Abstract
Phylogenetic and stable-isotope analyses implicated two methanogen-like archaeal groups, ANME-1 and ANME-2, as key participants in the process of anaerobic methane oxidation. Although nothing is known about anaerobic methane oxidation at the molecular level, the evolutionary relationship between methane-oxidizing archaea (MOA) and methanogenic archaea raises the possibility that MOA have co-opted key elements of the methanogenic pathway, reversing many of its steps to oxidize methane anaerobically. In order to explore this hypothesis, the existence and genomic conservation of methyl coenzyme M reductase (MCR), the enzyme catalyzing the terminal step in methanogenesis, was studied in ANME-1 and ANME-2 archaea isolated from various marine environments. Clone libraries targeting a conserved region of the alpha subunit of MCR (mcrA) were generated and compared from environmental samples, laboratory-incubated microcosms, and fosmid libraries. Four out of five novel mcrA types identified from these sources were associated with ANME-1 or ANME-2 group members. Assignment of mcrA types to specific phylogenetic groups was based on environmental clone recoveries, selective enrichment of specific MOA and mcrA types in a microcosm, phylogenetic congruence between mcrA and small-subunit rRNA tree topologies, and genomic context derived from fosmid sequences. Analysis of the ANME-1 and ANME-2 mcrA sequences suggested the potential for catalytic activity based on conservation of active-site amino acids. These results provide a basis for identifying methanotrophic archaea with mcrA sequences and define a functional genomic link between methanogenic and methanotrophic archaea.
Anaerobic methane production and consumption play critical roles in carbon cycling in marine sediments (26). These processes are enabled by related groups of methanogenic archaea and methane-oxidizing archaea (MOA) (reviewed in references 31 and 32). All known methanogens express the enzyme methyl coenzyme M reductase (MCR), which catalyzes the terminal step in biogenic methane production (reviewed in references 8, 27, and 30). Currently the presence of MCR is considered a diagnostic indicator of methanogenesis (8, 16, 18, 27, 30). The genomes of all methanogenic archaea encode at least one copy of the mcrA operon (reviewed in reference 7, 27, 30). Composed of two alpha (mcrA), beta (mcrB) and gamma (mcrG) subunits, the mcrA holoenzyme catalyzes heterodisulfide formation between coenzyme M and coenzyme B from methyl-coenzyme M and coenzyme B and the subsequent release of methane (5). Functional constraints on its catalytic activity have resulted in a high degree of MCR amino acid sequence conservation, even between phylogenetically distant methanogenic lineages (18, 27). This conserved primary structure has been used to develop degenerate PCR primers for recovering naturally occurring mcrA fragments from a variety of environments (16, 18). The resulting mcrA sequence data have been employed as a proxy for methanogen diversity (16, 18).
Diagenetic modeling and geochemical studies have predicted and identified the process of anaerobic methane oxidation in anoxic marine environments (1, 14, 19, 25, 26, 33). Subsequent culture-independent biochemical and molecular studies revealed the lipid biomarker and genetic signatures of methanogen-related archaeal communities associated with anaerobic methane oxidation (4, 12, 21-24). Currently no archaeon capable of anaerobic methane oxidation has been isolated in pure culture, but coupled fluorescent in situ hybridization and isotopic analysis have linked two specific groups of MOA, ANME-1 and ANME-2, to the process of anaerobic methane oxidation (22). The specific molecular mechanisms underlying anaerobic methane oxidation remain obscure. One possibility suggested by phylogenetic and biochemical considerations is that MOA have co-opted the methanogenic pathway, reversing key steps to enable methane oxidation anaerobically. To begin testing this hypothesis, we attempted to isolate mcrA genes from MOA by a variety of approaches, including PCR surveys of naturally occurring populations, enrichment cultures, and genomic library screening.
MATERIALS AND METHODS
Site description and sampling.
Sediment push cores were collected at Eel River Basin with the ROV Ventana and at Monterey Canyon with the ROV Tiburon. Benthic sediments collected from Monterey Bay were incubated for 24 weeks on a continuous-flow anaerobic sediment incubator (AMIS) (10a). The AMIS incubator was designed to provide anaerobic, methane-saturated, sulfate-containing seawater to the sediments to foster the enrichment of MOA. Enrichments with the AMIS bioreactor led to the growth of MOA in both sediments collected from seeps (seep sediments) as well as sediments collected away from seeps (nonseep sediments). Prior to AMIS incubation, MOA were not detected in nonseep sediment following small-subunit (SSU) 16S rRNA-directed PCR screening and in situ hybridization with MOA group-specific probes (10a). After incubation, MOA were detectable, and 1-g subsamples of these incubated sediments were taken for analysis in this study. Piston cores were collected at Blake Ridge off the R/V Cape Hatteras. For microbiological analysis, 0.5 g of sediment was diluted in 1 ml of 1× phosphate-buffered saline-ethanol and stored at −20°C until processed.
Sediment DNA extraction.
From 0.25 to 0.5 g of sediment in 1× phosphate-buffered saline-ethanol was diluted in 1500 μl of 1× phosphate-buffered saline and sonicated for 30 s at 30 A (Sonics and Material Inc., Danbury, Conn.) on ice. Samples were layered over a 50% Percoll (Sigma)-1× phosphate-buffered saline continuous gradient and centrifuged for 15 min at 4,800 rpm in an HS-4 rotor at 4°C. DNA for subsequent PCR amplification was extracted from sediment pellets following Percoll gradient centrifugation with a Fast soil prep kit (MoBio, San Diego, Calif.). Final elution volumes varied between 30 and 50 μl of TE (10 mM Tris, 1 mM EDTA, pH 7.5).
Fosmid library construction and screening.
Fosmids were prepared by a modification of a previously described protocol (28). Pooled supernatant from 10 g of Eel River T201 Percoll layered sediment was filtered onto 3-μm-pore-size polycarbonate filters and either frozen at −20°C or processed immediately for high-molecular-weight DNA extraction (Hallam et al., unpublished data). Briefly, high-molecular-weight DNA was end-repaired and separated on 0.8% agarose in 1× TAE overnight at 30 V. Then 40- to 50-kb fragment pools were gel purified and cloned into the vector pEpiFOS (Epicentre) according to the manufacturer's instructions. Ligated DNA was packaged with the Epicentre MaxPlax lambda packaging extract and used to transfect Escherichia coli DH10B cells (Bethesda Research Laboratories). Transfected cells were selected on Luria-Bertani (LB) agar containing chloramphenicol.
The resulting clones were picked into 96-well (n = 37) microtiter dishes containing LB supplemented with chloramphenicol and 7% glycerol and stored at −80°C. For screening purposes each 96-well plate was individually pooled, and plasmid DNA was extracted by standard alkaline lysis procedures (28). Fosmid subclone libraries were generated with the Topo Shotgun kit (Invitrogen, Carlsbad, Calif.). Briefly, high-molecular-weight DNA was nebulized to 1 to 3 kb, end-repaired, and cloned into the Topo blunt-end cloning vector pCR4. Ligated DNA was used to transform electrocompetent E. coli Top10 cells. Transformants were selected on LB containing 50 μg of kanamycin per ml under blue-white selection. The resulting clones were robotically picked at the Joint Genome Institute into 384-well microtiter dishes containing LB with 50 μg of kanamycin per ml plus 7% glycerol and stored at −80°C.
SSU rRNA and mcrA gene amplification.
SSU rRNA sequences were PCR amplified from environmental and fosmid DNA extracts with archaeon-specific primers A20_F (5′-TTCCGGTTGATCCYGCCRG) and A958_R (5′-YCCGGCGTTGAMTCCAATT). mcrA group a and c to e sequences were PCR amplified with universal mcrA primers ME1 (5′-GCMATGCARATHGGWATGTC) and ME2 (5′-TCATKGCRTAGTTDGGRTAGT) (11). mcrA group b was amplified with specific primer pair AOM39_F (5′ GCTGTGTAGCAGGAGAGTCA) and AOM40_R (5′ GATTATCAGGTCACGCTCAC). PCR conditions for both target sequences were identical. The 50-μl amplification reaction mixtures contained 1 μl of template DNA, 41.5 μl of 1× buffer, 1 μl each of 10 μM forward and reverse primer, 2.5 U of TaqPlus Precision polymerase (Stratagene, La Jolla, Calif.), and 5 μl of 10 mM stock deoxynucleoside triphosphate mixture. Amplifications were carried out with the following profile: 94°C for 3 min, then 36 cycles of 94°C for 40 s, 55°C for 1.5 min, and 72°C for 2 min, followed by a final extension at 72°C for 10 min.
Clone library construction and sequencing.
SSU rRNA and mcrA amplicons were visualized on 1% agarose gels in 1× TBE and purified directly with the Qiaquick PCR purification kit (Qiagen, Valencia, Calif.). Purified products from fosmid screening were sequenced directly (see below). Purified products from environmental samples and AMIS microcosm enrichment were cloned into the pCR4-Topo vector with a Topo TA cloning kit for sequencing (Invitrogen, Carlsbad, Calif.) and transformed by chemical transformation into TOP10 one shot cells according to the manufacturer's instructions. Transformants were transferred to 96-well plates containing 180 μl of LB containing 50 μg of kanamycin per ml and 7% glycerol and stored at −80°C.
Plasmid DNA was purified from glycerol stocks with the Montage Plasmid Miniprep96 kit (Millipore, Bedford, Mass.) following the manufacturer's protocol and stored at −20°C. Plasmid insert sequence data were collected on an ABI Prism 3100 DNA sequencer (Applied Biosystems Inc, Foster, Calif.) with Big Dye chemistry (PE Biosystems, Foster, Calif.) according to the manufacturer's instructions. Plasmids were sequenced bidirectionally with M13F and M13R primers. SSU rRNA and mcrA amplicons from fosmid screening were sequenced bidirectionally with the A20 and A958 and the AOM39 and AOM40 primer pairs, respectively. Sequences were edited manually from traces with Sequencher software version 4.1.2 (Gene Codes Corporation, Ann Arbor, Mich.).
Phylogenetic analysis.
Phylogenetic analyses of SSU and mcrA sequences were preformed on sequences from known MOA groups, and representatives from the primary lines of descent within methanogenic groups. SSU rRNA sequence data were compiled with ARB software (www.arb-home.de) and aligned with sequences from the GenBank database with the FastAligner program. Aligned sequences were visually inspected for conservation of secondary structure features and manually edited when necessary. SSU rRNA trees were based on comparison of 541 nucleotides. Deduced amino acid sequences for environmental mcrA clones were determined from 684 bp of overlapping nucleotide sequence and aligned with the Clustal method implemented in MegAlign (DNA Star, Madison, Wis.). mcrA trees were based on comparison of 276 amino acids with the exception of Methanosaeta concilii (157 amino acids). Phylogenetic trees for both SSU rRNA and mcrA genes were generated with distance and parsimony methods implemented in PAUP version 4.0b10 (29). SSU rRNA sequence distances were estimated with the Kimura two-parameter model. Bootstrapping for distance and parsimony was accomplished with 1,000 replicates per tree with heuristic search methods.
Gene phylogenies for mcrA and mrt operon subunits were constructed from representative methanogenic lineages and two fosmids containing the complete mcrA operon from MOA groups ANME-1 and ANME-2 (Gzfos17A3 and Gzfos35D7). Alignments based on complete protein sequences were generated with the Clustal method implemented in MegAlign (DNA Star, Madison, Wis.). Unrooted trees for both mcrA and mrt subunits were generated with distance and parsimony methods implemented in PAUP version 4.0b10 (29). Bootstrapping for distance and parsimony was accomplished with 1,000 replicates per tree with heuristic search methods.
Nucleotide sequence accession numbers.
mcrA and rRNA gene sequences were submitted to GenBank and have been assigned accession numbers AY324362 to AY324373 and AY324374 to AY324382, respectively. In addition, genomic DNA sequences from GZfos17A3 and GZfos35D7 containing the complete mcrA operon were submitted to GenBank and have been assigned accession numbers AY327048 and AY327049, respectively.
RESULTS
Detection of PCR-amplified mcrA sequence in anaerobic methane oxidation-associated samples.
Samples were collected from marine sediments that were known to be active in anaerobic methane oxidation and that contained methanotrophic archaeal groups in different proportions. The AMIS bioreactor was also incorporated in the study because it enriched for the growth of specific MOA types. PCR-derived clone libraries containing a conserved region of the mcrA locus were generated (see Materials and Methods) and compared among and between anaerobic methane oxidation-associated environmental samples, methane-oxidizing microcosm enrichment (10a), and genomic libraries from purified MOA cell preparations (Table 1 and Materials and Methods).
TABLE 1.
Sample origin and environmental data
| Location and sample type | Sample | Core depth (cm) | Water depth (m) | Coordinates (°N, °W) | Description |
|---|---|---|---|---|---|
| Eel River | T201 | 4-7 | 550 | 40.48, 124.36 | Clam patch, high CH4 fosmid library |
| GZfos | |||||
| Monterey Canyon | |||||
| Seep | F17.1 | 0-25 | 955 | 36.77, 122.08 | Clam patch, high CH4 |
| Nonseep | C4.1 | AMIS microcosma | |||
| Blake Ridge | PC26 | 76 | 2,707 | 31.53, 75.45 | Hemipelagic sediment |
AMIS microcosm cultured from nonseep reference core collected at least 25 m away from any known seep site.
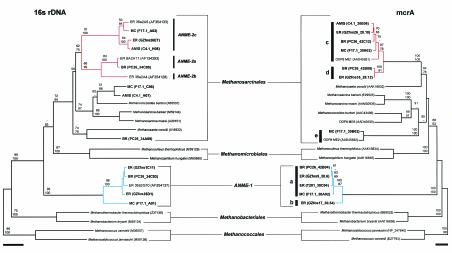
A total of 93 deduced mcrA clones fell into five distinct groups, a to e (Table 2 and Fig. 1). Nucleotide sequence heterogeneity within each group was moderate, with the majority of polymorphic sites falling in third-codon positions. Amino acid sequence identity between representative group a and group b sequences was 92.5%, and 93.3% between representative group c and group d sequences. Based on these observations, group a, group b, group c, and group d were collapsed into groups a-b and c-d, respectively. Group a-b was on average 44.9% (±1.3%) identical to group c-d. Group e was on average 45.6% (±1.9%) identical to group a-b, and 59.6% (±0.3%) identical to group c-d. Compared to mcrA sequences from well-characterized methanogenic groups, mcrA group a-b was on average 44.6% (±2.9%) identical, group c-d was 63.6% (±1.8%) identical, and group e was 58.1% (±5.2%) identical.
TABLE 2.
Archaeal 16s rDNA and mcrA clone recovery from MOA-containing sediments and fosmid DNA librarya
| DNA | Group | Recovery (no. of clones)
|
Total no. of clones | ||||
|---|---|---|---|---|---|---|---|
| Eel River
|
Monterey Canyon F17.1 | AMIS microcosm C4.1 | Blake Ridge PC26 | ||||
| T201 | Gzfos | ||||||
| Archaeal 16s rDNA | ANME-1 | 62 | 8 | 1 | 3 | 74 | |
| ANME-2 | |||||||
| ANME-2a | 1 | 41 | 42 | ||||
| ANME-2b | 9 | 9 | |||||
| ANME-2c | 20 | 1 | 27 | 43 | 91 | ||
| Methanogen-like | 1 | 1 | 22 | 24 | |||
| Other | 6 | 49 | 31 | 5 | 91 | ||
| mcrA group | a | 13 | 4 | 9 | 25 | 51 | |
| b | + | 6 | + | ND | 6 | ||
| c | 3 | 4 | 3 | 14 | 5 | 29 | |
| d | 1 | 3 | 4 | ||||
| e | 3 | 3 | |||||
T201 environmental sequences and the Gzfos library were both derived from the same purified cell preparation of MOA from Eel River T201 sediment (see text). Environmental samples and fosmid library were screened with universal mcrA primers ME-1 and ME-2 and ANME-1 type b-specific mcrA primers AOM39 and AOM40 (see text). +, positive amplification with AOM39 and AOM40 primer set. mcrA group represents conceptual translation product. ND, not determined.
FIG. 1.
Distance comparison of SSU rRNA and mcrA-based phylogenetic trees of environmental clones and primary methanogenic lineages. Bootstrap values are based on 1,000 replicates each (neighbor joining on top and parsimony on bottom) and are shown for branches with greater than 50% support. Methanocaldococcus spp. were used as the out group reference. ER, Eel River; MC, Monterey Canyon; AMIS, microcosm; BR, Blake Ridge. Boldface identifies clones identified and sequenced in this study. Red highlights ANME-2 group members, and blue highlights ANME-1 group members. Scale bars represent 0.05 nucleotide or amino acid substitution per site.
Common mcrA groups were shared between locations. However, in two instances, a single group was associated with a specific sample or location. mcrA group e was detected solely in Monterey Canyon seep sediment, and only mcrA group c was detected in methane-oxidizing microcosm enrichment (Table 2). Taken together, these results indicate that mcrA genes are readily detectable in anaerobic methane oxidation sediments and that their sequence diversity is not uniformly distributed.
In order to assess mcrA amplification bias associated with degenerate primer screening (17) and to identify large-insert DNA clones (i.e., fosmids) for genomic analyses, PCR-derived mcrA clone recovery from Eel River environmental sample T201 was compared to clone recovery from a fosmid library constructed from T201 purified cell preparations. T201 provided a 4:1 clone ratio between group a and group c, compared to a 1:1 ratio in fosmid clones, despite a clear bias in ANME-1 SSU rRNA representation (Table 2). This pattern was repeated for other environmental samples containing both the a and c groups, suggesting a consistent amplification bias toward group a by the degenerate mcrA primers ME1 and ME-2 (Table 2).
Environmental distribution and phylogeny of MOA-associated mcrA sequences.
Recent studies suggest that mcrA can substitute for SSU rRNA sequences in determining phylogenetic relationships between methanogenic archaea (18). To test this assertion and determine the phylogenetic affiliation of MOA-associated mcrA groups, SSU rRNA sequence recovery was compared to mcrA sequence recovery in the same samples. Most samples contained a high proportion of ANME-1 and ANME-2 SSU rRNA clones relative to other archaeal groups, although clone recovery varied between sites. Eel River T201, although dominated by ANME-1 sequences, also contained ribotypes corresponding to ANME-2a/b and ANME-2c (Table 2). Monterey Canyon sample F17.1, although dominated by ANME-2c sequences, also contained an ANME-1-like and a Methanococcoides-like sequence (Table 2). Similarly, methane-oxidizing microcosm C4.1 (10a) enriched from Monterey Canyon sediment was dominated by sequences corresponding to ANME-2c, but also contained a methanogen-like sequence most similar to Methanococcoides spp. (Table 2). Nonetheless, the vast majority of SSU rRNA clones recovered from these Monterey Canyon and Eel River samples designated other (data not shown) failed to group with any bona fide methanogen group (Table 2 and data not shown). In contrast, Blake Ridge PC26, although dominated by ANME-2a, also contained a common methanogen-like ribotype corresponding to Methanosaeta spp. (Table 2 and Fig. 1).
Phylogenetic trees constructed for mcrA and SSU rRNA sequences recovered from the same samples exhibited a high degree of congruence, enabling tentative assignment of MOA-associated mcrA groups to specific archaeal lineages (see Materials and Methods). The mcrA group a-b was associated with the ANME-1 lineage, while group c-d was associated with the ANME-2 lineage (Fig. 1). ANME-2a/b and ANME-2c rRNA genes formed a monophyletic group within the Methanosarcinales (Fig. 1). Similarly, mcrA group c-d, along with environmental sample ODP8-ME1 (3), formed a monophyletic cluster most closely related to Methanosaeta concilii (Fig. 1). The mcrA group e fell within the Methanosarcinales. (Fig. 1). The absence of ANME-1 in the methane-oxidizing microcosm enrichment (10a) provided independent support for the associations inferred from SSU rRNA and mcrA tree topologies. Consistent with this observation, all but one SSU rRNA clone and all mcrA clones retrieved from the methane-oxidizing enrichment fell within the ANME-2c subdivision and mcrA group c, respectively (Table 2 and Fig. 1). The mcrA group e, recovered only in Monterey Canyon, putatively fell within the Methanococcoides along with ODP8-ME2 and ODP8-ME6, based on SSU rRNA tree topology (Table 2 and Fig. 1).
To better define the evolutionary origin of MOA-associated mcrA groups described above, sequence alignments containing both methyl coenzyme M reductase (mcrA) and methyl coenzyme M reductase II (mrt) subunits from Methanocaldococcus jannaschii, Methanococcus vannielii, Methanthermobacter thermoautotrophicus, and Methanosarcina mazei were compared to the homologous genes in MOA-associated environmental PCR clones and fosmids. Sequence identity between the common region of mcrA and mrtA in M. jannaschii and M. thermoautotrophicus was 80.2% and 76.6%, respectively. The mrtA sequence identity between M. jannaschii and M. thermautotrophicus was 84.7%. In contrast, ANME-1-affiliated mcrA group a-b was on average 47.3% (±1.3%) identical to M. jannaschii and 48.6% (±1.3%) identical to M. thermoautotrophicus mrtA. The ANME-2-affiliated mcrA group c-d was on average 57.5% (±0.9%) identical to M. jannaschii and 57.9% (±3.7%) identical to M. thermoautotrophicus mrtA.
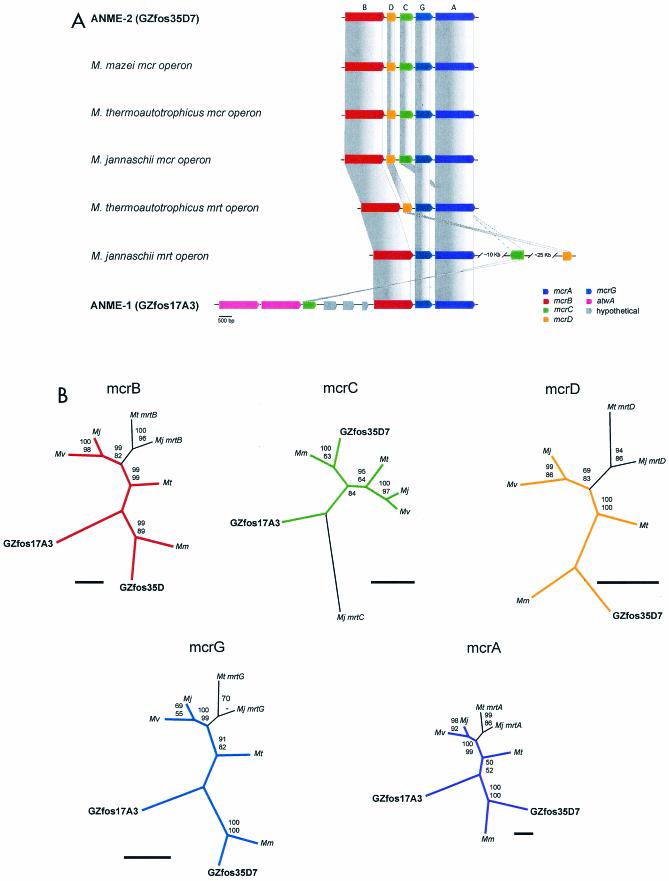
The mrt locus is found in both the Methanococcales and Methanobacteriales lineages and represents a second, genetically distinct mcrA operon with alternative subunit composition and order (Fig. 2A) (16, 27). In contrast, members of the Methanosarcinales contain only the mcrA locus, based on analysis of several completed genomes (9, 27). From a phylogenetic perspective, mrtA sequences form a related group within the Methanococcales mcrA lineage (16, 18). The subunit composition and order of ANME-2-associated fosmid GZfos35D7 resembled the mcrA structure found in all methanogenic lineages (mcrBDCGA) (Fig. 2A). In contrast, the subunit composition and order of ANME-1-associated fosmid GZfos17A3 appeared to diverge from canonical mcrA structure by the loss of mcrD and rearrangment of mcrC (Fig. 2A). No mcrD homolog could be deduced from the entire range of 37,609 contiguous bp carried by GZfos17A3.
FIG.2.
(A) Schematic depiction of mcrA and mrt operon structure for MOA-associated fosmids and major methanogenic lineages. The mcrA operon typically consists of mcrBDCGA. The mrt operon structure varies between lineages. Scale bar, 500 bp. (B) Gene trees for mcrA and mrt subunits depicted in A. Abbreviations for methanogenic species harboring these genes are Mj, Methanocaldococcus jannaschii; Mv, Methanococcus vannielii; Mt, Methanothermobacter thermautotrophicus; and Mm, Methanosarcina mazei. Accession numbers for sequences used in the analyses are shown in parentheses for mcrB-Mj (NP_247836.1), mcrB-Mv (P07956), mcrB-Mt (NP_276296.1), mcrB-Mm (NP_633268.1), mcrC-Mj (NP_247838.1), mcrC-Mv (P07960), mcrC-Mt (NP_276294.1), mcrC-Mm (NP_633266.1), mcrD-Mj (NP_247837.1), mcrD-Mv (P07957), mcrD-Mt (NP_276295.1), mcrD-Mm (NP_633267.1), mcrG-Mj (NP_247839.1), mcrG-Mv (P07963), mcrG-Mt (NP_276293.1), mcrG-Mm (NP_633265.1), mcrA-Mj (NP_247840.1), mcrA-Mv (E27793), mcrA-Mt (NP_276292.1), mcrA-Mm (NP_633264.1), mrtB-Mj (NP_247045.1), mrtB-Mt (NP_276260.1), mrtC-Mj (NP_247058.1), mrtD-Mj (NP_247083.1), mrtD-Mt (NP_276259.1), mrtG-Mj (NP_247046.1), mrtG-Mt (NP_276258.1), mrtA-Mj (NP_247047.1), and mrtA-Mt (NP_276257.1). Bootstrap values are based on 1,000 replicates each (neighbor joining on top and parsimony on bottom) and are shown for branches with greater than 50% support. Trees are unrooted. Scale bars represent 50 amino acid substitutions.
Although the pattern of rearrangement observed for the GZfos17A3 mcrA operon resembles that of mrt operons in M. jannaschii and M. thermoautotrophicus, analysis of homology between individual GZfos17A3 subunits is consistent with mcrA affiliation (Fig. 2B). The mrt subunits from M. jannaschii and M. thermoautotrophicus were most closely related to the respective mcrA paralogs and formed subdivisions within each group (Fig. 2B). MOA-associated mcrA subunits grouped consistently within the major mcrA groups and not with mrt subdivisions (Fig. 2B). In addition, all mcrA subunits from ANME-2-associated fosmid GZfos35D7 grouped with their M. mazei cognates, consistent with the SSU rRNA and mcrA phylogeny shown in Fig. 1 (Fig. 2B).
Conservation of active-site amino acids in MOA mcrA groups.
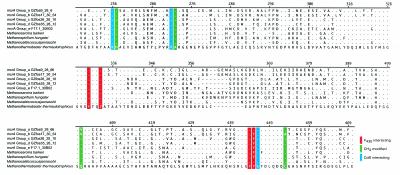
The conservation of primary structure relevant to catalytic activity in MOA mcrA groups was evaluated by comparing conserved amino acid residues among groups a to e and several methanogenic lineages (Fig. 3). The crystal structure of mcrA from Methanothermobacter thermoautotrophicus was used as a reference (6). Within the coverage area of MOA-associated mcrA groups, the structure of mcrA from M. thermoautotrophicus exhibits 11 conserved amino acids involved in active-site function. These include five methyl-modified amino acids, H257, R271, Q400, C452, and G445, four cofactor F430 interacting amino acids, F330, Y333, F443, and Y444, and two additional coenzyme B binding amino acids, K256 and A272 (6).
FIG. 3.
Amino acid alignment of representative environmental mcrA types and primary methanogenic lineages. Methanothermobacter thermoautotrophicus was used as the reference sequence (GenBank U10036). Position numbers correspond to the reference sequence. Amino acid identity at a given position is denoted by dots, and gaps are marked by dashes. Conserved amino acids are coded by color according to predicted CH3 modification (green), F430 binding (red), or coenzyme B interaction (blue).
A second mcrA crystal structure determined for a phylogenetically distant methanogen, Methanosarcina barkeri, reveals marked conservation of these amino acids and their modifications except for the Q147 methyl modification and a substitution, Y444 to F (10). In contrast, among the many conserved sites, MOA-associated mcrA group a contained two amino acid substitutions, converting Q400 to V and R270 to K. Group b contained four amino acid substitutions, converting Q400 to V, C452 to A, R270 to K, and Y444 to F. Groups c to e contained a single amino acid substitution, converting Y444 to F. Given that mcrA from M. barkeri is catalytically active, the Y444 to F substitution common to groups b to e appears to be neutral in terms of protein function, a supposition supported by parallel substitution in other methanogenic lineages (Fig. 3 and data not shown).
MOA-associated mcrA groups a to d contained a high proportion of cysteine residues compared to other methanogenic lineages. Within the conserved coverage area, cysteine accounted for 1.79% of residues in M. thermoautotrophicus, 1.31% in M. barkeri, 0.9% in Methanocaldococcus jannaschii, and 1.3% in Methanospirillium hungatei (Fig. 3). In contrast, cysteine accounted for 3.95% of residues in mcrA group a, 3.07% in group b, 3.57% in group c, 4.02% in group d, and 2.65% in group e.
DISCUSSION
Identification of mcrA in MOA.
In total, our data suggest that mcrA genes are present in MOA groups and discernible from those of other methanogenic archaea. Two mcrA groups (a and b) correspond to ANME-1, and two mcrA groups (c and d) correspond to ANME-2, identifications that are based on four types of evidence. First, variation in the representation of ANME-1 and ANME-2 ribotypes in three separate natural MOA populations, Monterey Canyon, Eel River, and Blake Ridge, suggested rough correlation between specific mcrA groups and specific ANME ribotypes. Second, congruence between mcrA and SSU rRNA tree topologies was indicative of mcrA group affiliation relative to SSU rRNA relationships among known methanogenic and methane-oxidizing archaeal lineages. Third, only mcrA group c sequences were recovered from a methane-oxidizing microcosm containing only a single MOA type, ANME-2c. Finally, fosmids representing mcrA groups a to d were identified from libraries derived from Eel River T201 cell preparations enriched for ANME-1 and ANME-2 (Hallam et al., unpublished data). The mcrA sequence identity within the methane-oxidizing ANME-1 and ANME-2 groups raises several questions relating to the origin, evolution, and function of methanogenic genes in MOA.
Evolution and divergence of MOA-associated mcrA genes.
The parallel distribution of ANME-1 and ANME-2 mcrA groups in two closely related subdivisions, a-b and c-d, raises the formal possibility of mcrA operon duplication or gene transfer within each lineage. For example, within the deeper branching Methanobacteriales and Methanococcales lineages, phylogenetic studies suggest that the mcrA operon paralog mrt originated in the Methanococcales and was subsequently transferred to a member of the Methanobacteriales (15, 20). Comparison of a-b and c-d MOA-associated mcrA groups to mrtA is inconsistent with the lateral gene transfer of this locus into MOA groups. MOA-associated mcrA sequences grouped consistently among themselves and with mcrA sequences from other lineages.
These observations are further supported by comparison of the complete mcrA operon from two fosmids representing mcrA group d (ANME-2, GZfos35D7) and b (ANME-1, GZfos17A3). The mcrA subunits from both fosmids consistently grouped with homologous mcrA subunits and not with the corresponding mrt subunits, with the possible exception of the mcrC subunit from GZfos17A3. In ANME-2, the canonical mcrA operon structure mcrBDCDGA was conserved and, combined with gene similarities, appears to reflect descent from a common Methanosarsinales ancestor, consistent with rRNA phylogenetic relationships. In contrast, in ANME-1 there was a significant deviation from this gene order and arrangement, more similar to that found in the M. jannaschii mrt structure, with mrcC located several kilobases upstream of mcrBGA. This may reflect functional as well as evolutionary differences between the ANME-1 and ANME-2 MOA groups.
Structure-function comparison of MOA and methanogen mcrA genes.
Comparison of active-site amino acids suggests that MOA-associated mcrA groups c-d and e may have the potential to catalyze the terminal step in methanogenesis. In contrast, mcrA group a-b harbors substitutions in universally conserved residues subject to methyl modification, including Q400 and C472. Residue Q400, located in the vicinity of cofactor F430, plays an important role in active-site geometry (6, 10). Although methyl modification of Q400 does not appear to be essential (10), substitution of residue Q400 with V in mcrA group a-b could alter active-site geometry and therefore protein function. The methyl moiety of C472, although outside the active site, forms two hydrophobic interactions with the side chains of H382 of McrB and L468 of McrA (6, 10). Changing C472 to A in mcrA group b may affect the geometry of these interactions.
Given these changes, the catalytic potential of group a-b is more uncertain from the standpoint of canonical methanogenesis. Despite the group a-b substitutions, the very presence of mcrA genes in MOA suggests activation of one or more elements in the methanogenic pathway. Although at present no known biological mechanism for anaerobic methane cleavage has been identified, biochemical models suggest that anaerobic activation of the methane C-H bond via mcrA could theoretically occur by the formation of an adduct between F430 (nickel porphyrin) and the radical mercaptoheptanoyl threonine phosphate (2, 13). This is consistent with physiological studies that suggest that methanogens are capable of simultaneous production and low-level oxidation of methane under anaerobic conditions (34, 35). Relevant here is the observation that both methane production and oxidation were equally inhibited by the substrate analog 2-bromoethanesulfonic acid, a potent inhibitor of mcrA function (34, 35).
Genomic potential of methanogenic pathway genes in MOA.
Our results show that MOA contain one of the essential and diagnostic genes of the methanogenic pathway, even though environmental evidence suggests that MOA consume but do not necessarily produce methane. The identification of these genes provides a means to identify ANME group members on the basis of mcrA sequence. Moreover, identification of MOA-associated mcrA groups defines a functional genomic link between methanogenic and putative reverse methanogenic archaea. Ongoing genomic analysis of fosmid libraries derived from Eel River T201 cell preparations have identified numerous operons containing methanogenic genes, including formylmethaneofuran dehydrogenases, formyltransferases, cyclohydrolases, F420-reducing dehydrogenases, and methyltransferases. These observations provide strong support for the hypothesis that MOA may have co-opted key elements of the methanogenic pathway to enable an anaerobic methanotrophic lifestyle (Hallam et al., unpublished data). Specific questions relating to methanogenic protein function in MOA await further genomic, biochemical, structural, and proteomic analysis.
Acknowledgments
Special thanks to Victoria Orphan, Bill Ussler, and Charlie Paull for providing Blake Ridge sediment core samples, to David Graham for providing valuable scientific commentary, to Lynne Christianson at MBARI and the Joint Genome Institute staff for providing technical support, to all the pilots of the ROVs Ventana and Tiburon and the crews on board the R/V Point Lobos, R/V Western Flyer, and R/V Cape Hatteras.
This study was supported by the David and Lucille Packard Foundation. Part of this work was performed under the auspices of the U.S. Department of Energy’s Office of Science, Biological and Environmental Research Program and the University of California, Lawrence Livermore National Laboratory, under contract no. W-7405-Eng-48, Lawrence Berkeley National Laboratory under contract no. DE-AC03-765F00098, and Los Alamos National Laboratory under contract no. W-7405-ENG-36.
REFERENCES
- 1.Barnes, R. O., and E. D. Goldberg. 1976. Methane production and consumption in anaerobic marine sediments. Geology 4:297-300. [Google Scholar]
- 2.Berkessel, A. 1991. Methyl-coenzyme M reductase: model studies on pentadentate nickel complexes and a hypothetical mechanism. Bioorg. Chem. 19:101-115. [Google Scholar]
- 3.Bidle, K. A., M. Kastner, and D. H. Bartlett. 1999. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia margin (ODP site 892B). FEMS Microbiol Lett. 177:101-108. [DOI] [PubMed] [Google Scholar]
- 4.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 5.Ellermann, J., R. Hedderich, R. Bocher, and R. K. Thauer. 1988. The final step in methane formation. Investigations with highly purified methyl-CoM reductase (component C) from Methanobacterium thermoautotrophicum (strain Marburg). Eur. J. Biochem. 172:669-677. [DOI] [PubMed] [Google Scholar]
- 6.Ermler, U., W. Grabarse, S. Shima, M. Goubeaud, and R. K. Thauer. 1997. Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science 278:1457-1462. [DOI] [PubMed] [Google Scholar]
- 7.Ferry, J. G. 1992. Biochemistry of methanogenesis. Crit. Rev. Biochem. Mol. Biol. 27:473-503. [DOI] [PubMed] [Google Scholar]
- 8.Ferry, J. G. 1999. Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol. Rev. 23:13-38. [DOI] [PubMed] [Google Scholar]
- 9.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grabarse, W., F. Mahlert, S. Shima, R. K. Thauer, and U. Ermler. 2000. Comparison of three methyl-coenzyme M reductases from phylogenetically distant organisms: unusual amino acid modification, conservation and adaptation. J. Mol. Biol. 303:329-344. [DOI] [PubMed] [Google Scholar]
- 10a.Girguis, P. R., V. J. Orphan, S. J. Hallam, and E. F. Delong. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69:AEM 515-03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 13.Hoehler, T. M., and M. J. Alperin. 1996. Anaerobic methane oxidation by a methanogen-sulfate reducer consortium: geochemical evidence and biochemical considerations, p. 326-333. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer, Dordrecht, The Netherlands.
- 14.Joye, S. B., T. L. Connell, L. G. Miller, R. S. Oremland, and R. S. Jellison. 1999. Oxidation of ammonia and methane in an alkaline, saline lake. Limnol. Oceanogr. 44:178-188. [Google Scholar]
- 15.Lehmacher, A., and H. P. Klenk. 1994. Characterization and phylogeny of mcrII, a gene cluster encoding an isoenzyme of methyl coenzyme M reductase from hyperthermophilic Methanothermus fervidus. Mol. Gen. Genet. 243:198-206. [DOI] [PubMed] [Google Scholar]
- 16.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 17.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by with defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 19.Martens, C. S., D. B. Albert, and M. J. Alperin. 1999. Stable isotope tracing of anaerobic methane oxidation in the gassy sediments of Eckernforde Bay, German Baltic Sea. Am. J. Sci. 299:589-610. [Google Scholar]
- 20.Nolling, J., A. Elfner, J. R. Palmer, V. J. Steigerwald, T. D. Pihl, J. A. Lake, and J. N. Reeve. 1996. Phylogeny of Methanopyrus kandleri based on methyl coenzyme M reductase operons. Int. J. Syst. Bacteriol. 46:1170-1173. [DOI] [PubMed] [Google Scholar]
- 21.Orphan, V. J., K. U. Hinrichs, W. Ussler, 3rd, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. Delong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 23.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pancost, R. D., J. S. Sinninghe Damste, S. de Lint, M. J. van der Maarel, and J. C. Gottschal. 2000. Biomarker evidence for widespread anaerobic methane oxidation in Mediterranean sediments by a consortium of methanogenic archaea and bacteria. The Medinaut Shipboard Scientific Party. Appl. Environ. Microbiol. 66:1126-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reeburgh, W. S. 1976. Methane consumption in Caraico Trench waters and sediments. Earth Planet. Sci. Lett. 28:337-344. [Google Scholar]
- 26.Reeburgh, W. S. 1996. Microbial growth on C1 compounds. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 27.Reeve, J. N., J. Nolling, R. M. Morgan, and D. R. Smith. 1997. Methanogenesis: genes, genomes, and who's on first? J. Bacteriol. 179:5975-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein, J. L., T. L. Marsh, K. Y. Wu, H. Shizuya, and E. F. DeLong. 1996. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 178:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swafford, D. L. 2000. Phylogenetic analysis with parsimony (and other methods), version 4.0b10. Sinauer Associates, Sunderland, Mass.
- 30.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. 1998 Marjory Stephenson Prize Lecture. Microbiology 144:2377-2406. [DOI] [PubMed] [Google Scholar]
- 31.Valentine, D. L. 2002. Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: a review. Antonie Van Leeuwenhoek 81:271-282. [DOI] [PubMed] [Google Scholar]
- 32.Valentine, D. L., and W. S. Reeburgh. 2000. New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2:477-484. [DOI] [PubMed] [Google Scholar]
- 33.Whiticar, M. J., E. Faber, and M. Schoell. 1986. Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation-isotope evidence. Geochim. Cosmochim. Acta 50:693-709. [Google Scholar]
- 34.Zehnder, A. J., and T. D. Brock. 1979. Biological energy production in the apparent absence of electron transport and substrate level phosphorylation. FEBS Lett. 107:1-3. [DOI] [PubMed] [Google Scholar]
- 35.Zehnder, A. J., and T. D. Brock. 1979. Methane formation and methane oxidation by methanogenic bacteria. J. Bacteriol. 137:420-432. [DOI] [PMC free article] [PubMed] [Google Scholar]