Abstract
Streptococcus oralis, a member of the mitis group of oral streptococci, is implicated in the pathogenesis of infective endocarditis and is the predominant aciduric non-mutans-group streptococcus in dental plaque. We undertook to identify the most abundant surface-associated proteins of S. oralis and to investigate changes in protein expression when the organism was grown under acidic culture conditions. Surface-associated proteins were extracted from cells grown in batch culture, separated by two-dimensional gel electrophoresis, excised, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry and liquid chromatography-tandem mass spectrometry. Putative functions were assigned by homology to a translated genomic database of Streptococcus pneumoniae. A total of 27 proteins were identified; these included a lipoprotein, a ribosome recycling factor, and the glycolytic enzymes phosphoglycerate kinase, fructose bisphosphate aldolase, glyceraldehyde-3-phosphate dehydrogenase, and enolase. The most abundant protein, phosphocarrier protein HPr, was present as three isoforms. Neither lactate dehydrogenase nor pyruvate oxidase, dominant intracellular proteins, were present among the proteins on the gels, demonstrating that proteins in the surface-associated pool did not arise as a result of cell lysis. Eleven of the proteins identified were differentially expressed when cells were grown at pH 5.2 versus pH 7.0, and these included superoxide dismutase, a homologue of dipeptidase V from Lactococcus lactis, and the protein translation elongation factors G, Tu, and Ts. This study has extended the range of streptococcal proteins known to be expressed at the cell surface. Further investigations are required to ascertain their functions at this extracellular location and determine how their expression is influenced by other environmental conditions.
Streptococcus oralis, a member of the mitis group of the viridans group streptococci, is a component of normal dental plaque, in which it forms a significant proportion of the aciduric microflora (5, 34). Aciduricity, the ability to grow under conditions of low pH, is considered a virulence determinant for bacteria associated with the initiation and progression of dental caries, although the precise role of S. oralis in this disease process has yet to be fully defined. In addition to the potential role of S. oralis in dental caries, it is well documented that S. oralis is associated with a range of extraoral diseases including endocarditis and infections in susceptible patients, including those who are immunologically compromised. Douglas and coworkers (13) documented that, out of 42 cases of infective endocarditis from which viridans group streptococci were isolated, the most common species were from the mitis group, namely, Streptococcus sanguis, S. oralis, and Streptococcus gordonii (isolation frequencies of 31.9, 29.8, and 12.7%, respectively). S. oralis is also the predominant species of the viridans group streptococci associated with septicemia in neutropenic patients (2) and neonates (33).
The cell wall envelope of gram-positive bacteria is host to a wide range of molecules, including many functionally and structurally important proteins, and mediates contact with the external environment, especially host tissues and fluids (23). Thus, many virulence determinants in pathogenic organisms are surface proteins. Many of the cell surface streptococcal proteins contain primary sequence signatures necessary to anchor them in the cell wall, including M proteins, which contain the LPXTG anchor motif (10). Recently, however, a surface protein from Streptococcus pyogenes that binds to immunoglobulin G but that, distinct from members of the M protein superfamily, is anchorless and contains no LPXTG motif has been characterized (18). Other streptococcal surface-associated proteins, notably the glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase and enolase, are anchorless and represent a new class of virulence factors (9) as they possess fibronectin- and plasmin-binding activities (26, 27), indicating that these proteins are bifunctional, having roles in addition to central metabolism when localized at the cell surface. The anchorless pneumococcal α-enolase also has dual functionality, as it binds plasminogen at the cell surface, which may facilitate bacterial penetration of the basement membrane (4). While genomic analysis can identify proteins likely to be surface localized based on the presence of anchor motifs, analysis of protein expression can characterize anchorless proteins.
A systematic investigation of the proteins expressed at the cell surface of S. oralis has yet to be conducted. In a study of S. gordonii, an opportunistic pathogen closely related to S. oralis, a shift from oral to blood pH (pH 6.2 to 7.3) was found to modulate the expression of a number of genes, including those coding for surface proteins (32). The aim of our study was to apply the proteomic methodologies two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) and protein identification using mass-spectrometric techniques (matrix-assisted laser desorption ionization-time of flight [MALDI-TOF] mass spectrometry [MS] and liquid chromatography-tandem MS [LC-MS/MS]) to the analysis of the predominant surface-associated proteins of S. oralis and to determine the effect of culture at neutral and low pH on their expression. The S. oralis genome has not been sequenced, but it is phylogenetically closely related to Streptococcus pneumoniae, with 16S rRNA genes exhibiting over 99% similarity (3, 19). The S. pneumoniae genome is now complete (30; http://www.tigr.org/), and we have previously demonstrated that the genomic data for this organism, in combination with peptide mass fingerprinting (PMF) using MALDI-TOF MS, can be used to identify S. oralis proteins (36).
MATERIALS AND METHODS
Bacterial isolate and culture conditions.
The bacterium used throughout this study, S. oralis strain 176N, is an aciduric strain isolated from the interproximal plaque of a child (36) and was stored at −70°C in cryovials (Protect; Technical Service Consultants Limited, Heywood, Lancashire, United Kingdom). When required, it was subcultured onto Columbia agar (Oxoid Limited, Basingstoke, Hampshire, United Kingdom) supplemented with 5% (vol/vol) defibrinated horse blood (TCS Microbiology, Botolph Claydon, Buckingham, United Kingdom). Cultures were incubated in an anaerobic cabinet (MK3 anaerobic workstation; Don Whitley Scientific Limited, West Yorkshire, United Kingdom) in an atmosphere of 10% CO2-10% H2-80% N2 at 37°C for 16 to 24 h. Cultures for protein extraction were grown in triplicate statically in brain heart infusion (BHI; Oxoid), pH 7.0, and BHI adjusted to pH 5.2 by the addition of disodium hydrogen orthophosphate and citric acid, each at a final concentration of approximately 20 mM. Media were inoculated with 5% (vol/vol) bacterial suspension from mid-exponential-phase cultures in BHI and incubated aerobically at 37°C until mid-exponential phase.
Preparation of S. oralis surface-associated proteins.
The cells were pelleted by centrifugation at 2,700 × g (20°C, 10 min), and proteins associated with the cell surface were extracted in accordance with the method of Kessler and Yagi (20). Cell pellets were washed twice in phosphate-buffered saline (PBS tablets; Sigma Chemical Company, Poole, Dorset, United Kingdom) and resuspended in PBS with 0.2% (wt/vol) N-dodecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate (Zwittergent; Sigma). The suspensions were incubated at 28°C with shaking at 80 rpm (Microtherm; Camlab Limited, Cambridge, United Kingdom) for 1 h, and the cells were pelleted at 6,000 × g (10 min) and 20°C. The supernatant containing the extracted cell surface proteins was diluted 1:4 with 50 mM Tris-HCl (pH 7.5), passed though a filter (0.2-μm-pore-size Acrodisc syringe filter; Pall Filtron Gelman Laboratory, Portsmouth, Hants, United Kingdom) to remove remaining cells, and dialyzed overnight against 2 liters of 50 mM Tris-HCl (pH 7.5) with protease inhibitors (Complete protease inhibitor cocktail tablets; Roche Diagnostics Limited, Lewes, East Sussex, United Kingdom). The dialyzed material was concentrated with a centrifugal concentrator at 12,000 × g (Nanosep 10 kDa Omega; Pall Filtron Gelman Laboratory).
To demonstrate that proteins in cell surface fractions did not arise as a result of cell lysis, whole-cell proteins, including the soluble intracellular pool, were extracted from aliquots of each culture as previously described (36). These preparations and the cell-associated proteins were analyzed by 2-D PAGE, as described below.
Analysis of bacterial proteins by 2-D PAGE.
Cell surface proteins from S. oralis were precipitated from the concentrated solution by the addition of 4 volumes of ice-cold acetone. The resulting pellet was collected by centrifugation (15,100 × g, 10 min) at 4°C. Soluble intracellular and cell surface protein fractions were resuspended in a solution containing 7 M deionized urea, 2 M thiourea, 2% Tergitol NP-40, 62 mM dithiothreitol, and 2% pH 3 to 10 carrier ampholytes (Bio-Rad Laboratories Ltd., Hemel Hempstead, Hertfordshire, United Kingdom) to give a protein concentration of approximately 1 μg/μl and used to rehydrate 7-cm pH 4 to 7 linear immobilized pharmalyte gradient (IPG) strips (Bio-Rad). Strips were rehydrated overnight under active conditions in a Protean isoelectric focusing cell (Bio-Rad) and focused for 20,000 to 25,000 V-h according to the manufacturer's instructions. Prior to loading on the second dimension, focused IPG strips were equilibrated sequentially in a buffer (Tris-HCl containing 6 M urea, 30% [vol/vol] glycerol, 2% sodium dodecyl sulfate [SDS]) containing 1% dithiothreitol or 2.5% iodoacetamide for 15 min each and applied to 12% SDS gels. SDS-PAGE was carried out with a Mini Protean II cell (Bio-Rad), and proteins were resolved at a constant voltage of 100 V over 2 h. Proteins were visualized following staining with colloidal Coomassie brilliant blue (CCBB) G (Sigma) (25). The Mrs of individual resolved proteins were determined by comparison with low-molecular-weight markers (Sigma), and pIs were deduced from the linearity of the IPG strips. Gels were scanned (300 dots per in.), and spot detection was carried out with 2-D Advanced software (version 5.1; Phoretix International, Newcastle upon Tyne, United Kingdom). Following the determination of spot boundaries, integrated optical densities (IOD) were measured for each protein and expressed as percentages of the total protein detected per gel. The sum of the individual IOD values was used to calculate total expression of the given polypeptide for proteins that occurred as isoforms. Three independent cultures for each growth condition were processed. For the comparison of the effect of culture at low pH on the expression of surface-associated proteins, these were considered to have altered expression if the mean percent IOD was up- or down-regulated >1.5-fold (i.e., >50% change in mean percent IOD). Statistically significant differences in protein expression levels were determined by Student's t test (P < 0.05). The most abundant and differentially expressed proteins were excised from gels and identified as described below. The most abundant proteins (greatest percent IOD) that were detected in intracellular fractions but that were absent in surface-associated protein pools were also analyzed in this way.
Identification of streptococcal proteins by peptide mass fingerprinting.
Individual spots were excised from 2-D gels, washed, dehydrated in acetonitrile (ACN), reduced, and treated with iodoacetamide to S-alkylate the proteins as described by Wilkins et al. (36). Gel pieces were swollen in a digestion buffer containing a final concentration of 50 mM NH4HCO3 and 12.5 ng of trypsin (sequencing grade modified trypsin; Promega UK, Southampton, Hampshire, United Kingdom)/μl, and enzymatic cleavage was carried out overnight at 37°C. Peptide extracts were applied to a ZipTip (Millipore Ltd., Watford, Hertfordshire, United Kingdom), rinsed in 0.1% trifluoroacetic acid (TFA; high-pressure liquid chromatography grade; Perbio Science UK Ltd., Chester, United Kingdom), and eluted in 1:1 ACN-0.1% TFA in one-third of the original volume. This solution was mixed 1:1 on the mass spectrometer sample plate with a saturated solution of α-cyano-4-hydroxycinnamic acid (99% purity: Sigma-Aldrich, Gillingham, Dorset, United Kingdom) in 70% ACN-0.03% TFA. Mass spectra were acquired on a MALDI-TOF mass spectrometer (Voyager Elite; Applied Biosystems, North Warrington, Cheshire, United Kingdom) in reflector mode with delayed extraction. Samples were irradiated with a nitrogen laser (337 nm) at ca. 1,000 U with a 3-ns pulse width and accelerating potential of 20 kV. All spectra were obtained as 300-shot averages. MALDI-TOF spectra were calibrated on Data Explorer software, and monoisotopic mass peaks were labeled by close external calibration with a peptide mixture containing des-arg1-bradykinin, angiotensin 1, and glu1-fibrinopeptide B (Applied Biosystems).
MS-Fit (University of California San Francisco Mass Spectrometry Facility; http://prospector.ucsf.edu/), installed locally, was used to identify proteins from peptide mass fingerprints. The scoring for missed cleavage sites was set at 0. All searches were performed against The Institute for Genomic Research (TIGR) annotated genomic database for S. pneumoniae TIGR4 (http://www.tigr.org/). Putative functions were usually assigned if the first entry returned had a high MOWSE (molecular weight search) score (28), ≥4 peptides were successfully matched to the database entry, ≥10% amino acid sequence coverage was demonstrated, and there was agreement (±10%) on the observed and theoretical Mrs and pIs of the protein. If no putative function was assigned to a protein in the database (i.e., hypothetical protein) or for clarification of its function, the amino acid sequence data were searched with BLAST (1) against all of the other proteins represented in the Comprehensive Microbial Resource (http://tigrblast.tigr.org/cmr-blast/) and against the nonredundant databases of all microorganisms at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Entrez).
Identification of streptococcal proteins by LC-MS/MS.
Low-mass proteins from 2-D gels that were not successfully identified by MALDI-TOF MS were analyzed by LC-MS/MS on a ProteomeX (Thermo Finnigan, Hemel Hempstead, Herts, United Kingdom). In-gel tryptic digests of gel-resolved proteins were generated as described above, and digest supernatants were dried in a vacuum centrifuge and resuspended in 10 μl of 0.1% formic acid. Chromatography of 5-μl aliquots of each sample was performed on a 100- by 0.18-mm BioBasic C18 column (ThermoHypersil-Keystone, Runcorn, Cheshire, United Kingdom). Peptides were eluted with aqueous ACN (5 to 65% ACN over 30 min) containing 0.1% formic acid at a flow rate of 2 μl/min. Spectra were acquired in data-dependent MS/MS mode with dynamic exclusion prior to analysis and comparison with the translated genomic sequence data for S. pneumoniae TIGR4 with TurboSEQUEST software (Thermo Finnigan). Putative functions were usually assigned if the first entry returned had a high score, ≥2 peptides were successfully matched to the database entry, ≥20% amino acid sequence coverage was demonstrated, and there was agreement (±10%) on the observed and theoretical Mrs and pIs of the protein.
Examination of identified S. oralis proteins for the presence of anchor motifs.
The translated sequences for all proteins assigned putative functions by PMF or LC-MS/MS were acquired from the S. pneumoniae TIGR4 genomic database at TIGR. The amino acid sequences of these proteins were searched for the presence of the LPXTG anchor and choline-binding motifs and for signal peptides as described by Tettelin et al. (30).
RESULTS
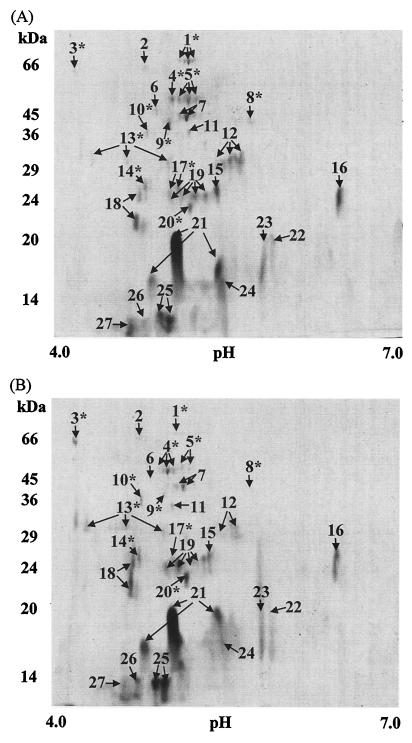
Surface-associated proteins extracted from cells of S. oralis cultured at pH 7.0 were separated by 2-D PAGE, and this resulted in a map in which 65 ± 12 well-resolved proteins were detected in the pH range of 4 to 7 when gels were CCBB stained (Fig. 1). This compared with 60 ± 9 proteins detectable for S. oralis cells grown to mid-exponential phase at pH 5.2. The variation in the number of proteins visualized arose as a result of the differences in detection of very low abundance proteins in each of the replicates. The most abundant and differentially expressed proteins were excised from both gels and subjected to further analysis by PMF and LC-MS/MS in order to assign putative functions (Table 1). Notably, the most abundant surface-associated protein identified from both pH 5.2- and 7.0-grown S. oralis cultures was the homologue of phosphocarrier protein HPr, which was present as three distinct isoforms. Other abundant surface-associated proteins included adenylate kinase, ribosome recycling factor, ribosomal proteins S6 and L7/L12, transcription elongation factor GreA, a lipoprotein, the electron transport protein thioredoxin, and the glycolytic enzymes phosphoglucomutase, phosphoglycerate kinase, enolase, fructose bisphosphate aldolase, and triosephosphate isomerase. None of these surface-associated proteins had altered expression at low pH. One protein (SP0845) was identified as a lipoprotein from the annotated genomic database. When the amino acid sequence data were searched with BLAST against all of the proteins in the Comprehensive Microbial Resource, the lipoprotein was found to have 65% sequence identity with a CD4+ T-cell-stimulating antigen precursor of S. pyogenes (SPy1228).
FIG. 1.
Surface-associated proteins of S. oralis separated by 2-D PAGE. Surface-associated proteins from pH 7.0- (A) and pH 5.2-grown (B) cultures were separated by isoelectric focusing in the pH range of 4 to 7 in the first dimension and by 12% SDS-PAGE in the second dimension. Resolved proteins were visualized following staining with CCBB, and the locations of molecular mass markers and pIs are shown. Spot numbering indicates those proteins identified by PMF and LC-MS/MS, and the numbers match the numbers in Table 1. *, proteins with altered expression at pH 5.2 compared with those extracted from cells cultured at pH 7.0; multiple arrows, proteins for which different isoforms were observed.
TABLE 1.
Identity of S. oralis surface-associated proteins separated by 2-D PAGE
| Functional category | Spot no.a | Putative functionb | Gene symbolc | Acc. no.d | Observed migratione
|
Theoretical migrationf
|
No. of peptides matchedg | Coverageh | Ratio mean spot volume 5.2/7.0i | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pI | Mr | pI | Mr | ||||||||
| Cellular processes and stress response | 20 | Superoxide dismutase (manganese dependent) | sodA | SP0766 | 5.1 | 24.9 | 4.9 | 22.4 | 4 | 26 | 2.23j |
| 25 | Thioredoxink | trx | SP1776 | 4.8 | 11.0 | 4.5 | 11.4 | 6 | 71 | NDEl | |
| Central and intermediary metabolism | 2 | Phosphoglucomutase | pgm | SP1498 | 4.7 | 64.5 | 4.7 | 62.7 | 7 | 17 | NDE |
| 4 | Dipeptidase | pepV | SP0623 | 4.9 | 50.7 | 4.8 | 50.8 | 6 | 12 | 2.02j | |
| 7 | Phosphoglycerate kinase | pgk | SP0499 | 5.1 | 47.0 | 4.9 | 41.9 | 9 | 24 | NDE | |
| 6 | Enolase | eno | SP1128 | 4.8 | 46.6 | 4.7 | 47.1 | 8 | 22 | NDE | |
| 8 | Glyceraldehyde-3-phosphate dehydrogenase | gap | SP2012 | 5.6 | 40.5 | 5.3 | 35.9 | 10 | 40 | 0.45j | |
| 10 | Manganese-dependent inorganic pyrophosphatase | ppaC | SP1534 | 4.7 | 35.0 | 4.6 | 33.5 | 6 | 26 | 2.67j | |
| 12 | Fructose bisphosphate aldolase | fba | SP0605 | 5.4 | 30.4 | 5.2 | 31.4 | 9 | 33 | NDE | |
| 19 | Triosephosphate isomerase | tpi | SP1574 | 5.0 | 25.8 | 4.8 | 26.5 | 5 | 27 | NDE | |
| Transport and binding | 21 | Phosphocarrier protein HPrk | ptsH | SP1177 | 4.9 | 17.5 | 4.5 | 8.9 | 4 | 41 | NDE |
| Purines, pyrimidines, nucleosides, and nucleotides | 15 | Adenylate kinase | adk | SP0231 | 5.2 | 27.4 | 5.0 | 23.7 | 8 | 30 | NDE |
| Transcription and translation | 1 | EF-G | fusA | SP0273 | 5.0 | 68.7 | 4.9 | 76.8 | 21 | 37 | 0.38j |
| 9 | EF-Tsk | tsf | SP2214 | 4.9 | 39.1 | 4.6 | 37.4 | 12 | 37 | 0.27j | |
| 17 | EF-Pk | efp | SP0435 | 4.9 | 24.5 | 4.6 | 20.6 | 8 | 54 | 0.33 | |
| 5 | EF-Tuk | tuf | SP1489 | 4.9 | 46.0 | 4.9 | 44.0 | 13 | 33 | 0.31 | |
| 16 | Ribosome recycling factor | frr | SP0945 | 6.3 | 23.7 | 6.0 | 20.7 | 6 | 30 | NDE | |
| 18 | Transcription elongation factor GreA | greA | SP1517 | 4.6 | 21.5 | 4.5 | 17.6 | 4 | 32 | NDE | |
| 24 | Ribosomal protein S6k | rpsF | SP1541 | 5.3 | 15.9 | 4.9 | 11.2 | 6 | 73 | NDE | |
| 27 | Ribosomal protein L7/L12k | rplL | SP1354 | 4.5 | 12.0 | 4.1 | 12.4 | 4 | 37 | NDE | |
| Cell envelope | 11 | Lipoprotein | SP0845 | 4.9 | 36.0 | 5.4 | 36.7 | 5 | 17 | NDE | |
| Other | 14 | Serine/threonine protein phosphatasek | SP1201 | 4.7 | 27.2 | 4.3 | 27.1 | 4 | 29 | 2.74j | |
| 23 | Bacterocin transport accessory proteink | bta | SP1499 | 5.7 | 17.2 | 5.7 | 12.9 | 4 | 42 | NDE | |
| Unknown | 3 | Conserved hypothetical protein | SP0868 | 4.2 | 60.5 | 4.8 | 46.3 | 4 | 10 | 3.26 | |
| 13 | Conserved hypothetical protein | SP1922 | 4.7 | 33.6 | 4.5 | 25.8 | 6 | 24 | 1.63 | ||
| 22 | Conserved hypothetical proteink | SP2202 | 5.8 | 15.9 | 5.2 | 10.2 | 3 | 48 | NDE | ||
| 26 | Conserved hypothetical proteink | SP0122 | 4.6 | 10.0 | 4.3 | 9.3 | 2 | 48 | NDE | ||
Spot number refers to the proteins labeled in Fig. 1.
Putative functions were assigned from the TIGR database for S. pneumoniae.
Gene symbol in the TIGR database for S. pneumoniae.
Accession number in the TIGR database for S. pneumoniae.
Calculated from data presented in Fig. 1. Mrs are in thousands.
As given in the TIGR database for S. pneumoniae.
Number of tryptic peptides observed contributing to percentage amino acid coverage.
The percentage of amino acid coverage (peptides observed/theoretical value from sequence data).
Ratio of IOD for each protein derived from cells cultured at pH 5.2 to that for the protein derived from cells cultured at pH 7.0 (mean; n = 3 for each growth condition).
Significantly different (P < 0.05) as calculated by Student's t test.
Proteins identified by LC-MS/MS. All other identifications were achieved by MALDI-TOF MS.
NDE, not differentially expressed.
Eleven of the S. oralis proteins identified in the surface-associated fraction exhibited altered levels of expression when the organism was grown at low pH. Following growth at pH 5.2, six proteins were up-regulated 1.5-fold or greater and five were down-regulated, where down-regulated proteins are indicated by a mean spot volume ratio, defined as the ratio of the IOD value for each protein derived from cells cultured at pH 5.2 to that for the protein derived from cells cultured at pH 7.0, of <0.67 (Table 1). The up-regulated proteins included two conserved hypothetical proteins with no assigned function in TIGR, spots 3 (SP0868) and 13 (SP1922). When the amino acid sequence for SP0868 was searched with BLAST against the National Center for Biotechnology Information nonredundant database, sequence homology (23%) with an iron-regulated ABC-type transporter membrane component from Clostridium acetobutylicum was found. The sequence for SP1922 contained a domain of unknown function, DUF28, found in bacterial and yeast proteins. The stress response protein superoxide dismutase, a serine/threonine protein phosphatase, a dipeptidase which had 67% sequence identity with a dipeptidase from Lactococcus lactis (encoded by pepV), and a manganese-dependent inorganic pyrophosphatase were significantly up-regulated (greater than 50% up-regulation) at low pH (P < 0.05). The manganese-dependent inorganic pyrophosphatase has 93% similarity to an intrageneric coaggregation-relevant adhesin from S. gordonii (accession number P95765). Elongation factor G (EF-G), EF-Tu, EF-Ts, EF-P, and glyceraldehyde-3-phosphate dehydrogenase were down-regulated (greater than 50% down-regulation) at low pH; EF-G, EF-Ts, and glyceraldehyde-3-phosphate dehydrogenase were down-regulated significantly (P < 0.05). Several of the proteins identified existed as multiple forms on the gels with differences in pI, namely, EF-G, EF-Tu, and EF-P; dipeptidase; triosephosphate isomerase; phosphoglycerate kinase; fructose bisphosphate aldolase; conserved hypothetical protein SP1922; phosphocarrier protein HPr; and thioredoxin. Other proteins, including transcription elongation factor GreA and phosphocarrier protein HPr, also exhibited different Mr forms.
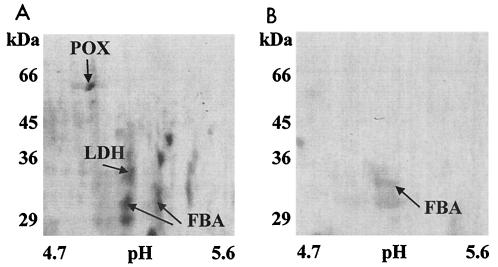
To demonstrate that no significant cellular lysis had occurred during the preparation of surface-associated proteins, the most abundant intracellular proteins of S. oralis grown at pH 5.2 were also identified. Among these, several proteins that were absent in surface-associated protein pools were visible. These included lactate dehydrogenase (pI 5.1, Mr 35,400) and pyruvate oxidase (pI 5.1, Mr 65,300) comprising 3.3 and 2.0%, respectively, of the total proteins. Neither of these proteins was identified among the surface-associated proteins excised from the gels (Fig. 2), and, in addition, no proteins with the pI and Mr of lactate dehydrogenase or pyruvate oxidase were observed on the 2-D gels of surface-associated proteins.
FIG. 2.
Cytoplasmic proteins were absent from the surface-associated protein preparation from pH 5.2-grown cultures separated by 2-D PAGE. Lactate dehydrogenase (LDH) and pyruvate oxidase (POX) were abundant in the whole-cell extract (A) but were absent on the 2-D gels of the surface-associated proteins (B). Fructose bisphosphate aldolase (FBA) was identified in both intracellular and surface-associated fractions.
To determine the presence of anchor motifs in the surface-associated proteins of S. oralis, the translated sequences for all proteins shown in Table 1 were acquired from TIGR. Interrogation of these sequences demonstrated that none contained LPXTG, signal peptide, or choline-binding motifs. These S. oralis proteins were therefore considered to belong to the anchorless class of surface proteins.
DISCUSSION
The S. oralis proteins described in this study were surface associated as neither lactate dehydrogenase nor pyruvate oxidase, which are abundant in the whole-cell extracts, was present on the 2-D gels. The most abundant surface-associated protein, identified from both acidic and neutral culture conditions, was the phosphocarrier protein HPr. HPr is a low-mass phosphocarrier protein of the bacterial phosphoenol pyruvate-sugar phosphotransferase system, the cell surface localization of which in other streptococcal species has been demonstrated (12, 14). In this location, HPr has a role not only in sugar transport but also in signal transduction which regulates catabolite repression. The presence of many proteins at the cell surfaces of gram-positive organisms can be inferred by the presence of specific anchor motifs in genomic sequence data (30). Using a proteomic approach, however, we have demonstrated the existence of additional surface-associated proteins, as none of the proteins identified in this study had anchor motifs suggestive of surface localization. Hughes et al. (17) identified the major surface proteins of Streptococcus agalactiae and similarly identified proteins which would not have been assigned to a cell surface location if a conventional genomically based approach had been adopted. Many of these anchorless proteins, which represent a new class of virulence determinants, are increasingly recognized as bifunctional (9); e.g., the glycolytic enzyme α-enolase from S. pneumoniae has plasminogen-binding activity (4). The mechanism by which such proteins are localized to the cell surface is largely unknown, but it is suggested that this may occur via a reassociation process (9). Here, we describe numerous novel anchorless proteins at the cell surface of S. oralis, suggesting that extraction with Zwittergent enriches for this class of proteins.
The oxidative stress proteins superoxide dismutase and thioredoxin were identified among the surface-associated proteins of S. oralis in this study. While it is likely that these surface proteins protect against oxidative stress in S. oralis, it has been proposed that, due to their expression on the outer surface of the cell, stress proteins and molecular chaperones may also act as microbial virulence factors, functioning as bacterial adhesins and promoting host tissue damage (21). The functions of these stress proteins in S. oralis and their potential roles in pathogenesis remain to be established. Homologues of EF-G, EF-Tu, EF-Ts, and EF-P were also identified among the surface-associated proteins of S. oralis, and these were down-regulated following growth at low pH. This is the first demonstration of elongation factors associated with the cell surface in streptococci. Elongation factors are members of a superfamily of regulatory GTP hydrolases (G proteins) that are implicated in the sorting and amplification of transmembrane signals and the direction of the synthesis and translocation of proteins (29). Recent work with Escherichia coli has suggested that EF-Tu and EF-G, in addition to their role in translation, possess chaperone properties and may be involved in processes other than translation, including protein folding and renaturation in the cytoplasm (7, 8).
Glycolytic enzymes, including glyceraldehyde-3-phosphate dehydrogenase and enolase, were identified among the surface-associated proteins of S. oralis described here. Glyceraldehyde-3-phosphate dehydrogenase and enolase have been identified at the cell surfaces of pathogenic streptococci, which raises questions as to their functions at this location (4, 26, 27). Glyceraldehyde-3-phosphate dehydrogenase and enolase lack an apparent wall anchor motif but bind serum components plasmin and plasminogen, which may assist these bacteria in generating an inflammatory response (26). In S. oralis, when the pH of the culture medium at which cells are grown is decreased to 5.2, the expression of surface-associated glyceraldehyde-3-phosphate dehydrogenase is down-regulated. The differential partitioning of glyceraldehyde-3-phosphate dehydrogenase has also been observed in S. gordonii under different pH conditions (24). Although the total enzyme activity associated with this protein remained relatively constant in S. gordonii, the protein was primarily associated with the cell surface following growth at pH 6.5. When the pH was altered experimentally to a more neutral pH, similar to that of human blood, however, glyceraldehyde-3-phosphate dehydrogenase was predominantly extracellular. The adaptive benefit of this increased secretion is not yet fully understood. Here, phosphoglycerate kinase and triosephosphate isomerase were also identified among the predominant surface-associated proteins of S. oralis. It has been demonstrated that phosphoglycerate kinase is present on the surfaces of group B streptococci, and sera directed against this protein protected neonatal animals from S. agalactiae infection (17). Surface-associated pneumococcal triosephosphate isomerase was also identified in a study of immunoreactive proteins identified by screening a genomic expression library with convalescent-phase serum (37).
We previously reported the detection and identification of a number of ABC transporters by examination of whole-cell proteins of S. oralis (36). In addition to these, a surface-associated protein (SP0868) not previously identified from the whole-cell proteins with homology to an iron-regulated ABC transporter was shown to be up-regulated at low pH in this study. The precise role of these proteins in the response to acid stress in streptococci is unclear, but Cvitkovitch et al. (11) used insertional mutagenesis to show that an ABC transporter made a significant contribution to the ability of Streptococcus mutans to grow at low pH. Immunization with components of ABC transporters also protects against S. pneumoniae infection in mice (6).
Among those proteins up-regulated at low pH were a peptidase, with 67% homology to the pepV gene product from L. lactis, and a protein encoding a manganese-dependent inorganic pyrophosphatase. The gene pepV encodes a dipeptidase which is involved in the final degradation of dipeptides and which may play a role in the degradation of the milk protein casein in lactococci (16), and a homologue has also recently been identified in the culture supernatant of S. gordonii FSS2, where it may also be involved in the acquisition of small peptides (15). The dipeptidase homologue identified here in S. oralis may also be involved in the degradation and acquisition of peptides. The pyrophosphatase has significant homology with an intrageneric adhesin from S. gordonii (35) and may facilitate increased coaggregation with other streptococcal species and other genera of bacteria at low pH, which has relevance for the persistence of S. oralis in the oral biofilm.
Several of the surface-associated proteins of S. oralis were present as isoforms, probably the result of posttranslational modifications. The phosphorylation and dephosphorylation of proteins in prokaryotes by the action of kinases and phosphatases, respectively, is now recognized as a key mechanism by which functional activity may be regulated. A surface-associated serine/threonine protein phosphatase, which is important in the phosphorylation of proteins and signal transduction in other organisms (22, 31), was found to be up-regulated at low pH in S. oralis, suggesting that it may be involved in posttranslational modifications of externalized, surface-associated proteins.
In this study we have identified the predominant surface-associated proteins of S. oralis and confirmed that specific proteins previously identified in other streptococcal species, involved in central and intermediary metabolism, stress response, and other functions, were also cell surface associated in this organism. We have also identified novel surface-associated proteins, including other glycolytic enzymes, not previously reported to be at the streptococcal surface. The expression of several of the proteins identified was influenced by the initial pH of the culture media, although the composition of the acidulant used in this study may have also contributed to modulation of the osmotic potential of the medium. In addition, culture at low pH results in reduced growth rates for S. oralis (36), which may have influenced protein expression. Nonetheless, we have detected numerous novel proteins and shown differential expression under conditions that have relevance to the biology of the organism in both oral cavity and in extraoral infections. The significance and function of these surface-associated proteins in the survival and pathogenicity of S. oralis are currently not known and require further investigation in future studies.
Acknowledgments
This study was supported in part by a Ph.D. Studentship awarded to Joanna C. Wilkins by the Pathological Society of Great Britain and Ireland.
We are grateful to Gary Woffendin of Thermo Finnigan (Hemel Hempstead, Hertfordshire, United Kingdom) for assistance with LC-MS/MS analyses.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beighton, D., A. D. Carr, and B. A. Oppenheim. 1994. Identification of viridans streptococci associated with bacteraemia in neutropenic cancer patients. J. Med. Microbiol. 40:202-204. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, R. W., J. A. Leigh, and M. D. Collins. 1991. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int. J. Syst. Bacteriol. 41:487-494. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273-1287. [DOI] [PubMed] [Google Scholar]
- 5.Brailsford, S. R., R. W. Byrne, S. Adams, L. Zoitopoulos, C. Allison, and D. Beighton. 1999. Investigation of the aciduric microflora of plaque. Caries Res. 33:290. [Google Scholar]
- 6.Brown, J. S., A. D. Ogunniyi, M. C. Woodrow, D. W. Holden, and J. C. Paton. 2001. Immunization with components of two iron uptake ABC transporters protects mice against systemic Streptococcus pneumoniae infection. Infect. Immun. 69:6702-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldas, T., S. Laalami, and G. Richarme. 2000. Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J. Biol. Chem. 275:855-860. [DOI] [PubMed] [Google Scholar]
- 8.Caldas, T. D., A. El Yaagoubi, and G. Richarme. 1998. Chaperone properties of bacterial elongation factor EF-Tu. J. Biol. Chem. 273:11478-11482. [DOI] [PubMed] [Google Scholar]
- 9.Chhatwal, G. S. 2002. Anchorless adhesins and invasins of gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 10:205-208. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cvitkovitch, D. G., J. A. Gutierrez, J. Behari, P. J. Youngman, J. E. Wetz, P. J. Crowley, J. D. Hillman, L. J. Brady, and A. S. Bleiweis. 2000. Tn917-lac mutagenesis of Streptococcus mutans to identify environmentally regulated genes. FEMS Microbiol. Lett. 182:149-154. [DOI] [PubMed] [Google Scholar]
- 12.Dixon, S., M. Haswell, D. Harrington, and I. C. Sutcliffe. 2001. Surface immunolocalisation of HPr in the equine pathogen Streptococcus equi. Syst. Appl. Microbiol. 24:486-489. [DOI] [PubMed] [Google Scholar]
- 13.Douglas, C. W., J. Heath, K. K. Hampton, and F. E. Preston. 1993. Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 39:179-182. [DOI] [PubMed] [Google Scholar]
- 14.Dubreuil, J. D., M. Jacques, D. Brochu, M. Frenette, and C. Vadeboncoeur. 1996. Surface location of HPr, a phosphocarrier of the phosphoenolpyruvate: sugar phosphotransfase system in Streptococcus suis. Microbiology 142:837-843. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein, J. M., D. Nelson, T. Kordula, J. A. Mayo, and J. Travis. 2002. Extracellular arginine aminopeptidase from Streptococcus gordonii FSS2. Infect. Immun. 70:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellendoorn, M. A., B. M. Franke-Fayard, I. Mierau, G. Venema, and J. Kok. 1997. Cloning and analysis of the pepV dipeptidase gene of Lactococcus lactis MG1363. J. Bacteriol. 179:3410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes, M. J., J. C. Moore, J. D. Lane, R. Wilson, P. K. Pribul, Z. N. Younes, R. J. Dobson, P. Everest, A. J. Reason, J. M. Redfern, F. M. Greer, T. Paxton, M. Panico, H. R. Morris, R. G. Feldman, and J. D. Santangelo. 2002. Identification of major outer surface proteins of Streptococcus agalactiae. Infect. Immun. 70:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawabata, S., Y. Tamura, J. Murakami, Y. Terao, I. Nakagawa, and S. Hamada. 2002. A novel, anchorless streptococcal surface protein that binds to human immunoglobulins. Biochem. Biophys. Res. Commun. 296:1329-1333. [DOI] [PubMed] [Google Scholar]
- 19.Kawamura, Y., X.-G. Hou, F. Sultana, H. Miura, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 20.Kessler, R. E., and Y. Yagi. 1983. Identification and partial characterization of a pheromone-induced adhesive surface antigen of Streptococcus faecalis. J. Bacteriol. 155:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewthwaite, J., A. Skinner, and B. Henderson. 1998. Are molecular chaperones microbial virulence factors? Trends Microbiol. 6:426-428. [DOI] [PubMed] [Google Scholar]
- 22.Mamoun, C. B., and D. E. Goldberg. 2001. Plasmodium protein phosphatase 2C dephosphorylates translation elongation factor 1β and inhibits its PKC-mediated nucleotide exchange activity in vitro. Mol. Microbiol. 39:973-981. [DOI] [PubMed] [Google Scholar]
- 23.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson, D., J. M. Goldstein, K. Boatright, D. W. S. Harty, S. L. Cook, P. J. Hickman, J. Potempa, J. Travis, and J. A. Mayo. 2001. pH-regulated secretion of a glyceraldehyde-3-phosphate dehydrogenase from Streptococcus gordonii FSS2: purification, characterization, and cloning of the gene encoding this enzyme. J. Dent. Res. 80:371-377. [DOI] [PubMed] [Google Scholar]
- 25.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie brilliant blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 26.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pancholi, V., and V. A. Fischetti. 1998. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 28.Pappin, D. J. C., P. Hojrup, and A. J. Bleasby. 1993. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 3:327-332. [DOI] [PubMed] [Google Scholar]
- 29.Sprang, S. R. 1997. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 66:639-678. [DOI] [PubMed] [Google Scholar]
- 30.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 31.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 32.Vriesema, A. J., J. Dankert, and S. A. Zaat. 2000. A shift from oral to blood pH is a stimulus for adaptive gene expression of Streptococcus gordonii CH1 and induces protection against oxidative stress and enhanced bacterial growth by expression of msrA. Infect. Immun. 68:1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West, P. W., R. Al-Sawan, H. A. Foster, Q. Electricwala, A. Alex, and D. Panigrahi. 1998. Speciation of presumptive viridans streptococci from early onset neonatal sepsis. J. Med. Microbiol. 47:923-928. [DOI] [PubMed] [Google Scholar]
- 34.Whiley, R. A., and D. Beighton. 1998. Current classification of the oral streptococci. Oral Microbiol. Immunol. 13:195-216. [DOI] [PubMed] [Google Scholar]
- 35.Whittaker, C. J., D. L. Clemans, and P. E. Kolenbrander. 1996. Insertional inactivation of an intrageneric coaggregation-relevant adhesin locus from Streptococcus gordonii DL1 (Challis). Infect. Immun. 64:4137-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkins, J. C., K. A. Homer, and D. Beighton. 2001. Altered protein expression of Streptococcus oralis cultured at low pH revealed by two-dimensional gel electrophoresis. Appl. Environ. Microbiol. 67:3396-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zysk, G., R. J. Bongaerts, E. ten Thoren, G. Bethe, R. Hakenbeck, and H. P. Heinz. 2000. Detection of 23 immunogenic pneumococcal proteins using convalescent-phase serum. Infect. Immun. 68:3740-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]