Abstract
Anaerobic methanotrophic archaea have recently been identified in anoxic marine sediments, but have not yet been recovered in pure culture. Physiological studies on freshly collected samples containing archaea and their sulfate-reducing syntrophic partners have been conducted, but sample availability and viability can limit the scope of these experiments. To better study microbial anaerobic methane oxidation, we developed a novel continuous-flow anaerobic methane incubation system (AMIS) that simulates the majority of in situ conditions and supports the metabolism and growth of anaerobic methanotrophic archaea. We incubated sediments collected from within and outside a methane cold seep in Monterey Canyon, Calif., for 24 weeks on the AMIS system. Anaerobic methane oxidation was measured in all sediments after incubation on AMIS, and quantitative molecular techniques verified the increases in methane-oxidizing archaeal populations in both seep and nonseep sediments. Our results demonstrate that the AMIS system stimulated the maintenance and growth of anaerobic methanotrophic archaea, and possibly their syntrophic, sulfate-reducing partners. Our data demonstrate the utility of combining physiological and molecular techniques to quantify the growth and metabolic activity of anaerobic microbial consortia. Further experiments with the AMIS system should provide a better understanding of the biological mechanisms of methane oxidation in anoxic marine environments. The AMIS may also enable the enrichment, purification, and isolation of methanotrophic archaea as pure cultures or defined syntrophic consortia.
Biologically mediated methane formation (methanogenesis) is the largest source of methane on the planet and has long been considered a carbon sink and terminal step in ecological carbon flow (13). However, over the last 25 years there has been compelling evidence suggesting that microbially mediated anaerobic oxidation of methane constitutes another major component of carbon cycling in these environments. Martens and Berner (22), Reeburgh (29), and Barnes and Goldberg (4) first posited the existence of a microbial community in anoxic marine sediments that was responsible for anaerobic oxidation of methane. The net reaction was described as SO42− + CH4 → HCO3− + HS− + 2H2O, where HS− is hydrogen bisulfide. This net reaction is exergonic at conditions found in situ, yielding ca. 25 kJ per mol of methane oxidized (21).
A subsequent field and laboratory study suggested that anaerobic oxidation of methane was mediated by a consortia of archaea and sulfate-reducing bacteria (17). Microbially mediated anaerobic oxidation of methane has been well correlated with characteristic geochemical profiles, namely the concomitant depletion of sulfate and methane in anoxic sediments (12, 19, 28, 38). Molecular phylogenetic analyses combined with 13C lipid isotopic determinations identified two major groups of methanogen-related archaea (ANME-1 and ANME-2) that apparently incorporate methane-derived carbon into cellular biomass and co-occur in sediments with sulfate-reducing bacteria (16). More recent studies have better phylogenetically characterized the archaeal groups involved in anaerobic oxidation of methane (8, 25, 33, 35), and studies with rRNA-targeted fluorescent in situ hybridization (FISH) (3, 10) have visually demonstrated a physical association between methane-oxidizing archaea (MOA) and sulfate-reducing bacteria (8). Single-cell phylogenetic identification coupled with microscopic mass spectrometric analysis directly suggest that members of ANME-1 and ANME-2 anaerobically consume methane (26). Furthermore, these studies suggested that individual MOA may mediate anaerobic oxidation of methane without tight spatial coupling to sulfate-reducing bacteria (27).
These prior studies have established a relation between specific microbial groups and geochemical profiles, however, the relation between the methane oxidation rates, the details of metabolic exchange between the syntrophic partners, in situ population growth, and environmental conditions that govern these processes remain poorly characterized. Experiments that address these issues have primarily been hindered by the lack of MOA isolates or cultures. Research on anaerobic oxidation of methane, however, has been advanced by employing both in situ isotopic tracers and serum vial incubations to examine the relation between anaerobic oxidation of methane and sulfate reduction (1, 11, 12, 15, 17, 24, 38), and support the hypothesis that MOA are likely to produce intermediates such as hydrogen, acetate, formate, or another compound (17, 31, 38). These and other experiments have also provided data on the metabolite stoichiometry and the effect of physical factors such as pressure and temperature on anaerobic oxidation of methane rate (15, 17, 24, 38).
The dynamics of anaerobic oxidation of methane may be quite different in situ, a constantly fluctuating environment, compared to static experimental systems such as serum vials. For example, static incubations may limit mass transfer for biosynthesis, result in the buildup of end products, and consequently restrict growth. To address some of these issues and foster the growth of MOA in laboratory settings, we developed a continuous-flow laboratory-based anaerobic methane incubation system (AMIS) to assess the conditions required for the enrichment of MOA in marine sediments. Our goals were to assess the relationship between environmental conditions and methane oxidation rates, and stimulate the growth and enrichment of MOA. The AMIS system differs substantially from previous attempts to cultivate MOA because is it designed to reestablish environmental conditions by minimally perturbing the sediments after collection, as well as provide metabolites and remove end products at environmentally relevant rates. Also, the AMIS facilitates ecophysiological experiments that would be exceedingly difficult to conduct in the field, in particular relating MOA population dynamics to environmental conditions.
In this study, we incubated both marine hydrocarbon seep sediments and marine nonseep (i.e., aerobic) sediments on the AMIS for 24 weeks. We measured methane oxidation rates and MOA population growth over the course of the incubation. This allowed us to evaluate the growth and metabolism of MOA in both seep and nonseep sediments both prior and subsequent to incubation on AMIS.
MATERIALS AND METHODS
Sediment sample collection.
During multiple expeditions in 2001 and 2002 on the R/V Point Lobos, we used the remotely operated submersible Ventana to search for undisturbed cold seep sites between 800 and 1,000 m in the Monterey Canyon. In December 2002, we surveyed a cold seep (36.77 N, 122.08 W), approximately 4 m in diameter, at a depth of 955 m. The seep zonation was typical of other seeps in Monterey Canyon, containing a Beggiatoa-like bacterial mat in the center of the seep encircled by numerous Calyptogena spp. clams (5).
Polycarbonate-sleeve push cores were used to collect sediment samples for incubation (6). The sleeves were 30 cm long and 5.25 cm in inner diameter, with 0.75-cm holes at 2.5-cm intervals along their length for subsampling sediments. Prior to the dive, the sampling ports were sealed with transparent, waterproof plastic tape to prevent leakage during sediment sample collection.
Sediment cores for laboratory incubation were removed from the center of the seep (core 1), the periphery of the seep (within the clam bed, core 2), and 25 m away from any visibly active seepage (core 4). The nonseep sediments were aerobic to 30 cm (217 ± 21 μM O2; N = 5), as determined by a modified Winkler titration assay (Hach Inc.). Although a strong sulfidic odor present throughout the sediment core suggested that the seep sediment cores were entirely anoxic, we used the aforementioned method to confirm that the bottom 15 cm of sediments was anoxic. Upon recovery, all push cores were kept sealed in their butyl rubber stoppers (size, 12.5 cm) and placed in an insulated cooler with icepacks. All sediment push cores arrived in the laboratory within 4 h of collection.
Sediment core subsampling.
After returning to the laboratory, each sediment core was placed into a nitrogen-purged glovebag and subsampled via the sampling ports at 2.5-cm intervals along the entire length of the core. Between 1.5 and 2.5 g of sediment was sampled from each port. At least 1 g of sediment from each interval was quickly frozen in a cryovial at −80°C for nucleic acid extraction. A sample from each interval was also placed into a 1.5-ml Eppendorf tube, mixed with an equal volume of 5% formalin in filter-sterilized seawater, incubated overnight at 4°C, centrifuged at 1,000 × g, transferred into 1:1 ethanol-phosphate-buffered saline as previously described (25), and stored at 4°C for fluorescent in situ hybridization (FISH). Then 5 g of sediment from the bottom of each push core was taken for methane oxidation rate measurements and temporarily stored in 60-ml glass syringes flushed with nitrogen and kept on ice. After sampling, the ports were sealed by compressing a 2.5-mm butyl rubber sheet (coated with high-vacuum grease) between the core sleeve and a Delrin plastic block machined to the sleeve's curvature. Stainless steel hose clamps held the assembly in place.
AMIS.
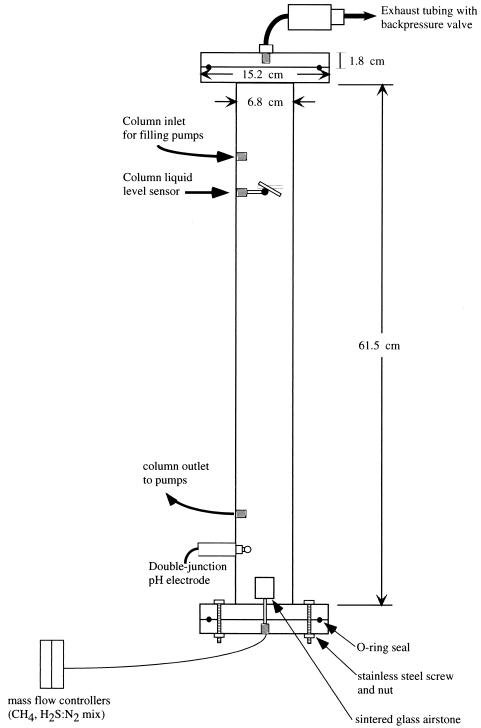
After subsampling, the cores were placed on the continuous-flow anaerobic sediment incubator system (anaerobic methane-oxidizing system [AMIS]). To mimic the chemistry of the porewater fluids found in situ, 0.2-μm filtered seawater (held in a 25-liter acrylic reservoir) was pumped by a diaphragm metering pump (Prominent Industries, Inc.) into a gas-tight acrylic seawater conditioning column (61.5 cm by 5.8 inner diameter; Fig. 1). A gas-tight polyethylene reed switch relay (Cole Parmer, Inc.) was used to control the metering pump and maintain a constant level in the column. Column seawater was bubbled with methane, hydrogen sulfide, and nitrogen gas to achieve dissolved concentrations of 1.5 mM, 510 μM, and 500 μM for each gas, respectively (dissolved gas concentrations were determined as previously described [14]). Mass flow controllers (Sierra Instruments, Inc.) regulated the gas flow into the column. Pressure in the column was maintained at 513 kPa by gas-tight spring-loaded polyvinylchloride backpressure valves (Ryan-Herco, Inc.). Seawater residence time in the conditioning column was 4 h.
FIG. 1.
Schematic of seawater conditioning column used to equilibrate filter-sterilized seawater with methane, hydrogen sulfide, and nitrogen gas. Gasses are bubbled into the bottom of the column through a sintered glass airstone. Filter-sterilized seawater is pumped into the top of the column, resides in the column for approximately 4 h, and is then pumped out near the bottom of the column. Seawater level in the column is controlled via liquid level relay. Conditioning column may also be used to adjust pH via a proportional pH controller.
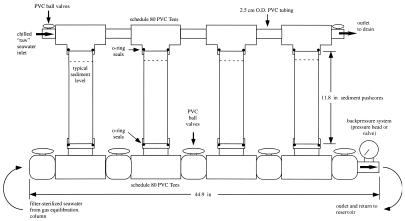
Seawater was then pumped out of the column by a diaphragm metering pump (Prominent Industries, Inc.) through a 0.2-μm capsule filter (Gelman Industries) and rigid polypropylene tubing (2.5-cm outer diameter, 1.5-cm inner diameter) into a schedule 80 polyvinylchloride (PVC) manifold on which four sediment cores can be incubated (hereafter referred to as the lower PVC manifold) (see Fig. 2). Female fittings (complementary to the core sleeve dimensions) with double Viton O-ring seals maintained a gas- and water-tight seal on the bottom of each sediment core sleeve. Gas-tight PVC ball valves were located on both the inlet and the outlet, allowing the manifold to be sealed to maintain the anaerobic conditions in the manifold during servicing. During operation, the backpressure in the lower PVC manifold was maintained at 200 kPa by a PVC backpressure valve with Viton seats (Ryan-Herco) or by running a length of PVC pipe vertically to create a seawater pressure head at the outlet. After circulating through the manifold, seawater returned to the 25-liter reservoir and was recirculated through the conditioning column.
FIG. 2.
Schematic of anaerobic sediment incubator manifold. The core sleeves are constructed from polycarbonate, seals on the sampling ports are 3-mm butyl rubber under Delrin plastic blocks machined to the core's curvature. The backpressure valve seals are Viton. All other components are schedule 80 polyvinylchloride and, where applicable, are cemented together with low-volatility solvents. Seawater enters the manifold and flows past all the sediment cores prior to returning to a 25-liter reservoir.
The upper PVC manifold consisted of PVC tees connected by rigid PVC tubing (Fig. 2). On the upper PVC manifold, each sediment core sleeve was also sealed to a PVC tee fitting with Viton O-rings. The upper PVC manifold was connected to a chilled seawater system (5°C) and could be sealed at both the inlet and outlet by gas-tight PVC ball valves. During operation, the entire manifold assembly and seawater reservoir were maintained in a coldroom at 5°C.
Sediment core incubation on the AMIS.
To prevent the sediments from settling into the manifold, a 5.25-cm nylon fiber mesh disk (Scotch-Brite, 3M Corp.), previously soaked for 1 week in a 1 M solution of filter-sterilized sodium sulfide, was placed on the bottom of each core to be incubated. The sediment cores were then sealed onto the lower and upper PVC manifold. The lower PVC manifold was purged with nitrogen gas until the manifold was filled with methane-saturated, sulfidic seawater. The lower PVC manifold ran continuously, whereas the upper PVC manifold was filled with chilled seawater and flushed once a week with fresh chilled seawater (5°C). The upper PVC manifold was sealed at one end, while the other end employed a seawater trap (PVC P-trap) to minimize exposure of the sediments to oxygen while allowing for advective flow from the bottom. Sediments were maintained on the AMIS for 24 weeks. Subsamples were taken for analysis once a month as described above.
Determining methane oxidation rates.
Five-gram sediment subsamples were taken from the bottom of each core prior to and at the end of incubation and placed into 20-ml borosilicate glass serum vials (Kimble Inc.). Two seep sediment subsamples that were poisoned with 2 ml of 10 M NaOH were used as controls. The vials were filled to the top with filter-sterilized anoxic seawater containing 1.2 mM methane and 400 μM hydrogen sulfide and sealed with crimp-topped butyl rubber stoppers. Samples were shaken and left to incubate for 96 to 108 h. A gas-tight glass syringe, 0.45-μm syringe filter, and disposable needle were used to slowly draw a 10-ml seawater sample from the serum vial. A membrane inlet mass spectrometer (Hiden Inc.) with a faraday cup/electron multiplier detector was used to analyze all seawater samples (14, 34).
Dissolved methane partial pressures of all samples was determined by integrating the signal of both the 15 and 16 m/z peaks for 4 min and subtracting the background signal attributable to water (determined by examining the 16, 17, and 18 m/z peaks of a vacuum-degassed water sample). A standard curve was calculated by analyzing a serial dilution of methane-saturated sterile seawater standards. Subsamples of each standard were then quantified with a Hewlett-Packard 5890 gas chromatograph with a molecular sieve column (Alltech, Inc.) and thermal conductivity detector. The methane concentrations before and after incubation were calculated from the standard curve (R2 = 0.995; P = 0.0001). After incubation, subsamples of sediments were weighed and stored at −80°C for nucleic acid analyses, and the remainder of the sediments were weighed before and after drying at 95°C for 36 h. The total volume of seawater in the vial was calculated by subtracting the volume of fresh sediment inserted into the vials. The mass of salts left after drying was subtracted from the total mass to calculate the dry mass of sediment. Specific methane oxidation rates were then calculated as the ratio of net methane oxidation rate to dry sediment mass per unit time.
Nucleic acid extraction and purification.
Prior to extraction, all sediment samples were weighed on an electronic balance (Mettler, Inc.). Sediment cell lysis and nucleic acid extraction from 0.5 to 1 g (wet weight) of sediment was conducted with a Fastprep bead beating machine (Bio 101, Inc.) and a soil extraction kit (MoBio Inc., San Diego, Calif.). The protocol for the nucleic acid extraction kit was modified by initially bead beating the sample with the Fastprep machine (speed 4.5 for 20 s), followed by two 5-min incubations at 70°C. In addition, the volumes of all solutions added or transferred during extraction were normalized among all samples. The remainder of the extraction procedure was carried out according to the manufacturer's instructions. The final volume of all extractions was 50 μl. This procedure typically produced DNA greater than 20 kbp in size. Nucleic acid extractions were then purified on small-scale cesium chloride density gradients in a refrigerated tabletop ultracentrifuge (Beckman Inc.).
archaeal SSU rRNA library construction, sequencing, and analysis.
Small subunit (SSU) rRNA genes were amplified by PCR with CsCl-purified nucleic acid extractions from sediments subsampled prior to and after incubation on the AMIS. PCRs (total volume, 50 μl) contained a 0.2 μM concentration of archaeon-targeted primers (A21f, 5′-TTCCGGTTGATCCYGCCGGA-3′, and A958r, 5′-YCCGGCGTTGAMTCCAATT-3′). Reaction mixtures also contained 5 μl of PCR buffer (containing 2 mM MgCl2; Promega, Inc.), 2.5 mM each deoxynucleotide triphosphate, and 0.025 U of Taq polymerase (in buffer B; Promega, Inc.). PCR amplifications were carried for 28 cycles, with an initial denaturation step of 2 min at 95°C and 25 cycles of PCR consisting of 30 s at 94°C, 30 s at 55°C, and 45 s at 72°C. A final 7-min extension at 72°C was added to facilitate A-tailing and subsequent cloning of amplified products.
To construct environmental rRNA clone libraries, amplicons were pooled from four reactions, cleaned, and concentrated with Microcon YM-100 (Millipore Inc.) spin filters. Amplicons were cloned into the pCR2.1 vector and transformed into chemically competent Escherichia coli according to the instructions of the manufacturer (Invitrogen, Inc.). Plasmids were purified via the Montage miniprep kit (Millipore, Inc.) and sequenced with BigDye chemistry (version 3.1) on an ABI 3100 capillary sequencer (ABI Biosystems, Inc.).
SSU rRNA sequences from clones were compared to a current database of genetic sequences (GenBank) with the Blast (basic local alignment search tool) program of the Ribosomal Database Project (2) to determine their approximate phylogenetic affiliation. Sequences were then aligned to the nearest neighbor and other known sequences with the automated alignment tool of the ARB program package (http://www.arb-home.de/). Phylogenetic trees for SSU rRNA genes were generated in PAUP* version 4.b10 (Sinauer Assoc., Inc.) with the distance and parsimony methods. SSU rRNA sequence distances were estimated with the Kimura two-parameter model. The tree was rooted to Methanococcus jannaschii. Bootstrapping for distance and parsimony was accomplished with 1,000 replicates per tree with heuristic search methods.
Quantifying nucleic acid recovery.
To quantify and compare the MOA biomass in sediments prior to and after incubation on the AMIS, the final nucleic acid yields were correlated to the total sediment mass initially used for nucleic acid extraction. Five-microliter subsamples of both the crude and CsCl-cleaned nucleic acid extractions were quantified with a PicoGreen double-stranded DNA quantification kit (Molecular Probes, Inc.), and a fluorescent imaging scanner (ABI Biosystems FluorImager). Both the crude extractions and CsCl-purified nucleic acid yields were correlated to the total sediment mass used in extraction via linear regression analysis (Girguis et al., unpublished data; Statview 5.0; Abacus Inc.).
Design and specificity of 5′ nuclease (TaqMan) primers and probe.
We designed PCR primers that targeted the ANME-2c group, AR468f, 5′-CGCACAAGATAGCAAGGG-3′, and AR736r, 5′-CGTCAGACCCGTTCTGGTA-3′. The TaqMan probe used targets a universal region homologous to the domain archaea (32) between these two primer pairs. Because the target organisms are closely related, there were no degeneracies in the primer pairs. ANME-2c-specific primers were checked for mismatches with the ARB software package (http://www.arb-home.de/) with a 16 rRNA SSU database of over 11,000 sequences. All probes and primers were screened and optimized for the requirements of TaqMan assays with Primer Express software (PE Biosystems Inc.).
To examine the possible cross-reactivity of the primers, we examined the formation of PCR products from Methanosarcina acetivorans genomic DNA, and plasmid DNA containing SSU rRNA gene fragments from representatives of major archaeal groups commonly found in seep sediments (25). Three-step PCRs were run for 35 cycles, and each reaction mixture contained 1 ml of AmpliTaq Gold buffer, 0.2 mM each deoxynucleoside triphosphate, 0.5 mM ANME-2c primer pair, 1.5 mM MgCl2, and 0.05 U of AmpliTaq Gold DNA polymerase. Reactions were run in a Perkin Elmer 9700 thermal cycler at 56 to 60°C annealing temperatures. Products were then quantified against DNA standards of known concentration with the PicoGreen double-stranded DNA quantification kit (Molecular Probes, Inc.) and an ImageQuant fluorescence detector (Applied Biosystems, Inc.)
Optimization of primer, probe, and MgCl2 concentrations.
All reactions were carried out in optical tubes or reaction trays (PE Biosystems, Inc.), and all tubes were sealed with optical caps. All reagents and template were delivered into the tubes with Microman M10 or M100 positive-displacement pipettes (Rainin, Emeryville, Calif.). All reactions were performed with a model 7700 sequence detection system (PE Biosystems) programmed with a soak step of 2 min at 50°C, allowing AmpErase uracyl N-glycosylase to hydrolyze residual PCR amplicons. An enzyme activation soak step (15 min at 95°C for AmpliTaq Gold or 2 min at 94°C for Platinum Taq) followed the initial soak step, followed by 40 cycles of 15 s for denaturation (94°C for Platinum Taq) and 1 min of annealing plus extension at 59°C.
To determine the primer concentrations yielding the minimal CT values and consequently the highest amplification efficiencies, a matrix of concentrations of forward and reverse primers were tested in TaqMan assays. Primer concentrations ranged from 100 to 1,500 nM. Annealing temperatures and MgCl2 concentrations were held constant at 59°C and 5 μM, respectively. The remaining parameters were as follows. In a final volume of 25 μl, reaction mixtures contained 1× Platinum Taq buffer and 0.025 U of Platinum Taq DNA polymerase (Life Technologies, Rockville, Md.), 200 mM each dATP, dCTP, and dGTP, 400 mM dUTP, and 0.25 U of AmpErase uracyl N-glycosylase (Perkin Elmer Biosystems).
To determine the probe concentration yielding the minimal CT values for a specific target, TaqMan assay reactions were performed with the optimized primer concentrations and a range of probe concentrations from 200 to 800 nM. Because the probe targets a broad range of archaea, we used both our target and nontarget plasmid DNA to assess the effect of nontarget DNA concentrations on probe efficiency.
To determine the optimal MgCl2 concentration, 5′-nuclease assay reactions were performed with the optimized primer and probe concentrations and a range of MgCl2 concentrations from 1 to 10 mM. The remaining reaction conditions were the same as used for the primer matrices. We chose the minimal MgCl2 concentrations that allowed high amplification efficiencies, and consistent, specific template amplification at different template concentrations.
Quantification of ANME-2c SSU rRNA from environmental and incubated sediments.
To produce a standard curve for quantification, plasmid DNA containing near full-length SSU rRNA genes from an ANME-2c archaeon (Eel-36a2A5, accession no. AF354133) was quantified against a known standard (Ebac31 [32]) with m13 forward and reverse primers with both the Ebac31 standards (pUC vector) and a serial dilution of our ANME-2c plasmid (pCR2.1 vector). After quantification, a serial dilution was made from the ANME-2c plasmid, and these preparations were used as the quantitative standard for all subsequent assays.
To determine the number of SSU rRNA copies in each subsample, CsCl-purified DNA extractions were used as template DNA in TaqMan assays. ANME-2c group-specific primers AR468f and AR736r were then used in conjunction with the ARCH519r fluorogenic probe on all samples analyzed. We collected and analyzed subsamples of sediment cores taken from the center and periphery of the seep study site, as well as nonseep sediments, before and after incubation on the AMIS. In addition, we analyzed 50 ml of AMIS-conditioned seawater to quantify the MOA found in the recirculating seawater system. As an additional control, we analyzed subsamples of a nonseep sediment pushcore that had been sealed with rubber stoppers, kept at the same temperature as the AMIS, and incubated for a period of 24 weeks.
The reagent concentrations for all quantitative assays were as follows. In a final volume of 25 μl, reaction mixtures contained 1× Platinum Taq buffer and 0.025 U of Platinum Taq DNA polymerase (Life Technologies, Rockville, Md.), 200 μM each dATP, dCTP, and dGTP, 400 mM dUTP, and 0.25 U of AmpErase uracyl N-glycosylase (Perkin Elmer Biosystems), 250 μM ANME-2c 468 forward primer, 1.5 mM ANME-2c reverse primer, 100 μM ARCH519r probe, and 5 mM MgCl2. TaqMan assays were run for 40 cycles, with an enzyme activation soak step (15 min at 95°C for AmpliTaq Gold or 2 min at 94°C for Platinum Taq), followed by 40 cycles of 15 s for denaturation (94°C for Platinum Taq) and 1 min of annealing plus extension at 59°C.
Fluorescent in situ hybridization.
Sediment subsamples stored in 50% ethanol-50% phosphate-buffered saline were diluted 1:25 by suspension in 0.2 μm of filter-sterilized seawater. Diluted samples were then briefly sonicated for 15 s at 32 A (Sonics and Materials Inc.) and then filtered onto 47-mm-diameter 0.2-μm-pore-size polycarbonate filters. Filters were washed and then hybridized with bacterial Desulfosarcina-Desulfococcus and archaeal ANME-2 group-specific fluorescently labeled probes (8) as previously described (25). Images were captured digitally with a spot-100 charge-coupled device camera (Diagnostic Instruments, Inc.).
Nucleotide sequence accession numbers.
The rRNA gene sequences were submitted to GenBank and have been assigned accession numbers AY323214 to AY323225.
RESULTS
Sediment core incubation on AMIS.
Sediment core incubations on AMIS resulted in changes in the color and texture of the sediment. In particular, nonseep sediments exhibited a dramatic shift in color from tan to black as the iron in the sediments was reduced by exposure to sulfide (ca. 510 μM in the recirculating seawater). This chemical reduction of the sediments served as a rough proxy for flow rate, and thus metabolite availability, through the sediments. By measuring the sediment reduction rate in the nonseep sediment, we estimated that sediment fluid flow rate during the AMIS incubations was approximately 75 cm year−1. In situ seep fluid flow rates are often ca. 10 cm year−1 (G. Aloisi, S. Bollwerk, K. Wallmann, A. N. Derkachev, E. Suess, and G. Bohrmann, Geophys. Res. Abstr. 5, abstr. no. 08718, 2003). In earlier incubations on the AMIS, in which we did not maintain a differential backpressure between the lower and upper manifold and relied instead on diffusion to provide methane to the sediments, we empirically determined the rate of chemical reduction solely from diffusion to be 15 cm year−1. In addition, strands of elemental sulfur, deposited by Candidatus sp. or Arcobacter sulfidicus (37), were found in the tops of some incubated cores only after incubation. Both seep and nonseep sediment cores (collected at the same time as core 1 and core 4) that were maintained at the same temperature but not incubated on AMIS showed no microbial mat growth on the tops of the sediment cores. Some core incubations also formed carbonate-like deposits along the bottom sediment surface.
Methane oxidation rates.
Before incubation on the AMIS, subsamples of core 1 and core 2 (at a depth of 20 cm) exhibited anaerobic methane oxidation rates similar to other anoxic marine sediments, ca. 30 nmol per gram of dry sediment per day (17, 24). There was high variability among the methane oxidation rates from the cores prior to incubation (Table 1). After incubation on AMIS, sediments from both core 1 and core 2 did not show any significant difference in methane oxidation rates (Table 1). The core 4 nonseep sediment subsamples had no detectable methane oxidation rates prior to incubation on the AMIS, but after incubation core 4 sediment exhibited anaerobic oxidation of methane rates approximately 50% of the rates observed in the incubated seep sediments (Table 1). All sediment cores incubated for at least 14 weeks on AMIS exhibited anaerobic oxidation of methane. The circulating seawater in the lower manifold exhibited anaerobic oxidation of methane rates comparable to the lowest measured AMIS sediment methane oxidation rates, and approximately 16% of the highest AMIS measured sediment oxidation rate. Killed sediment samples did not exhibit statistically significant methane oxidation rates (Table 1).
TABLE 1.
Comparison of methane oxidation rate and MOA populationsa
| Sediment | Mean methane oxidation rate (nmol/g of sediment per day) ± SE (low rate, high rate) | Mean no. of ANME 2c-specific 16s rRNA/g of sediment ± SE |
|---|---|---|
| Core 1 before incubation | 32.9 ± 13.9 (9.20, 45.3) | 4.73 (±1.65) × 105 |
| Core 1 after incubation | 28.7 ± 3.57 (23.8, 33.6) | 8.16 (±2.29) × 105 |
| Core 2 before incubation | 27.7 ± 10.2 (15.2, 43.2) | 5.11 (±2.89) × 105 |
| Core 2 after incubation | 29.0 ± 6.80 (21.0, 38.5) | 4.13 (±0.98) × 105 |
| Core 4 before incubation | 0.44 ± 2.10 (−2.82, 2.30) | N/D |
| Core 4 after incubation | 19.2 ± 9.60 (9.80, 35.2) | 2.93 (±0.32) × 105 |
| AMIS circulating seawater | 7.65 ± 3.18 | 1.37 (±2.90) × 103 |
A comparison was made of methane oxidation rate and ANME-2c MOA populations (ANME 2c-specific 16s rRNA copy number per gram of sediment) from marine sediments in Monterey Canyon before and after 24 weeks of incubation on the AMIS sediment incubation system. Core 1, anoxic sediments collected from the center of a seep; core 2, anoxic sediments collected from the periphery of the seep, within the clam field; core 4, oxic sediments collected away from any visible seepage. Five independent samples were used to determine all mean ± standard error values. Low rate and high rate are the lowest and highest methane oxidation rates measured for that sediment core. The AMIS circulating seawater values are expressed per milliliter of seawater. N/D, not detected
SSU rRNA archaeal diversity.
To survey archaea associated with cores incubated on AMIS, before and after incubation, four different archaeal SSU rRNA gene libraries were examined. Two of the libraries were derived from subsamples of the freshly collected seep and nonseep sediment cores that were collected for incubation on the AMIS (core 1 before and core 4 before, respectively). The other two libraries were constructed from these same sediments after incubation on the AMIS (core 1 after and core 4 after, respectively). In all cases, the nucleic acids were recovered from sediments in the bottom 2 cm of each sediment core, as these sediments were in continual contact with recirculating methane-saturated seawater in the AMIS lower manifold.
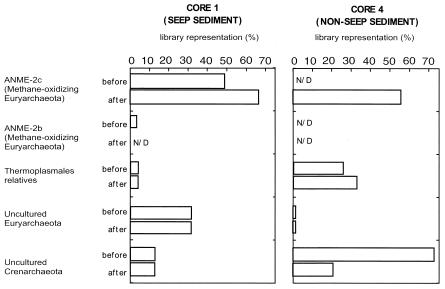
SSU rRNA clones in each library were sequenced to determine the archaeal phylogenetic diversity in the sediments. The representations of the different phylotypes found in each library are diagrammed in Fig. 3. Predominant phylotypes in each library were identified and the recovery of their rRNA from libraries was quantified (Fig. 3). Incubation on the AMIS led to shifts in the representation of rRNA clones recovered in core 4 sediments. The core 4 before library was dominated by Crenarchaeota, with the dominant phylotype (at 27% library representation) most similar to a sequence previously recovered from marine sediments, CRA8-27cm (36). The remaining clones were most similar to other sediment-associated uncultivated Euryarchaeota and Crenarchaeota. No SSU rRNA sequences affiliated with ANME-1 or ANME-2 MOA or any methanogen-like archaeon were recovered from the core 4 before sample. SSU rRNA sequences recovered from the core 4 after library were most similar to ANME-2c environmental clones Eel-36a2a5, Eel-36a2E1, and SB-17a1D3 recovered from Eel River and Santa Barbara basin sediments (25), while sequences related to the environmental crenarchaeal clone CRA8-27cm diminished in representation from 72 to 21% (36). One recovered sequence most similar to Methanolobus tindarus (Methanosarcinales) was recovered from the core 4 after library.
FIG. 3.
Representation (percent SSU rRNA/total clone number) of ANME-2c and ANME-2b group methane-oxidizing archaea, Thermoplasmales-type Euryarchaeota, and all other Eury- and Crenarchaeaota recovered from clone libraries before and after incubation on AMIS. Total number of clones in each library: core 1 before, 114; core 1 after, 93; core 4 before, 82; core 4 after, 75. All sequences were aligned with the ARB software package (http://www.arb-home.de/), identity was preliminarily assessed via Blast (2) and confirmed by maximum-likelihood analysis with the PAUP software package (Sinauer Associates, Inc.). N/D, not detected.
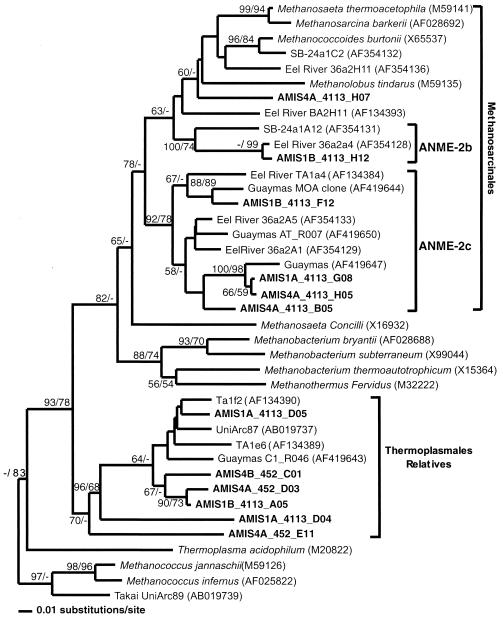
Incubation on the AMIS did not result in any significant changes in the diversity of recovered rRNA clones from the core 1 sediments. The core 1 before library was dominated by phylotypes related to Eel36a-2A5, Eel 36a2A1, and a Guaymas clone similar to ANME-2c MOA (33), all members of the ANME-2c group of MOA (Fig. 3 and 4). Four percent of recovered clones in the core 1 before library were most similar to Eel 36a2a4, a member of the ANME-2b group of MOA (Fig. 4). Sequences similar to archaeon VC2.1 (30) and Crenarchaeota CRA8-27cm (36) and archaeon 33-P92A98 (18) were also dominant representatives in the library (Fig. 3). The core 1 after library was dominated by the same phylotypes, with an increase in the representation of 16 SSU rRNA sequences most related to ANME-2c (25) from 24 to 32% (Fig. 4).
FIG. 4.
Phylogenetic tree illustrating the relationships of 16S rRNA Euryarchaeal clone sequences from Monterey Canyon seep and nonseep sediments, both prior to and following incubation (sequences in boldface) to selected cultured and environmental euryarchaeotal sequences in the database. Clone designations ending in B are sequences from sediment subsamples from cores 1 to 4, as indicated, before incubation on the AMIS, and those ending in A are sequences from sediment subsamples after incubation on the AMIS. Phylogenetic tree was generated in PAUP version 4.0b10 (Sinauer Associates, Inc.) with distance methods, and sequence distances were estimated with the Kimura two-parameter model. The tree was rooted to Methanococcus jannaschii. Bootstraps shown are for distance and parsimony, respectively. The scale bar represents a difference of 0.01 substitution per site.
We also constructed a library with primers specific to the domain Bacteria (25) to examine potential changes in the bacterial populations found in core 4 before and after incubation on AMIS. Preliminary data based on rRNA gene recovery suggest that Desulfosarcina-like bacteria were low in abundance in core 4 before incubation, but increased dramatically after incubation on the AMIS (data not shown).
Nucleic acid extraction and quantitative PCR.
Because the SSU rRNA clone libraries constructed from Monterey Canyon seep sediments suggested that ANME-2c were the predominant MOA, we developed TaqMan primers specific to the ANME-2c group (ANME-1 archaea were absent from all libraries, and ANME-2b appeared in low relative abundance; Fig. 4). ANME-2c-specific primers were tested against all dominant archaeal groups represented in both the current clone libraries and those described previously (25). Representatives of the ANME-1 and ANME-2ab archaeal groups, as well as Desulfosarcina bacteria, did not cross-react with our primers. However, an environmental sequence Eel-36a2H11 (AF354136),related to Methanococcoides (25), did exhibit low levels of cross-reactivity. In our TaqMan analyses, amplification of templates of Methanococcoides relatives were only detected in the last five cycles of the reaction (outside the range of cycles used in analysis) and were consistently 3 orders of magnitude less per unit template in comparison to our target ANME-2c template. Methanococcoides relatives have been detected in some seep environments (25), but were not recovered in our 16 SSU clone libraries from Monterey Canyon (Fig. 4).
To examine changes in abundance of ANME-2c MOA resulting from incubation on the AMIS, we measured the ANME-2c copy number per gram of sediment in subsamples of cores 1, 2, and 4 taken from the bottom 2 cm of sediment (Table 1). Prior to incubation on the AMIS, core 1 and core 2 sediments had approximately 4.6 × 105 to 5.4 × 105 copies of ANME-2c rRNA per g of sediment (Table 1). Core 2 showed no statistical difference in the ANME-2c copy number per g of sediment after incubation on the AMIS. Core 1, however, had a significant increase in the ANME-2c copy number per gram of sediment after incubation on the AMIS (from 4.73 to 8.16 × 105 per g of sediment; P < 0.001) (Table 1). Core 4 showed a significant increase in the ANME-2c copies per gram of sediment (from undetectable to 2.93 × 105 ± 0.32 × 105 per g of sediment postincubation; P < 0.001) (Table 1), whereas this group was not detectable prior to incubation. After a 24-week incubation, a small but detectable number of ANME-2c MOA (1.37 × 103 ± 2.90 × 103 copies per ml of seawater; Table 1) were also found in the lower PVC manifold seawater. Subsamples of the nonseep sediment pushcore that had been sealed and maintained for 24 weeks at the same temperature as AMIS were also analyzed, and no ANME-2c copies were detected either before or after the 24-week period.
FISH.
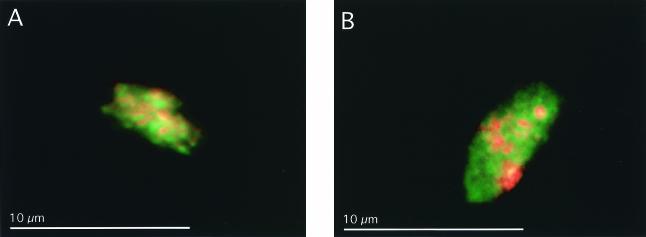
Whole-cell hybridizations revealed oval aggregations of MOA and sulfate-reducing bacteria, about 7 μm in length, in a freshly collected seep sediment as well core 4 sediments after incubation on the AMIS (Fig. 5). Although easily identifiable aggregations were rare, the use of specific FISH probes revealed an association of ANME-2 MOA and sulfate-reducing bacteria similar to those previously reported from other environments (8).
FIG. 5.
Whole-cell fluorescent in situ hybridization of methane-oxidizing consortia found in Monterey Canyon seep sediments (A) and in Monterey Canyon nonseep sediments after incubation on AMIS (B). Sediments were fixed in 4% formalin and transferred into a 1:1 ethanol-phosphate-buffered saline solution for storage. Green-stained cells correspond to Desulfosarcina-Desulfococcus bacteria (DSS658), and red-stained cells correspond to the archaeal ANME-2 group (EelMSMX932). Separate images were overlaid to represent the structure of the aggregate. Scale bar, 10 μm. No consortia were detectable in Monterey Canyon nonseep sediments before incubation on the AMIS.
DISCUSSION
Sustaining methane-oxidizing archaeal metabolism and growth with the AMIS.
The combined results of sediment core incubation experiments demonstrated that the continuous-flow AMIS incubation system promoted the metabolism, as well as the growth, of MOA populations indigenous to marine seep sediments. Our results also demonstrate that AMIS promoted MOA growth and metabolism in nonseep sediments as well (Table 1). We believe that incubation on AMIS fostered MOA metabolism and growth by promoting increased mass transfer to sediments via advective fluid flow.
At the end of the incubation, the circulating seawater in the lower PVC manifold also demonstrated significant, albeit low, methane oxidation rates (Table 1). Based on the observed methane oxidation and the quantitative PCR biomass proxy, we suggest that MOA were enriched in the circulating seawater during the later stages of incubation and may have been enriched in our manifold. Alternatively, some MOA may have been entrained from the sediments incubated on the manifold. Further studies are required to clarify these suppositions.
Although measuring anaerobic methane oxidation rates by sediment subsamples provided evidence for the presence of MOA in the incubated sediment cores, they cannot be used to make quantitative inferences about the population changes before and after incubation. Attempts to use FISH quantitatively on Monterey Canyon sediments have so far proven unreliable. As an alternative and less subjective approach to quantification, we used quantitative PCR to compare ANME-2c MOA rRNA copy number per gram of sediment before and after incubation. These data showed MOA in the core 4 nonseep sediments increasing from undetectable numbers to densities representing 40 to 60% of the MOA found in seep sediments and confirm that the MOA were reproductively viable and metabolically active when incubated on AMIS (Table 1). The ANME-2c archaea were the dominant MOA in the core 1 seep sediments before and after incubation and were the only MOA detected in the core 4 nonseep sediment after incubation (although there was evidence for the presence of ANME-2b as well as ANME-2c phylotypes within the core 1 libraries; Fig. 3). The absence of MOA groups other than ANME-2c in the nonseep core after incubation may be due to a “founder effect” caused by seed populations of ANME-2c archaea from the seep sediments, or may be due to differential viability and enrichment of different MOA groups at one atmosphere.
It is worth noting that both methane oxidation rates and ANME-2c copy number per gram of sediment measured from sediment subsamples often showed a high degree of variability, both before and after incubation, in both seep and nonseep sediments (Table 1). We believe this variability is inherent to deep-sea sediments, due to the natural variations in physicochemical features, such as sediment grain size that may affect mass-transfer of metabolites to microbial populations, and the resulting differences in porewater chemistry and microbial populations at fine scales.
We observed aggregations of MOA and Desulfosarcina-related bacteria in both freshly collected Monterey seep sediments, as well as in core 4 sediments after incubation on AMIS (Fig. 5). These observations suggest that incubation on the AMIS fostered the growth and association of MOA with sulfate-reducing bacteria, in physical associations similar to those observed in other environments (8). In addition to the preliminary evidence from SSU libraries, these results suggest that parallel increases in both MOA and SRB populations occurred in the sediment after incubation on AMIS.
rRNA genes recovered from core 4 nonseep sediments after incubation on AMIS exhibited a marked shift in the diversity of rRNA recovered in cloning experiments, from dominance by SSU rRNA most similar to Crenarchaeota to a marked shift towards ANME-2c SSU rRNA, respectively (Fig. 3). In addition, the total diversity of recovered SSU rRNA clones from this library appeared higher prior to incubation (Fig. 3). This shift may reflect an ecological succession occurring during AMIS incubation, in which the dominant nonseep microbial species are replaced by MOA phylotypes typically found in seep sediments (Fig. 3 and 4). Validating this hypothesis will require further incubations and experimental work on AMIS. Also, we cannot conclusively determine if the ANME-2c population found in the nonseep sediment after incubation arose from a seed population carried to the core by the flowing seawater system (during the first two weeks when we did not filter-sterilize the manifold seawater) or if they arose autochthonously. A nonseep sediment control core, however, that had been sealed for 24 weeks without incubating on AMIS did not contain detectable populations of ANME-2c rRNA, suggesting that the flow of methane and sulfate through the sediments (and not anoxia) were responsible for the observed MOA enrichments.
Environmental relevance of anaerobic oxidation of methane enrichments at one atmosphere.
Thermodynamic calculations demonstrate that the Gibbs free energy yield for anaerobic oxidation of methane at one atmosphere of methane pressure is low, and that high ambient methane concentrations and close physical associations (with respect to syntrophy) can produce energetic yields sufficient to support biosynthesis (17). Consequently the means by which MOA are capable of growing at one atmosphere pressures remains poorly understood. It is commonly suggested that high rates of anaerobic oxidation of methane observed in the deep oceans requires high dissolved methane concentrations that can only be attained at very high pressures (24), however there is ample evidence for anaerobic oxidation of methane near one atmosphere in both terrestrial and aquatic ecosystems (11, 17, 20, 21, 23, 28). The results presented here provide strong evidence that metabolically active MOA maintained in anoxic seep sediments at one atmosphere were capable of sustaining methane oxidation at rates sufficient to support relatively rapid growth. Our results demonstrate that MOA populations were enriched over a relatively short period of time (in nonseep sediments) during incubation on AMIS.
Although it is difficult to rectify the growth of MOA at one atmosphere with the low energetic yields from methane oxidation at one atmosphere, our results suggest that biological mechanisms may have evolved that facilitate rapid methane oxidation rates to provide sufficient energy for biosynthesis and growth. Jointly, the observed methane oxidation rates and MOA biomass proxy suggest a high methane turnover rate per unit MOA cell, although further experiments are required to provide a more quantitative assessment. In total, the data presented here provides further support that MOA flourishing near one atmosphere may be adapted to maintain high methane oxidation rates per cell to attain the energy necessary for growth and reproduction. Empirical data of microbial populations and isotopic distributions, as well as theoretical models are also consistent with the growth of MOA near one atmosphere (1, 7, 31, 35).
Insights into the cultivation of mixed communities.
The cultivation of microorganisms in consortia allows the examination of processes that are not governed by a single microbial phenotype (9). The AMIS system reported here selectively enriched for anaerobic oxidation of methane communities by mimicking many of the major physicochemical conditions found in situ with minimal perturbation to the spatial arrangements of both physical and microbial associations. Although the AMIS methane concentrations (ca. 1.5 mM in AMIS), and hydrostatic pressure (ca. 2 atmospheres in AMIS) differed from deep water environments with respect to methane and pressure, in most other respects including temperature, Redox potential, sulfide and sulfate concentrations, pH, porosity, and flow, the AMIS recreated the ambient environment with high fidelity.
In principle, the AMIS is similar to a Winogradsky column, with the notable exception that metabolites, including energy sources and carbon substrates, are actively delivered through the sediments and to the microbial communities, thus reducing limitations imposed by diffusion. The AMIS also provides a means of conducting perturbation studies to better address ecophysiological questions and even allows examination of functional genes that are involved in methane metabolism, for instance, the identification of mcrA genes from MOA (15a). Our results indicate that the combined use of physiological and quantitative molecular genetic approaches provides a useful approach to studying community metabolism. The further development and implementation of bioreactors like AMIS that approximate the dynamic gradients and environmental conditions found in nature promises to enhance our understanding of natural microbial populations and processes.
Acknowledgments
Special thanks to James Childress for generously loaning equipment, Marcelino Suzuki for extensive assistance with quantitative PCR and collard green recipes, and Tori Hoehler for invaluable commentary and critique. Thanks to Christina Preston, Lynne Christianson, and Shana Goffredi for assistance and guidance with sequencing and Jose de la Torre for assistance with phylogenetic analysis. As always, special thanks to the crew and pilots of the RV Point Lobos and the ROV Ventana.
This work was supported by the Monterey Bay Aquarium Research Institute and the Packard Foundation.
REFERENCES
- 1.Alperin, M. J., and W. S. Reeburgh. 1985. Inhibition experiments on anaerobic methane oxidation. Appl. Environ. Microbiol. 50:940-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, R. O., and E. D. Goldberg. 1976. Methane production and consumption in anoxic marine sediments. Geology 4:297-300. [Google Scholar]
- 5.Barry, J. P., H. G. Greene, D. L. Orange, C. H. Baxter, B. H. Robison, R. E. Kochevar, J. W. Nybakken, D. L. Reed, and C. M. McHugh. 1996. Biologic and geologic characteristics of cold seeps in Monterey Bay, California. Deep Sea Res. I Oceanogr. Res. Papers 43:1739-1762. [Google Scholar]
- 6.Barry, J. P., R. E. Kochevar, and C. H. Baxter. 1997. The influence of pore-water chemistry and physiology on the distribution of vesicomyid clams at cold seeps in Monterey Bay: Implications for patterns of chemosynthetic community organization. Limnol. Oceanogr. 42:318-328. [Google Scholar]
- 7.Bian, L., K.-U. Hinrichs, X. Tianmin, S. C. Brassell, N. Iversen, H. Fossing, B. B. Joergensen, and J. M. Hayes. 2001. Algal and archaeal polyisoprenoids in a recent marine sediment: molecular isotopic evidence for anaerobic methane oxidation. Geochem. Geophys. Geosystems 2, paper number 2000GC000112. (Online.)
- 8.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. Microscopic identification of a microbial consortium apparently mediating anaerobic methane oxidation above marine gas hydrate. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 9.Caldwell, D. E., G. M. Wolfaardt, D. R. Korber, S. Karthikeyan, J. R. Lawrence, and D. K. Brannan. 2002. Cultivation of Microbial Consortia and Communities, p. 92-100. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 10.DeLong, E. F., G. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for identification of single microbial cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 11.Devol, A. H. 1983. Methane oxidation rates in the anaerobic sediments of Saanich Inlet. Limnol. Oceanogr. 28:738-742. [Google Scholar]
- 12.Devol, A. H., and S. L. Ahmed. 1981. Are high rates of sulfate reduction associated with anaerobic methane oxidation? Nature 291:407-408. [Google Scholar]
- 13.Ferry, J. G. 1993. Methanogenesis: ecology, physiology, biochemistry & genetics. Chapman & Hall, New York, N.Y.
- 14.Girguis, P. R., R. W. Lee, N. Desaulniers, J. J. Childress, M. Pospesel, H. Felbeck, and F. Zal. 2000. Fate of nitrate acquired by the tubeworm Riftia pachyptila. Appl. Environ. Microbiol. 66:2783-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen, L. B., K. Finster, H. Fossing, and N. Iversen. 1998. Anaerobic methane oxidation in sulfate-depleted sediments: effects of sulfate and molybdate additions. Aquat. Microb. Ecol. 14:195-204. [Google Scholar]
- 15a.Hallam, S. J., P. R. Girguis, C. M. Preston, P. N. Richardson, and E. F. Delong. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69: AEM 514-03, companion paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinrichs, K.-U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 17.Hoehler, T. M., M. J. Alperin, D. B. Albert, and C. S. Martens. 1994. Field and laboratory studies of methane oxidation in an anoxic marine sediment: Evidence for a methane-sulfate reducer consortium. Global Biogeochem. Cycles 8:451-463. [Google Scholar]
- 18.Huber, J. A., D. A. Butterfield, and J. A. Baross. 2002. Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol. 68:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iversen, N., and B. B. Joergensen. 1985. Anaerobic methane oxidation rates at the sulfate-methane transition in marine sediments from Kattegat and Skagerrak (Denmark). Limnol. Oceanogr. 30:944-955. [Google Scholar]
- 20.Martens, C. S., D. B. Albert, and M. J. Alperin. 1999. Stable isotope tracing of anaerobic methane oxidation in the gassy sediments of Eckernfoerde Bay, German Baltic Sea. Am. J. Sci. 299:589-610. [Google Scholar]
- 21.Martens, C. S., and R. A. Berner. 1977. Interstitial water chemistry of Long Island Sound sediments. Limnol. Oceanogr. 22:10-25. [Google Scholar]
- 22.Martens, C. S., and R. A. Berner. 1974. Methane production in the interstitial waters of sulfate-depleted marine sediments. Science 185:1167-1169. [DOI] [PubMed] [Google Scholar]
- 23.Munson, M. A., D. B. Nedwell, and T. M. Embley. 1997. Phylogenetic diversity of archaea in sediment samples from a coastal salt marsh. Appl. Environ. Microbiol. 63:4729-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nauhaus, K., A. Boetius, M. Kruger, and F. Widdel. 2002. In vitro demonstration of anaerobic oxidation of methane coupled to sulfate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4:296-305. [DOI] [PubMed] [Google Scholar]
- 25.Orphan, V. J., K.-U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. DeLong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 27.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeburgh, W. S. 1980. Anaerobic methane oxidation; rate depth distributions in Skan Bay sediments. Earth Planet Sci. Lett. 47:345-352. [Google Scholar]
- 29.Reeburgh, W. S. 1976. Methane consumption in Cariaco Trench waters and sediments. Earth Planet Sci. Lett. 28:337-344. [Google Scholar]
- 30.Reysenbach, A. L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a mid-Atlantic Ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorenson, K. B., K. Finster, and N. B. Ramsin. 2001. Thermodynamic and kinetic requirements in anaerobic methane oxidizing consortia exclude hydrogen, acetate, and methanol as possible electron shuttles. Microb. Ecol. 42:1-10. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small subunit rRNA genes in mixed microbial populations employing 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teske, A., K.-U. Hinrichs, V. Edgcomb, A. de Vera Gomez, D. Kysela, S. P. Sylva, L. S. Mitchell, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, K. L., D. Lloyd, J. Benstead, and S. H. Lloyd. 1998. Diurnal oscillations of gas production and effluxes (CO2 and CH4) in cores from a peat bog. Biol. Rhythm Res. 29:247-259. [Google Scholar]
- 35.Thomsen, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wirsen, C. O., S. M. Sievert, C. M. Cavanaugh, S. J. Molyneaux, A. Ahmad, L. T. Taylor, E. F. DeLong, and C. D. Taylor. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zehnder, A. J., and T. D. Brock. 1980. Anaerobic methane oxidation: occurrence and ecology. Appl. Environ. Microbiol. 39:194-204. [DOI] [PMC free article] [PubMed] [Google Scholar]