Abstract
Sequencing the glnA genes of two chemically induced Azospirillum brasilense glutamine synthetase mutants revealed an Arg→Cys mutation, corresponding to the glutamate binding site, in one mutant and an Asp→Asn mutation, corresponding to the ammonium binding site, in the second mutant. The phenotypic changes in these mutants are discussed in relation to their genotypes.
Azospirillum brasilense is a gram-negative nitrogen-fixing soil bacterium, well known for its ability to colonize plant roots and to increase plant productivity (16). The main ammonium-assimilating pathway in this bacterium is the glutamine synthetase (GS; EC 6.3.1.2), glutamate synthase (EC 1.4.4.13) pathway (25). GS, a key enzyme in the nitrogen metabolism of both prokaryotes and eukaryotes, catalyzes the condensation of ammonium and glutamate to yield glutamine. This ATP-consuming process is regulated both transcriptionally (7) and posttranslationally by adenylylation of the GS in nitrogen excess conditions (4, 15, 25). Presently, only one GS (GSI type) has been found in A. brasilense, and no inactivating insertion mutations of glnA (encoding GS) have yet been reported. Previously isolated nitrosoguanidine-induced mutants 7028 and 7029 were used in this study (9). Both mutants have reduced GS activity. Mutant 7029, which has less than 1.5% GS activity compared to the wild type, lacks nitrogenase activity, while 7028, which still has ∼18% GS activity compared to the wild type, has constitutive nitrogenase activity (i.e., fixing nitrogen in the presence of ammonia) (9). [14C]methylammonium uptake, which is used as a measure of the activity of the AmtB ammonium transporter, is impaired in both mutants (19). Plasmids carrying the wild-type glnA gene complement both mutants for glutamine auxotrophy, wild-type nitrogen fixation, and [14C]methylammonium uptake (4; A. Van Dommelen, unpublished results).
Measuring ammonium excretion.
Since release of combined nitrogen is a key trait for agronomically useful diazotrophic bacteria and since impairment of ammonium assimilation generally correlates with ammonium excretion (17, 20, 22), whether strains 7028 and 7029 excrete ammonium was tested. A. brasilense strains Sp7 (wild type) (18), 7028, and 7029 were grown in liquid MMAB medium (24) supplemented with 100 mg of glutamine/liter and containing 8 mM KNO3 as the nitrogen source. The ammonium concentration in the supernatant of exponentially grown cells was measured as described by Chaney and Marbach (5). Concentrations up to 2 mM ammonium were observed for strain 7029. For strain 7028 a maximum concentration of 1 mM ammonium was measured, whereas, for the wild-type strain Sp7, no ammonium excretion was detected (detection limit of the assay: 25 μM). Complementing the GS mutants with plasmid pAB462, containing the wild-type glnA gene (4), resulted in loss of ammonium excretion (data not shown).
Identification of the glnA mutations in 7028 and 7029.
Since the presence of functional glnB and glnA promoters in mutants 7028 and 7029 had been established previously (7), it was decided to determine the glnA coding sequence in both mutants. Two primers annealing outside the published Sp7 glnA sequence (GenBank accession no. M26107.1) (3) and two high-fidelity PCR polymerases (Vent [Biolabs] and High Fidelity [Roche]) were used to amplify the glnA regions of 7028 and 7029. Three independent amplification products from each mutant were sequenced. Primer 1 (5′GTGAATTCTTGGGAAAGGCATGACATAACG3′) anneals 80 bp upstream of the glnA coding region, and primer 2 (5′GTGAATTCGGGCGGACACCGGAATCCG3′) anneals 20 bp downstream of the glnA coding region. Both primers contained an EcoRI restriction site at their 5′ ends to facilitate cloning the amplified fragments. Four differences between the sequenced glnA coding regions and the published A. brasilense Sp7 glnA sequence were found (Table 1). The observed changes can be divided in two groups: differences found in both mutant strains and differences found in only one of the mutant strains. The two differences found in both GS mutants, compared to the M26107.1 glnA sequence, are also found in the A. brasilense glnA sequence submitted by Chen and coworkers (GenBank accession no. AF323964.1). Therefore, they probably reflect an inaccuracy in the published M26107.1 sequence. These changes occur in regions not known to be of importance in the GS reaction mechanism. The CG→GC substitution at positions 368 and 369 results in a Gly at position 123 of the amino acid sequence, which corresponds to the GS sequences of related bacteria, such as Rhodospirillum rubrum, Rhodobacter sphaeroides, Rhodobacter capsulatus, and other proteobacteria, such as Azorhizobium caulinodans, Sinorhizobium meliloti, Rhizobium leguminosarum, Azotobacter vinelandii, and Pseudomonas aeruginosa. The CTG insertion at position 1243 generates a Leu residue that is conserved in GS of prokaryotes (8).
TABLE 1.
Differences in nucleotide and deduced amino acid sequence between the published Sp7 glnA gene (M26107.1) and the glnA sequences of mutants 7028 and 7029
| Nucleotide position(s) | Nucleotide(s) in:
|
Amino acid position | Amino acid in
|
Strain(s) | ||
|---|---|---|---|---|---|---|
| Published sequence | This study | Published sequence | This study | |||
| 139 | G | A | 52 | D | N | 7029 |
| 368, 369 | CG | GC | 123 | A | G | 7028, 7029 |
| 964 | C | T | 322 | R | C | 7028 |
| 1243 | —a | CTG | 415 | — | L | 7028, 7029 |
—, no nucleotide or amino acid at this position.
The sequence difference found only in mutant 7028 (C964T) changes a large positively charged Arg, corresponding to a conserved residue of the glutamate binding site, to a small, nonpolar Cys. It has been proposed that the guanidine group of the conserved Arg residue forms hydrogen bonds with the α-carboxylate group of glutamate (12).
In mutant 7029, the G139A mutation changes a conserved Asp into an uncharged Asn at a site corresponding to the proposed NH4+ binding site (1, 13). The negative charge of the conserved Asp is crucial in the GS catalytic mechanism since it facilitates the deprotonation of NH4+ in order to generate the active species NH3, which will attack the γ-glutamyl phosphate formed in the GS biosynthetic reaction (11). This Asp residue also increases the affinity of GS for ammonium binding.
Resistance of GS mutant 7028 to GS inhibitor MetSox.
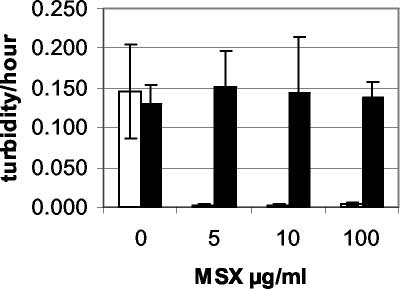
Since the potent GS inhibitor methionine sulfoximine (MetSox) is known to bind at the same site as glutamate (8), the question of whether there was any effect on the sensitivity of mutant 7028, which has an altered glutamate binding site, to this antimetabolite was investigated. Wild-type Sp7 and GS mutant 7028 were grown in liquid MMAB medium (24) with 20 mM ammonium as the nitrogen source and different concentrations of MetSox. Strain 7028 contained plasmid pAB462, with the wild-type glnA gene (4), to ensure glutamine-independent growth. Growth was measured in a Bioscreen C growth analyzer (Labsystems) with a white band filter. Concentrations up to 100 μg of MetSox/ml did not significantly alter growth of strain 7028, while 5 μg of MetSox/ml was enough to completely inhibit wild-type growth (Fig. 1).
FIG. 1.
Growth of wild-type Sp7 (white) and GS mutant 7028 (containing plasmid pAB462) (black) in minimal medium with 20 mM ammonium as the nitrogen source and different concentrations of MetSox (MSX). The data are means and standard errors from five replicates.
Measuring the GS biosynthetic reaction with hydroxylamine as a substrate.
Although the GS biosynthetic reaction with ammonium as a substrate is strongly reduced (9) in both GS mutants, when the biosynthetic GS reaction was measured as described by Bender and coworkers (2), activity in mutant 7029 was found to be twice as high as wild-type activity (data not shown). In this reaction ammonium is replaced by hydroxylamine and the concentration of γ-glutamyl hydroxamate formed is determined spectrophotometrically according to the reaction
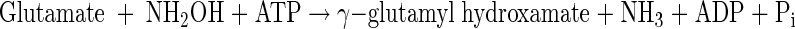 |
The enhanced reaction with this alternative substrate most likely reflects a better binding of the uncharged hydroxylamine when the negatively charged Asp of the ammonium binding site is replaced by an uncharged Asn. Mutant 7028 was found to have 15% of the wild-type biosynthetic GS activity when the method of Bender and coworkers (2) was used. This corresponds to the activity measured by Gauthier and Elmerich (9) with ammonium as a substrate.
Influence of GS mutation on the expression of the Ntr-regulated amtB gene.
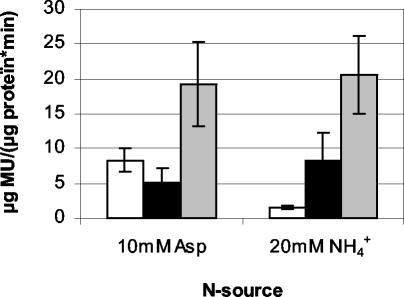
The NtrB-NtrC two-component regulatory system has a central role in nitrogen regulation in many bacteria (14). The A. brasilense amtB gene, encoding an ammonium transporter, is activated by this two-component regulatory system in nitrogen-limiting conditions (21). To monitor amtB expression, pFAJ302 containing a pamtB-gusA translational fusion (21) was conjugated into the wild type and GS mutants. After an overnight preculture in rich medium, cells were grown for 4 h in MMAB medium supplemented with 100 mg of glutamine/liter and the nitrogen source indicated in Fig. 2. β-Glucuronidase activity was measured fluorometrically by monitoring the cleavage of 4-methylumbelliferyl-β-d-glucuronide (Sigma-Aldrich N.V.) to 4-methylumbelliferone and glucuronic acid (10, 23). Results are shown in Fig. 2. Both GS mutants fail to repress amtB transcription in the presence of 20 mM NH4Cl. This could indicate that in the wild type the glutamine formed by GS signals a high nitrogen level in the nitrogen sensory cascade of A. brasilense. It has also been proposed that glutamine is the signaling molecule for the ammonium switch-off of the A. brasilense nitrogenase (26). In this respect it is interesting that de Zamaroczy and coworkers (7) found that the glnA promoters of GS mutants 7028 and 7029 also fail to respond to high levels of ammonium. This change was not due to a change in the glnA promoter since de Zamaroczy and coworkers (7) found no difference in nucleotide sequence between the glnA promoters of both GS mutants and the wild type.
FIG. 2.
Expression of a pamtB-gusA fusion (pFAJ302) in wild-type Sp7 (white) and glnA mutants 7028 (black) and 7029 (grey) grown in minimal medium supplemented with the indicated nitrogen source. MU, 4-methylumbelliferone. The data are means and standard deviations from four replicates.
Although the GS mutants lack [14C]methylammonium uptake, which reflects the activity of the amtB gene product (19), amtB is expressed in the conditions used for the [14C]methylammonium uptake assay (MMAB supplemented with 10 mM aspartate as the nitrogen source) (Fig. 2). Perhaps the lack of [14C]methylammonium uptake that was observed (19) is due to the presence of excreted ammonium (which is taken up with a much higher affinity than [14C]methylammonium) or a negative posttranslational regulation of the AmtB transporter. In Escherichia coli and Azotobacter vinelandii, the PII-like signal transduction protein GlnK binds to AmtB and negatively regulates AmtB transport activity (6).
Conclusion.
In this paper, alterations of the ammonium and glutamate binding site of GS were found in two ammonium-excreting Azospirillum mutants. Since in Azospirillum GS is not switched off during plant association, as it is in, e.g., Rhizobium (17), protein engineering of GS offers the possibility to generate ammonium-excreting Azospirillum strains, as exemplified by the two mutants analyzed in this study.
Acknowledgments
We thank C. Elmerich and M. de Zamaroczy for kindly providing strains 7028 and 7029 and plasmid pAB462. We are also grateful to M. de Zamaroczy for constructive discussion and useful suggestions.
A.V.D. is the recipient of a postdoctoral fellowship from the Onderzoeksfonds K. U. Leuven. Part of this research was funded by a grant (GOA/98/Vanderleyden) from the Flemish Government.
REFERENCES
- 1.Alibhai, M., and J. J. Villafranca. 1994. Kinetic and mutagenic studies of the role of the active site residues Asp-50 and Glu-327 of Escherichia coli glutamine synthetase. Biochemistry 33:682-686. [DOI] [PubMed] [Google Scholar]
- 2.Bender, R. A., K. A. Janssen, A. D. Resnick, M. Blumenberg, F. Foor, and B. Magasanik. 1977. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J. Bacteriol. 129:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozouklian, H., and C. Elmerich. 1986. Nucleotide sequence of the Azospirillum brasilense Sp7 glutamine synthetase structural gene. Biochimie 68:1181-1187. [DOI] [PubMed] [Google Scholar]
- 4.Bozouklian, H., C. Fogher, and C. Elmerich. 1986. Cloning and characterization of the glnA gene of Azospirillum brasilense Sp7. Ann. Inst. Pasteur Microbiol. 137B:3-18. [DOI] [PubMed] [Google Scholar]
- 5.Chaney, A. L., and E. P. Marbach. 1962. Modified reagents for determination of urea and ammonia. Clin. Chem. 8:130-132. [PubMed] [Google Scholar]
- 6.Coutts, G., G. Thomas, D. Blakey, and M. Merrick. 2002. Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 21:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Zamaroczy, M., A. Paquelin, and C. Elmerich. 1993. Functional organization of the glnB-glnA cluster of Azospirillum brasilense. J. Bacteriol. 175:2507-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberg, D., H. S. Gill, G. M. Pfluegl, and S. H. Rotstein. 2000. Structure-function relationships of glutamine synthetases. Biochim. Biophys. 1477:122-145. [DOI] [PubMed] [Google Scholar]
- 9.Gauthier, D., and C. Elmerich. 1977. Relationship between glutamine synthetase and nitrogenase in Spirillum lipoferum. FEMS Microbiol. Lett. 2:101-104. [Google Scholar]
- 10.Jefferson, R. A. 1987. Assaying chimeric genes in plants: the gus gene fusion system. Plant. Mol. Biol. Rep. 5:387-405. [Google Scholar]
- 11.Liaw, S. H., and D. Eisenberg. 1994. Structural model for the reaction mechanism of glutamine synthetase, based on five crystal structures of enzyme-substrate complexes. Biochemistry 33:675-681. [DOI] [PubMed] [Google Scholar]
- 12.Liaw, S. H., C. Pan, and D. Eisenberg. 1993. Feedback inhibition of fully unadenylylated glutamine synthetase from Salmonella typhimurium by glycine, alanine, and serine. Proc. Natl. Acad. Sci. USA 90:4996-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liaw, S. H., I. Kuo, and D. Eisenberg. 1995. Discovery of the ammonium substrate site on glutamine synthetase, a third cation binding site. Protein Sci. 4:2358-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrick, M. J., and R. A. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okon, Y., S. L. Albrecht, and R. H. Burris. 1976. Carbon and ammonia metabolism of Spirillum lipoferum. J. Bacteriol. 128:592-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okon, Y., and J. Vanderleyden. 1997. Azospirillum-plant association. A case study for molecular mechanisms in phytostimulation, crop yield improvement, and rhizosphere ecology. ASM News 63:366-370. [Google Scholar]
- 17.Patriarca, E. J., R. Tatè, and M. Iaccarino. 2002. Key role of bacterial NH4+ metabolism in Rhizobium-plant symbiosis. Microbiol. Mol. Biol. Rev. 66:203-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarrand, J. J., N. R. Krieg, and J. Döbereiner. 1978. A taxonomic study of the Spirillum lipoferum group, with the description of a new genus, Azospirillum gen. nov., and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can. J. Microbiol. 24:967-980. [DOI] [PubMed] [Google Scholar]
- 19.Van Dommelen, A., E. Van Bastelaere, V. Keijers, and J. Vanderleyden. 1997. Genetics of Azospirillum brasilense with respect to ammonium transport, sugar uptake and chemotaxis. Plant Soil 194:155-160. [Google Scholar]
- 20.Van Dommelen, A. 1998. Ph.D. thesis. Katholieke Universiteit Leuven, Heverlee, Belgium.
- 21.Van Dommelen, A., V. Keijers, J. Vanderleyden, and M. de Zamaroczy. 1998. (Methyl)ammonium transport in the nitrogen-fixing bacterium Azospirillum brasilense. J. Bacteriol. 180:2652-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Dommelen, A., R. De Mot, and J. Vanderleyden. 2001. Ammonium transport, unifying concepts and unique aspects. Aust. J. Plant Physiol. 28:1-9. [Google Scholar]
- 23.Van Dommelen, A., V. Keijers, E. Somers, and J. Vanderleyden. 2002. Cloning and characterisation of the Azospirillum brasilense glnD gene and analysis of a glnD mutant. Mol. Genet. Genomics 266:813-820. [DOI] [PubMed] [Google Scholar]
- 24.Vanstockem, M., K. Michiels, J. Vanderleyden, and A. Van Gool. 1987. Transposon mutagenesis of Azospirillum brasilense and Azospirillum lipoferum: physical analysis of Tn5 and Tn5-Mob insertion mutants. Appl. Environ. Microbiol. 53:410-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westby, C. A., C. S. Enderlin, N. A. Steinberg, C. M. Joseph, and J. C. Meeks. 1987. Assimilation of 13NH4+ by Azospirillum brasilense grown under nitrogen limitation and excess. J. Bacteriol. 169:4211-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, Y., R. H. Burris, P. W. Ludden, and G. P. Roberts. 1994. Posttranslational regulation of nitrogenase activity in Azospirillum brasilense ntrBC mutants: ammonium and anaerobic switch-off occurs through independent signal transduction pathways. J. Bacteriol. 176:5780-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]