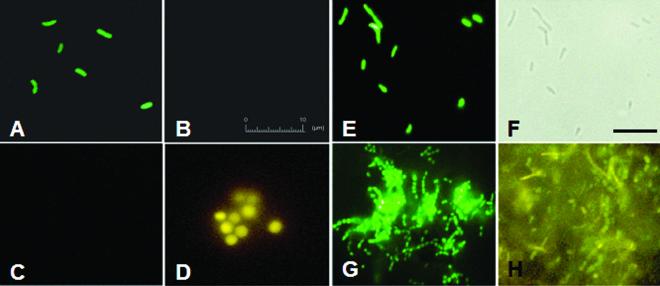
Pure cultures of E. coli and Methanosarcina acetivorans harvested during stationary phase were fixed with 4% paraformaldehyde for 2 h as described previously (22). The fixed cells were hybridized overnight at room temperature with DNA MB Bact0338 and with a DNA MB specific to Archaea [TAMRA5′-CGTGCTCCCCCGCCAATTCCTAGCACG-3′-DABCYL, S-D-Arch-0915-a-A-(1+)20(+6); archaeal domain probe sequence is in bold (Hristova et al., Abstr. 100th Gen. Meet. Am. Soc. Microbiol.) (hereafter referred to as DNA MB Arch0915)]. The samples were then visualized by using fluorescence microscopy with appropriate filter sets. E. coli and M. acetivorans cells fluoresced (Fig. 3A and D, respectively), indicating that DNA MBs hybridized to their target 16S rRNA molecules and opened their stem-loop structures. E. coli and M. acetivorans cells did not fluoresce when incubated with DNA MB Arch0915 (Fig. 3B) and DNA MB Bact0338 (Fig. 3C), respectively, suggesting the specificity of the two DNA MBs. In addition, bacterial cells (green) were distinguished from archaeal cells (yellow) in a mixed community obtained from a laboratory-scale upflow anaerobic sludge blanket (UASB) reactor (4a) (Fig. 3H). However, the background fluorescence observed with this environmental sample was substantial.
FIG. 3.
Epifluorescence micrographs showing the use of DNA MBs and PNA MB to detect bacterial and archaeal cells. Pure culture of E. coli (A and B) and M. acetivorans (C and D) were fixed by using 4% paraformaldehyde and were incubated with DNA MB Bact0338 (A and C) and DNA MB Arch0915 (B and D). The 10-μm bar in panel B also applies for panels A, C, and D. Fluorescence micrograph of pure culture of E. coli hybridized with PNA MB Bact0338 (E) and corresponding phase-contrast image (F). Fluorescence micrographs of biomass from UASB reactor incubated with PNA MB Bact0338 (G) and DNA Bact0338 and Arch0915 MBs (H). The bar in panel F corresponds to 10 μm and also applies for panels E, G, and H.
Fixed E. coli cells were hybridized overnight at room temperature with PNA MB Bact0338 (Fig. 3E and F). The fluorescence intensities of 50 cells from images collected during this hybridization experiment and during the hybridiziation experiment with E. coli cells and the DNA and PNA Bact0338 MBs were quantified with program ImageJ (NIH Image [http://rsb.info.nih.gov/ij/index.html]). The fluorescence intensities (mean plus or minus standard deviation) were 52.5 ± 6.0 arbitrary units and 155.2 ± 17.2 arbitrary units for hybridizations with DNA and PNA MBs, respectively. Thus, the use of the PNA MB resulted in a much higher hybridization response than the use of the DNA MB. To confirm this observation and to evaluate hybridization kinetics, we harvested E. coli cells in mid-log phase and performed hybridizations at 37°C for different incubation times between 5 min and 120 min for the PNA and DNA Bact0338 MBs in their respective optimized hybridization conditions. Hybridization mixtures were analyzed with an EPICS XL-MCL flow cytometer (Beckman Coulter, Inc., Miami, Fla.) (Fig. 4). The hybridization signal obtained with the PNA MB increased sharply during the first 15 min of incubation and reached a maximum after 60 min, indicating very fast hybridization kinetics. In contrast, the DNA MB hybridization signal increased much more slowly. Furthermore, the signal-to-noise ratio obtained with the PNA MB was about 18 times higher than that for the DNA MB after 2 h of hybridization, confirming our qualitative visual observation (Fig. 3A and E) and the results obtained for the quantification of rRNA in solution (Fig. 2). This observation again indicated that PNA MBs are more suitable than DNA MBs for obtaining quantitative results when rRNA is the target.
FIG. 4.
Flow cytometry analysis of hybridization kinetics for mid-log-phase-harvested cells of E. coli hybridized for different time periods with PNA and DNA Bact0338 MBs. a.u. indicates arbitrary units.
PNA MB Bact0338 was used to hybridize a sample obtained from a laboratory-scale UASB reactor (4a). Bacterial cells exhibited bright fluorescence (Fig. 3G), and by comparison of phase-contrast and fluorescence micrographs, bacterial cells were distinguished easily from archaeal cells in this complex microbial community (data not shown). The signal-to-noise ratio of this hybridization was much better than for the DNA MB hybridization of a similar sample (compare Fig. 3G and H). The higher signal-to-noise ratio may be due to a variety of reasons, including reduction of nonspecific binding of PNA MBs to environmental matrices, improved target accessibility of the PNA MB, and better cell permeability of the PNA MB.
Although the above results suggest that the PNA MB Bact0338 detects only target cells, they did not confirm its specificity in the presence of cells containing 16S rRNA with few mismatches. Therefore, we obtained cultures of Verrucomicrobium spinosum (DSM 4136) and Planctomyces limnophilus (DSM 3776), which have two (5′ - - A - - - - - - - - U - - - - - - 3′) and three (5′ - - A - - - - - - - - U - - - U - - 3′) mismatches with the Bact0338 target site, respectively (4). Cells of E. coli (perfect match with Bact0338 target site), V. spinosum, and P. limnophilus were harvested during stationary phase and were fixed with 4% paraformaldehyde for 2 h (22). The fixed cells were hybridized overnight at room temperature with 100 nM concentrations of DNA and PNA Bact0338 MBs in their corresponding hybridization buffers, containing different concentrations of formamide ranging from 0 to 70%. Each hybridization was performed in triplicate. Hybridization mixtures were analyzed with an EPICS XL-MCL flow cytometer as described above, and results for the PNA MB are plotted in Fig. 5. Hybridization signals for E. coli cells were maximal when the concentration of formamide ranged between 20 and 40% and decreased slightly for higher formamide concentrations. Hybridization signals for V. spinosum and P. limnophilus cells reached their maximum for formamide concentrations between 10 and 20% and 0 and 10%, respectively, and decreased significantly for formamide concentrations above 20 and 10%, respectively. The maximum hybridization intensities obtained with the DNA MB were three to five times lower than for the corresponding signals with PNA MB, and the trends observed for PNA MB hybridization for different formamide concentrations described above were not obvious for DNA MB hybridization (data not shown). The results of this formamide and specificity study indicate that, at the appropriate stringency, PNA MBs can be used to specifically detect target cells, while DNA MBs lack specificity.
FIG. 5.
Formamide and specificity study for PNA MB Bact0338. Flow cytometry analysis of E. coli (EC, perfect match), V. spinosum (VS, two mismatches), and P. limnophilus (PL, three mismatches) cells hybridized with 50 nM PNA MB Bact0338 in hybridization buffer containing different concentrations of formamide (0 to 70% [vol/vol]). a.u. indicates arbitrary units. Each value shown is the mean plus or minus standard deviation of the mean of three replicates.
Our FISH results show that DNA and PNA MBs can be used successfully to detect rRNA in whole cells without a wash step. Furthermore, we demonstrated that FISH with PNA MBs is preferred because hybridization signals were obtained in a few minutes compared to several hours to overnight for DNA MBs and traditional FISH probes. In addition, the signal-to-noise ratio and the specificity were much better for PNA MBs than for DNA MBs.
Quantitative FISH for the study of degradation of rRNA with PNA MB.
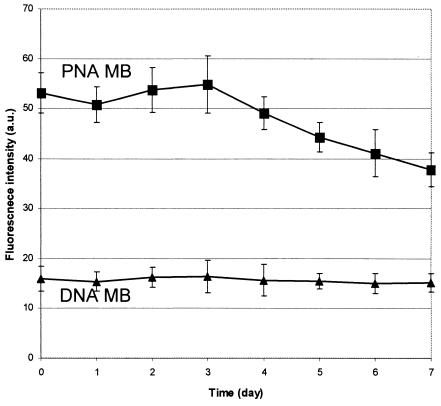
E. coli cells were harvested during stationary phase and were washed with phosphate-buffered saline (0.9% NaCl-10 mM sodium phosphate [pH 7.2]). The pellets were kept at room temperature, and two aliquots were collected each day for 7 days and fixed as described above. All aliquots were incubated overnight at room temperature with PNA and DNA Bact0338 MBs, each in their optimized hybridization buffers and in triplicate. The hybridization mixtures were analyzed with flow cytometry (Fig. 6). The average fluorescence intensity of freshly harvested E. coli cells (day 0) incubated with the PNA MB was about three times higher than that of the signal obtained with the DNA MB. We did not determine why there was a smaller difference in signal between the PNA MB and DNA MB hybridization observed in this experiment (Fig. 6) than in the kinetic experiment described above (Fig. 4). This difference may be due to lower levels of rRNA in cells harvested during stationary phase versus mid-log phase; i.e., an increase in cellular rRNA content may result in a nonproportional increase in hybridization response when PNA MBs are used in FISH due to the better target accessibility of PNA MBs.
FIG. 6.
Study of degradation of cellular E. coli rRNA. Flow cytometry analysis of stationary-phase-harvested cells of E. coli, deprived for different numbers of days of nutrients hybridized with PNA and DNA Bact0338 MBs. a.u. indicates arbitrary units. Each value shown is the mean plus or minus standard deviation of the mean of three replicates.
The fluorescence intensity obtained with the PNA MB began to decrease for the sample fixed 4 days after harvesting, and a 30% drop in fluorescence intensity was observed for the sample fixed 7 days after harvesting (Fig. 6). These results suggest that the cellular rRNA content of E. coli cells harvested in stationary phase starts to decrease 4 days after being deprived of nutrients and that at least 30% of the original rRNA is degraded after being deprived of nutrients for 7 days. The degradation rates for E. coli rRNA determined with traditional FISH (17, 24) were rather low and were in the same range as the degradation rate determined here by using FISH with PNA MBs and flow cytometry. Studies from the 1970s on E. coli RNA degradation showed that rRNA was more stable than mRNAs and that the degradation of E. coli rRNA started only after ribosomal monosomes were separated into their subunits (3, 13). Recent studies have shown that the rRNA degradation rates for different microorganisms varied considerably (7, 8, 12, 14, 17, 24). It should be noted that most past studies used methods to quantify isolated rRNA directly and thus provide information on the degradation of intact rRNA molecules. In contrast, the use of oligonucleotide probe hybridizations does not allow evaluation of degradation of intact rRNA molecules but only provides information about the degradation of the target site of the MBs used. The method for rRNA quantification presented here can be used to rapidly and accurately determine degradation rates for different target sites for various organisms. This information should help to determine when rRNA-based methods can be used to evaluate decreases in metabolic activity.
In this study, we successfully applied DNA and PNA MBs for the detection of rRNA in solution and in whole cells. The use of MBs allows the omission of wash steps, which makes it possible to detect and identify microbial cells in miniaturized systems, such as microfluidic devices (35), for which it is difficult to apply wash steps when real-time detection is an objective. In addition, the omission of wash steps saves time during the total analysis. When quantification of rRNA concentrations in solution or of cellular rRNA levels is necessary, PNA MBs provide results superior to those of DNA MBs. In addition, hybridization kinetics and sensitivity were significantly better with PNA MBs. In FISH, the failure to detect target cells (false-negative results) or to obtain extremely weak fluorescence signals may be due to low cellular ribosome levels (18), which is common for slowly growing populations in a wide range of environments (11, 34). Our results suggest that the use of PNA MBs instead of traditional FISH probes or DNA MBs would be helpful to detect these cells. As shown in previous FISH studies (2, 9, 10), probes targeting regions with low accessibility sometimes provide very low or undetectable hybridization signals. PNA MBs might also provide a solution for this limitation.
Acknowledgments
We thank William Metcalf for providing M. acetivorans cells and Dominic Frigon for help with data analysis.
This work was supported by the National Science Foundation (BES 00-86696).
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apirion, D. 1975. The fate of mRNA and rRNA in Escherichia coli. Brookhaven Symp. Biol. 26:286-306. [PubMed] [Google Scholar]
- 4.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 4a.Daugherty, B. J., L. T. Angenent, R. Agbisit, K. Rausch, M. Tumbleson, and L. Raskin. 2002. Using biological processes to recover sulfur from corn wet milling industry waste streams. In WEFTEC 2002: Proceedings of the 75th Annual Water Environment Federation Technical Exhibition and Conference. Water Environment Federation, Alexandria, Va.
- 5.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 6.Egholm, M., O. Buchardt, L. Christensen, C. Behrens, S. M. Freier, D. A. Driver, R. H. Berg, S. K. Kim, B. Norden, and P. E. Nielsen. 1993. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 365:566-568. [DOI] [PubMed] [Google Scholar]
- 7.Fegatella, F., and R. Cavicchioli. 2000. Physiological responses to starvation in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 66:2037-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flardh, K., P. S. Cohen, and S. Kjelleberg. 1992. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J. Bacteriol. 174:6780-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs, B. M., F. O. Glockner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn, D., R. I. Amann, W. Ludwig, A. D. Akkermans, and K. H. Schleifer. 1992. Detection of micro-organisms in soil after in situ hybridization with rRNA-targeted, fluorescently labelled oligonucleotides. J. Gen. Microbiol. 138:879-887. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, D., L. M. Shih, and Y. C. Zee. 1994. Degradation of rRNA in Salmonella strains: a novel mechanism to regulate the concentrations of rRNA and ribosomes. J. Bacteriol. 176:4761-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan, R., and D. Apirion. 1975. The fate of ribosomes in Escherichia coli cells starved for a carbon source. J. Biol. Chem. 250:1854-1863. [PubMed] [Google Scholar]
- 14.Kerkhof, L., and P. Kemp. 1999. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 30:253-260. [DOI] [PubMed] [Google Scholar]
- 15.Kourentzi, K. D., G. E. Fox, and R. C. Willson. 2001. Microbial detection with low molecular weight RNA. Curr. Microbiol. 43:444-447. [DOI] [PubMed] [Google Scholar]
- 16.Lima, W. F., B. P. Monia, D. J. Ecker, and S. M. Freier. 1992. Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry 31:12055-12061. [DOI] [PubMed] [Google Scholar]
- 17.Lisle, J. T., S. C. Broadaway, A. M. Prescott, B. H. Pyle, C. Fricker, and G. A. McFeters. 1998. Effects of starvation on physiological activity and chlorine disinfection resistance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4658-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593. [Google Scholar]
- 19.Ortiz, E., G. Estrada, and P. M. Lizardi. 1998. PNA molecular beacons for rapid detection of PCR amplicons. Mol. Cell. Probes 12:219-226. [DOI] [PubMed] [Google Scholar]
- 20.Perry-O'Keefe, H., S. Rigby, K. Oliveira, D. Sorensen, H. Stender, J. Coull, and J. J. Hyldig-Nielsen. 2001. Identification of indicator microorganisms using a standardized PNA FISH method. J. Microbiol. Methods 47:281-292. [DOI] [PubMed] [Google Scholar]
- 21.Perry-O'Keefe, H., H. Stender, A. Broomer, K. Oliveira, J. Coull, and J. J. Hyldig-Nielsen. 2001. Filter-based PNA in situ hybridization for rapid detection, identification and enumeration of specific micro-organisms. J. Appl. Microbiol. 90:180-189. [DOI] [PubMed] [Google Scholar]
- 22.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittmann, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raskin, L., B. E. Rittmann, and D. A. Stahl. 1996. Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl. Environ. Microbiol. 62:3847-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richardson, R. 2001. Investigations of mixed microbial communities involved in bioremediation using combinations of molecular, microscopy-based, and selective culturing techniques. Ph.D. dissertation. University of California at Berkeley, Berkeley.
- 25.Schofield, P., A. N. Pell, and D. O. Krause. 1997. Molecular beacons: trial of a fluorescence-based solution hybridization technique for ecological studies with ruminal bacteria. Appl. Environ. Microbiol. 63:1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stender, H., A. Broomer, K. Oliveira, H. Perry-O'Keefe, J. J. Hyldig-Nielsen, A. Sage, B. Young, and J. Coull. 2000. Rapid detection, identification, and enumeration of Pseudomonas aeruginosa in bottled water using peptide nucleic acid probes. J. Microbiol. Methods 42:245-253. [DOI] [PubMed] [Google Scholar]
- 27.Stender, H., M. Fiandaca, J. J. Hyldig-Nielsen, and J. Coull. 2002. PNA for rapid microbiology. J. Microbiol. Methods 48:1-17. [DOI] [PubMed] [Google Scholar]
- 28.Stender, H., T. A. Mollerup, K. Lund, K. H. Petersen, P. Hongmanee, and S. E. Godtfredsen. 1999. Direct detection and identification of Mycobacterium tuberculosis in smear-positive sputum samples by fluorescence in situ hybridization (FISH) using peptide nucleic acid (PNA) probes. Int. J. Tuberc. Lung Dis. 3:830-837. [PubMed] [Google Scholar]
- 29.Stender, H., K. Oliveira, S. Rigby, F. Bargoot, and J. Coull. 2001. Rapid detection, identification, and enumeration of Escherichia coli by fluorescence in situ hybridization using an array scanner. J. Microbiol. Methods 45:31-39. [DOI] [PubMed] [Google Scholar]
- 30.Teske, A., N. B. Ramsing, K. Habicht, M. Fukui, J. Kuver, B. B. Jorgensen, and Y. Cohen. 1998. Sulfate-reducing bacteria and their activities in cyanobacterial mats of solar lake (Sinai, Egypt). Appl. Environ. Microbiol. 64:2943-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theron, J., and T. E. Cloete. 2000. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit. Rev. Microbiol. 26:37-57. [DOI] [PubMed] [Google Scholar]
- 32.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 33.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 34.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 35.Xi, C., M. Balberg, J. Selby, S. Boppart, and L. Raskin. 2002. Use of molecular beacons to study mixing and hybridization in microfluidic devices, 541-544. In A. Dittmar and D. Beebe (ed.), Proceedings of the 2nd Annual International IEEE/EMBS Special Topic Conference on Microtechnology in Medicine and Biology. Institute of Electrical and Electronic Engineers, Piscataway, N.J.