Abstract
The complete nucleotide sequence of the 13-kb plasmid pRV500, isolated from Lactobacillus sakei RV332, was determined. Sequence analysis enabled the identification of genes coding for a putative type I restriction-modification system, two genes coding for putative recombinases of the integrase family, and a region likely involved in replication. The structural features of this region, comprising a putative ori segment containing 11- and 22-bp repeats and a repA gene coding for a putative initiator protein, indicated that pRV500 belongs to the pUCL287 subfamily of theta-type replicons. A 3.7-kb fragment encompassing this region was fused to an Escherichia coli replicon to produce the shuttle vector pRV566 and was observed to be functional in L. sakei for plasmid replication. The L. sakei replicon alone could not support replication in E. coli. Plasmid pRV500 and its derivative pRV566 were determined to be at very low copy numbers in L. sakei. pRV566 was maintained at a reasonable rate over 20 generations in several lactobacilli, such as Lactobacillus curvatus, Lactobacillus casei, and Lactobacillus plantarum, in addition to L. sakei, making it an interesting basis for developing vectors. Sequence relationships with other plasmids are described and discussed.
Lactobacilli belong to the lactic acid bacteria (LAB) group. The genus comprises several species important for the food industry, among which is Lactobacillus sakei, one of the most important bacterial species involved in meat fermentation and preservation. L. sakei belongs to the dominant microflora naturally occurring on vacuum-packed meat and is also used in Western Europe as starter cultures for fermented products (especially sausage) because of its ability to rapidly ferment carbohydrates in the meat environment (11, 26).
Most lactobacilli harbor one or more plasmids, the size of which can vary from 1.2 to 150 kb (for a review, see reference 50). Different functions have already been found on these plasmids, among which are genes for lactose metabolism, bacteriocin synthesis, exopolysaccharide production, or DNA restriction-modification (R-M). With the development of molecular biology and functional genomics of lactobacilli, interest is growing for the characterization of replicons themselves as potential useful (food-grade) vectors. Two modes of DNA replication are used by circular bacterial plasmids, namely, rolling circle (RC) and theta. RC plasmids have been assumed to be the most widespread in gram-positive bacteria. However, recent years have seen the characterization of a large number of theta-replicating plasmids from gram-positive, and especially from LAB, hosts. Several mechanisms are known for the initiation of theta replication. Theta replicons can be classified according to their dependence on three key components. These are plasmid-encoded initiator Rep proteins necessary for strand opening and/or interaction with other components of the initiation complex, origins of replication (generically termed ori) with specific DNA structural organization for strand opening and initiator-protein binding, and host-encoded DNA polymerase I for nascent strand DNA synthesis. At least five classes of theta replicons have been recognized (10, 37). Class A plasmids are independent of DNA polymerase I. They encode their own Rep proteins and possess chromosomal-like origins of replication (often called oriA for these plasmids), with an AT-rich region generally containing repeats and Rep-binding iterons (15). oriA may also exhibit DnaA boxes, which allow for the participation of the host-encoded DnaA initiation protein in plasmid replication, in combination with the Rep protein. A number of plasmids from gram-negative hosts were grouped in class A, and different subfamilies could be distinguished according to the degree of sequence similarity of their Rep proteins (for example, pSC101 or RK2) (15). Rep-encoding plasmids in gram-positive bacteria were recognized as prototypes for class D (pAMβ1/pIP501 type, a particular case for which an oriA-like structure is present but not essential for replication) or, on the basis of absence of similarity with Rep proteins of known families, as prototypes for newly described families (yet unclassified), namely: pUCL22 (20), a family also referred to as pCI305 or pWV02, which is especially widespread in lactococci (45) and probably belongs to class A (37); pLS32, to which belong, for example, plasmids pLH1 (49) and pCI2000 (27) from LAB and which is also possibly related to class A (27); and pUCL287, which replicates in Enterococcus and lactobacilli (4). In contrast to RC plasmids, which are rather unstable and small (<12 kb) (25, 29), theta-type plasmids replicate by means of a double-stranded rather than a single-stranded replication intermediate, which results in better structural stability, allowing for the insertion of large DNA fragments. This property is useful for the construction of cloning vectors.
Only a small fraction of the Lactobacillus plasmids—and to our knowledge, none of the L. sakei plasmids—that have been described in the literature have been entirely sequenced. Such complete sequences bring interesting insights into plasmid-borne functions in lactobacilli, genetic exchange between bacteria, and replicons that are potentially useful for the development of genetic tools. The latter is an aim of our research engaged in functional genomics of L. sakei (30). Both RC and theta types of replicons have been reported in lactobacilli. In L. sakei, RC plasmids closely related to those of Lactobacillus curvatus (31) and Lactobacillus plantarum (9) have been identified, and the partial sequence of a potential theta-replicating plasmid is known, namely, pSAK1 (GenBank Z50862), belonging to the pLS32 family (49). In this paper, we present the analysis of the complete sequence of a new L. sakei plasmid. Its relationship with other plasmids, a first characterization of its replicon (which belongs to the pUCL287 family of theta replicons), and its ability to be maintained in several lactobacilli are reported.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Lactobacilli were grown in static MRS liquid cultures containing 1% glucose (16) at 30 or 37°C for Lactobacillus casei. All the strains of L. sakei and L. curvatus (except two from ATCC) were from the INRA collection (5). L. sakei 23K, G3AF, 160K, and 64F are laboratory strains, are totally (23K and 160K) or partially (64F and G3AF) cured of their native plasmids, and have been previously described (6). L. sakei RV332, containing plasmid pRV500, was originally isolated from vacuum-packed beef meat (12) and subsequently identified as a member of the B2 group by random amplified polymorphic DNA analysis (5). Escherichia coli TG1, used for cloning and preparation of sequencing templates, was grown in Luria-Bertani medium at 37°C and aerated by vigorous shaking (44). The respective media were solidified by the addition of 1.5% agar for plating. Where appropriate, antibiotics were added as follows: ampicillin at 100 mg · liter−1 or erythromycin at 150 mg · liter−1 for E. coli and erythromycin at 5 mg · liter−1 for lactobacilli.
DNA manipulations, plasmid construction, and bacterial transformations.
Common DNA manipulation methods were performed as described elsewhere (35, 44). Total and plasmid DNA was extracted from lactobacilli as described by Anderson and McKay (1). L. sakei crude-cell lysates were obtained by extended lysozyme treatment. When needed, plasmid DNA was purified on Qiagen plasmid purification columns according to the instructions of the manufacturer. Plasmid pRV500 was purified from L. sakei RV332 by CsCl gradient centrifugation after extraction by the method of Anderson and McKay (1).
pRV566 was obtained by cloning the KpnI-EcoRI fragment of pRV500, which spans the iterons, repA, and the downstream open reading frame (ORF), into pRV300, a plasmid containing the replicon colE1 of E. coli and a gene conferring resistance to erythromycin (34).
All lactobacilli were transformed by electroporation by using the procedure previously described for L. sakei (6). E. coli was transformed as described by Dower et al. (17). Plasmids pIL253 (46) and pGhost4 (7), purified from E. coli, were used as standards to compare transformation efficiencies in Lactobacillus strains. For Southern hybridization experiments, total pRV566 was used as a probe after restriction digestion and labeling with the ECL kit from Amersham.
Sequence analysis of pRV500.
BglII, EcoRI, or KpnI fragments of pRV500 were subcloned into pBluescriptSKII+ (Stratagene) and used as starting points for sequence determination. The primer-walking method, directly using pRV500 DNA as template, completed sequencing. Sequencing reactions were performed with the ABI dye terminator sequencing reagents according to the instructions of the manufacturer. Sequencing gels were run on an ABI377 sequencer. Fragment assembly and sequence analysis were performed with the Artemis package from the Sanger Center, the GCG software package of the University of Wisconsin, and the Clustal program.
Determination of plasmid copy number and stability tests.
Plasmid quantity in total DNA extracted from strains of L. sakei 23K or RV332 containing pRV500 or pRV566 plasmids was estimated in dot blot experiments in which the hybridization signal of known amounts of sample total DNA was compared to that of known amounts of purified plasmid DNA. By knowing the respective sizes of chromosomal and plasmid DNA, the copy number of plasmid could then be deduced from the determined quantity of plasmid in the samples (21). The experiment was repeated three times with two different total DNA extractions for each strain. Two different quantities of total DNA samples were deposited on each membrane (e.g., 0.5 μg and 1 μg). An equivalent amount (e.g., 0.5 μg) of chromosomal DNA (from strain 23K) was added to purified plasmid used as standard to mimic the conditions of total DNA of the samples. The probe was either a ClaI restriction fragment of 0.71 kbp or a fragment of 0.67 kbp generated by PCR amplification with oligonucleotides AGTAGTATTATAAAGTTGACT and ATAAGGGAGTTAATTCTTC and containing the 5′ or 3′ parts of repA of pRV566, respectively. The probe did not give any signal with total DNA of L. sakei strain 23K without plasmid. For testing segregational stability of pRV566, strains harboring the plasmid were grown at 30°C in 100 ml of MRS without antibiotic selection. Reinoculations of 100 μl of a 10−3 dilution of the culture (representing at least 3 × 104 CFU) into 100 ml of fresh MRS were performed every 24 h. In these conditions, each phase of growth corresponded to approximately 20 generations. At the end of each phase of growth, diluted aliquots were spread on MRS- and erythromycin-containing MRS plates, and colonies were scored. The experiment was done twice.
Nucleotide sequence accession number.
The GenBank accession number of pRV500 sequence is AF438419.
RESULTS
Sequence analysis of the single plasmid of L. sakei RV332.
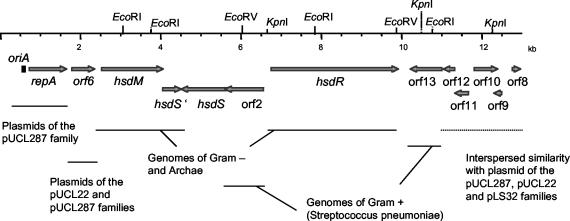
Plasmid DNA was isolated from L. sakei strain RV332 by each of the following methods, namely, separation of total DNA by centrifugation through CsCl gradient and separation of crude cellular lysates through plasmid isolation columns. A single plasmid, which we named pRV500, was detected. Its size was estimated to be 12.5 to 13.5 kb by restriction fragment analysis. This was confirmed by the determination of the complete circular sequence of 12,959 nucleotides. A total of 13 ORFs preceded by potential ribosome binding sites were identified (Fig. 1). The coding sequences represent a total of 11,139 nucleotides, or 86% of the size of the plasmid. Overall, the GC content of the plasmid (38.1%) is lower than that of the chromosome of L. sakei (41.3%) (S. Chaillou, personal communication).
FIG. 1.
Physical map of pRV500 and sequence relationships with databases. The 13 ORFs of pRV500 are depicted by open arrows in the direction of their putative transcription. Direct repeats (iterons) located in the putative origin of replication oriA are shown. Segments having significant nucleotide sequence similarity or coding for products showing amino acid identity above 30% with sequences in databases are indicated.
To attribute a function to the deduced products of the ORFs, they were compared to gene products in databases (Table 1). We could identify a region (oriA, repA) that we assumed was involved in replication of a theta-type plasmid and which is detailed in the next section. Four gene products were similar to proteins of bacterial type I restriction-modification systems. Each of the subunits required for this system to be functional was identified in pRV500: the methylase (HsdM), the restriction (HsdR), and the specificity (HsdS) subunits. An additional truncated gene, hsdS′, potentially encoding the N-terminal part of an additional S subunit, was also found. Best hits for these gene products were observed with bacteria other than LAB and especially with gram-negative bacteria. The GC content of the R-M genes was higher (from 38.6 to 43.4%) than in the other parts of the plasmid (Table 1).
TABLE 1.
General features of the ORFs in pRV500 and putative functions for their products
| ORF | Position | Size (amino acids) | GC content (%) | Proposed function | Organism and protein or plasmid with protein database similarity (SPTREMBL or Swissprot accession no.)a | % Amino acid identityb |
|---|---|---|---|---|---|---|
| Orf1 (hsdR) | 6759-9923 | 1,054 | 41.5 | Type I R-M system: restriction subunit | Campylobacter jejuni, HsdR (q9pmc0) | 43 |
| Xylella fastidiosa, R-M I endonuclease (q9p9y1) | 42 | |||||
| Methanosarcina mazei, MM0431 (q8pzr1) | 40 | |||||
| Orf2 | 6571-5612 | 319 | 38.3 | Recombinase of the integrase family (Int/rec) (tyrosine DNA recombinase) | Streptococcus pneumoniae TIGR4, integrase/recombinase SP0890 (q97rd2) | 53 |
| Staphylococcus aureus MW2, XerC (q8nwz8) | 28 | |||||
| Lactobacillus leichmannii, XerC recombinase (q48733) | 26 | |||||
| Orf3 (hsdS) | 5616-4491 | 375 | 40.5 | Type I R-M system: specificity subunit | Pasteurella haemolytica, HsdS (P95511) | 25 |
| Campylobacter jejuni, HsdS (q8rj16) | 30 | |||||
| Orf4 (hsdS) | 4039-4494 | 151 | 38.4 | Type I R-M system: specificity subunit (truncated) | Klebsiella pneumoniae, HsdS (034140) | 50 |
| Methanosarcina mazei, type I R-M specificity subunit MM0430 (q8pzr2) | 48 | |||||
| Salmonella enterica, Sty SBLI (P72419) | 45 | |||||
| Orf5 (hsdM) | 2517-4049 | 510 | 43.3 | Type I R-M system: methylase | Xylella fastidiosa, type I R-M methylase (q9p9x8) | 55 |
| Methanosarcina mazei, type I R-M MM2198 (q8puy0) | 53 | |||||
| Campylobacter jejuni, HsdM (q9pmb6) | 52 | |||||
| Orf6 | 1782-2351 | 189 | 35.3 | Unknown | Plasmid pJW563, Lactococcus lactis, OrfX (q48655)c | 58 |
| Plasmid pSRQ700, Lactococcus lactis, OrfX (q93k29)c | 35 | |||||
| Plasmid pUCL287, Tetragenococcus halophilus, RepBd | 30 | |||||
| Orf7 (repA) | 721-1656 | 311 | 34.4 | Initiator protein (replication) | Plasmid pMD5057, Lactobacillus plantarum, RepA (AAN40880)d | 94 |
| Plasmid pUCL287, Tetragenococcus halophilus, RepA (q51863)d | 79 | |||||
| Plasmid pLKS, Lactobacillus plantarum, RepA (q9s0j9)d | 66 | |||||
| Orf8 | 12747-12959 | 70 | 31.2 | Unknown | Plasmid pRC18, Lactobacillus casei, hypothetical protein (q8vual)d | 92 |
| Plasmid pLP9000, Lactobacillus plantarum, hypothetical protein (q8kjs5) | 74 | |||||
| Plasmid pMD136, Pediococcus pentosaceus, Orf7 (q9X3b8)c | 43 | |||||
| Orf9 | 12487-12287 | 66 | 35.8 | Unknown | No similarity | |
| Orf10 | 11803-12390 | 195 | 38.3 | Putative integrase/recombinase (Int/rec) (tyrosine DNA recombinase) | Plasmid pLP9000, Lactobacillus plantarum, putative integrase/recombinase (q8kjs4) | 94 |
| Plasmid pRC18, Lactobacillus casei, putative integrase/recombinase (q8vua2)d | 94 | |||||
| Plasmid pLH1, Lactobacillus helveticus, Orf195a integrase/recombinase (O50346)e | 90 | |||||
| Orf11 | 11664-11314 | 116 | 35.3 | Unknown | Plasmid pSRQ800 Lactococcus lactis, Orf3 (O07029)c | 86 |
| Orf12 | 11317-11033 | 94 | 36.1 | Unknown—HTH motif | Plasmid pSRQ800 Lactococcus lactis, Orf4 (O07030)c | 85 |
| Orf13 | 10999-10211 | 262 | 31.6 | Unknown | Streptococcus pneumoniae R6, hypothetical protein SP0558 (q97s55) | 38 |
Only three selected hits among E values above 1 E-05 (blast program) are indicated.
% identity is on whole length except where values are indicated in bold.
Theta-type plasmid belonging to the pUCL22 family.
Theta-type plasmid belonging to the pUCL287 family.
Theta-type plasmid belonging to the pLS32 family.
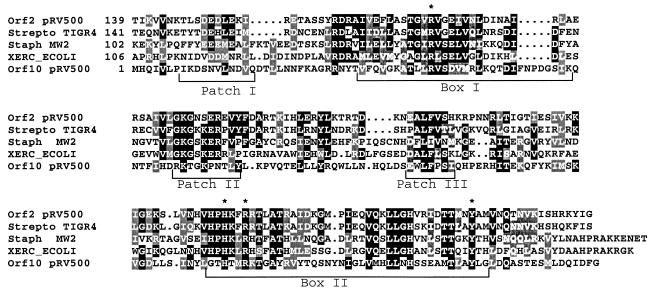
Orf2 and Orf10 gene products were similar to site-specific DNA recombinases. Two major families of site-specific recombinases have been described, namely, tyrosine recombinases, also known as integrases/recombinases (Int/rec), and serine recombinases, also called resolvases/invertases. They mediate break-and-join recombination but are structurally diverse and functionally versatile (promoting integration, excision, inversion, and transposition) and, as such, may be involved in different biological functions (41, 47). By similarity search, Orf2 and Orf10 would belong to the Int/rec family. Moreover, both Orf2 and Orf10 exhibited the conserved regions and tetrad signature of the catalytic domain of Int/rec (Fig. 2). Orf10 consists only of the catalytic domain and does not possess an additional N-terminal domain.
FIG. 2.
Blocks of conserved residues between gene products of pRV500 and site-specific recombinases of the integrase family. Numbers at the left of the alignment indicate the coordinates of the first residue shown for each sequence. Residues identical or similar in at least half of the different sequences are shaded black or gray, respectively. See Table 1 for accession numbers for recombinases of Streptococcus pneumoniae TIGR4 and of Staphylococcus aureus MW2. Blocks indicated as patches and boxes are reported according to the results of Nunes-Düby et al. (42). The tetrad signature of the family is indicated by asterisks above the four relevant residues.
Orf6, 8, 11, 12, and 13 of pRV500 showed amino acid similarity to gene products of unknown function (or controversial function; see the discussion for Orf6) carried by several plasmids (Table 1). Only one ORF of pRV500 (Orf9) showed no detectable similarity to gene products in databases. However, this region does possess some nucleotide similarity with other plasmids, as discussed below.
Replication features of pRV500 deduced from sequence analysis and relationship with other replicons.
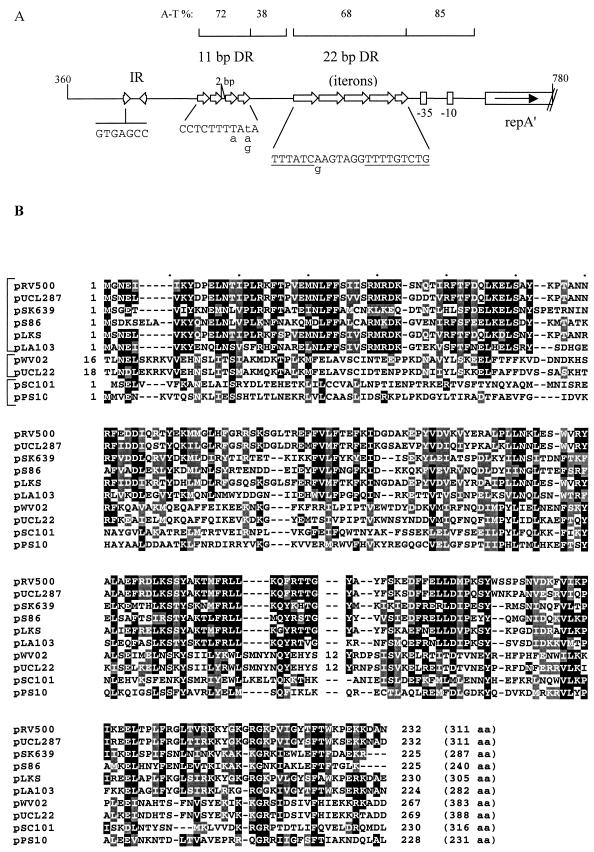
The gene likely to code for an initiator protein was identified in pRV500 by similarity to gene products in databases and was named repA. RepA of pRV500 is highly similar (>80% amino acid identity) to the initiator protein identified in plasmid pUCL287 of Tetragenococcus halophilus, the prototype of a family of theta-replicating plasmids of gram-positive bacteria (4). Two kinds of repeated sequences, lying upstream of repA, were also identified in pRV500 (Fig. 3A), namely, 11-bp direct repeats (four times; coordinates 472 to 517) and 22-bp iterons, tandemly repeated four and one-half times (coordinates 555 to 654). These features meet the typical configuration of the pUCL287 family of theta-replicating plasmids (3).
FIG.3.
(A) Structural organization of the ori region of pRV500. Coordinates of the schematized fragment are indicated at each end. The percentage of A-T composition of particular segments is indicated, and −35 and −10 boxes of the putative promoter of repA are represented. Each unit of identified direct (DR) or inverted (IR) repeats is depicted by an arrow. The sequence of the repeat units is indicated below. Lowercase letters give alternative sequence found in one of the units. Underlined letters indicate the nucleotides conserved in the iterons of pEF418 (AF408195) and pUCL287 (X75607). The motif TTGTCTGTTTAT, obtained by the juxtaposition of the repeated units, was also found to be present in the iterons of pMD5057 (AF440277), pLKS (AB035265), pRC18 (AF200347), and pLA105 (D49554) but not in those of pLA103 (D55703) or pSK639 (U40259). (B) Conservation of an N-terminal-to-central region in Rep proteins of three subfamilies of class A theta plasmids. Numbers at the left and at the right of the alignment indicate the coordinates of the first and last residues shown, respectively. The total number of residues for each protein is given in parentheses. Residue shading is as described in the legend for Fig. 2. Respective database accession numbers for pUCL287 to pPS10, in the order shown in the figure, are Q51863, P95741, Q47808, Q9S0J9, Q52180, Q07137, Q48681, P22308, and Q52546.
Such a structural configuration is also typical for class A theta plasmids. The ori region of these plasmids generally comprises 22-bp sequences iterated three to five times (constituting the Rep-binding iterons) and an upstream AT-rich region including tandemly repeated sequences (3, 28, 36, 45) (Fig. 3A). A certain sequence conservation in the ori segments and repeats was observed in highly related lactococcal replicons (45). Inside the pUCL287 family, sequence conservation of the oriA segment is high between pRV500 and pUCL287 itself, with the 11-bp tandem repeats, for example, being identical. For the 22-bp repeats, a stretch of conserved nucleotides was noticed among some pUCL287-type plasmids (Fig. 3A). Additional features such as DnaA boxes were not found in pRV500, suggesting that the host protein is not involved in pRV500 replication initiation. The level of similarity between Rep proteins allowed different subfamilies to be distinguished. Sequence relationships may be detectable only by multiple alignments, as for the pUCL22 family and class A theta plasmids from gram-negative hosts, for which a conserved N-terminal-to-central region was eventually recognized (24). We found that Rep proteins of the pUCL287 family can also be included in a multiple alignment, as shown in Fig. 3. Blocks of amino acids in the central region of these proteins belonging to different subfamilies seem to be rather well conserved. In the N-terminal region, the conservation is high inside the pUCL287 subfamily but rather weak with the two others.
Sequence conservation among different plasmids from LAB hosts.
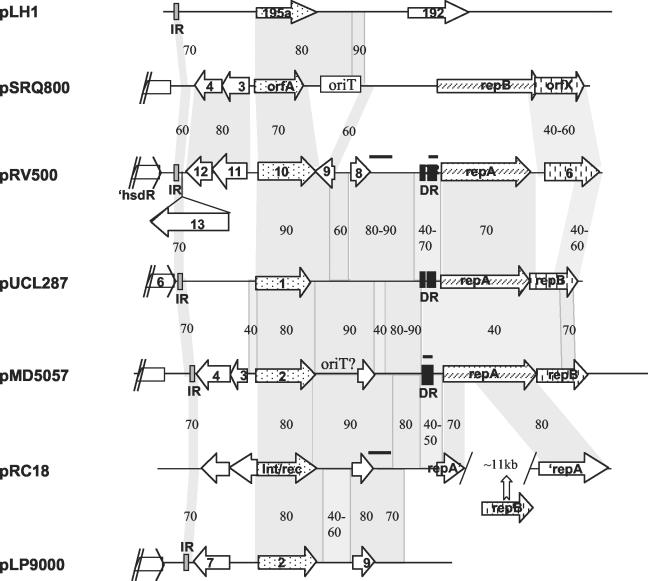
We identified sequence conservation among different plasmids around the replication region of pRV500 and around the gene for a putative Int/rec (orf10), as depicted in Fig. 4. First, orf6 downstream of repA in pRV500 showed nucleotide and/or amino acid similarity to ORFs carried by various theta-replicating plasmids (Table 1 and Fig. 4). Interestingly, the similarity is higher (up to 50% amino acid identity) with ORFs in plasmids of the pUCL22 subfamily than with the corresponding ORFs of pUCL287 or other plasmids of this subfamily (around 30% amino acid identity). The role of these ORFs is still unknown, but they are most probably not essential for plasmid replication in L. lactis (45). These ORFs (when present) seemed to be often located in the same unit of transcription and downstream of the rep genes coding for the initiator proteins; however, some plasmids appear to have experienced major genetic rearrangements, leading in pRC18, for example, to a split repA gene and a different location for the repB gene (Fig. 4). A DNA region surrounding orf10, a putative Int/rec gene, showed interspersed sequence conservation among plasmids isolated from LAB (Fig. 4). Apart from the recombinase gene, very few functions encoded by these regions have been identified, and a significant part seems to be noncoding. Thus, sequence conservation often appears more detectable at the nucleotide level. An origin of transfer (oriT) (Fig. 4), which is absent in pRV500, has been recognized in some of the plasmids (8, 14). A conserved feature is a palindromic sequence of about 40 bp, located downstream of orf13 in pRV500; it is identical at more than 90% where indicated in plasmids (Fig. 4) and could serve as a terminator for the upstream ORFs, as in pLH1 (49).
FIG. 4.
Sequence conservation among plasmids from LAB. Lfasta and Lalnview were used to detect and visualize nucleotide identity between plasmid sequences as schematized here (http://pbil.univ-lyon1.fr/lfasta.php). Gray shading and figures indicate sequence conservation and the percentage of nucleotide identity between two DNA segments. Segments with 100% nucleotide identity between pRV500 and pMD5057 or pRC18 are indicated by horizontal thick lines above the sequence lines. For easier visualization of sequence conservation, the DNA segment corresponding to orf13 of pRV500, which shares no similarity with other plasmids, was positioned underneath. Black and gray boxes represent stretches of direct repeats (DR) and palindromic sequences (IR), respectively. These palindromic sequences (about 40 bp) have higher conservation (>90% nucleotide identity) than that indicated according to the Lfasta program for the slightly larger region. orf genes putatively coding for recombinases and initiator proteins are indicated with dotted and hatched arrows, respectively, while those sharing amino acid similarity with orf6 of pRV500 are represented with arrows with vertical bars. Relevant portions of plasmids are schematized at the same scale here; for pLH1, pMD5057, and pLP9000, they are in the reverse direction relative to the entry in the database. Accession numbers for plasmid sequences (complete) were as follows: pLH1, AJ222725; pSRQ800, U35629; pUCL287, X75607; pMD5057, AF440277; pRC18, AF200347; pLP9000, AY096005.
Construction of a shuttle plasmid able to replicate in L. sakei and in E. coli.
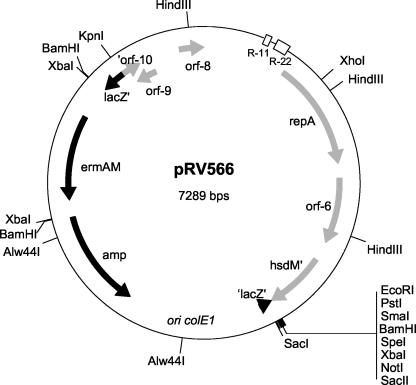
To experimentally prove its functionality for plasmid replication in L. sakei, a DNA fragment of pRV500, predicted to contain the replicon, was isolated and subcloned to produce the pRV566 shuttle plasmid (Fig. 5). The pRV300 moiety cannot replicate in L. sakei and does not have any DNA sequences homologous to the L. sakei chromosome, thus preventing insertion of the plasmid through homologous recombination into the chromosome (34). pRV566 was first established in E. coli and then successfully transferred by electroporation into our laboratory strain L. sakei 23K. The maintenance of pRV566 in L. sakei indicated that the 3.7-kb KpnI-EcoRI fragment contained a functional plasmid replicon for this host.
FIG. 5.
pRV566, a shuttle vector able to replicate in E. coli and several lactobacilli. ORFs originating from pRV500 and pRV300 are represented in gray and black, respectively. R-11 and R-22 indicate the position of 11- and 22-bp directed repeats, respectively; ermAM and amp code for erythromycin and ampicillin resistance, respectively.
To test whether the L. sakei replicon harbored by the KpnI-EcoRI fragment could autonomously replicate in E. coli, pRV566 was deleted from the colE1 replicon by using Alw44I and SmaI (Fig. 5). The ligation mixture could easily transform L. sakei 23K to erythromycin resistance, but no E. coli transformants could be obtained. The colE1-deleted plasmid obtained in L. sakei (which we named pRV217) could be reintroduced in L. sakei by electroporation but could not transform E. coli TGI. This indicated that the colE1 replicon was indeed responsible for the maintenance of pRV566 in E. coli and that the L. sakei replicon does not support plasmid replication in E. coli.
Host range of pRV566 in lactobacilli.
In order to investigate the host range of pRV566 among lactobacilli, its transformation efficiency was measured for several strains of L. sakei, L. curvatus, L. plantarum, and L. casei (Table 2). pRV566 DNA, prepared from E. coli or from L. sakei 23K, could be introduced by electroporation into 8 of the 13 strains tested with erythromycin resistance as a selective pressure. At least one strain of each Lactobacillus species mentioned above could be transformed by pRV566. The presence of pRV566 as a replicative plasmid was demonstrated in all transformed strains by Southern blot analysis of isolated plasmid DNA with pRV566 as a probe (not shown).
TABLE 2.
Transformation efficiency and stability of pRV566 in lactobacilli
| Recipient strain | No. of endogenous plasmids | Transformability with pRV566 with DNA prepared froma:
|
Stability of pRV566 with the indicated no. of generations without selective pressureb
|
|||||
|---|---|---|---|---|---|---|---|---|
| E. coli TG1 | L. sakei 23K | 20 | 40 | 60 | 80 | 100 | ||
| L. sakei ATCC15521 | ≥2c | 3 × 101 | 2 × 101 | 100 | 95 | 100 | 90 | 95 |
| L. sakei G3AF | 1c,d | 0 (8 × 105)h | 0 | |||||
| L. sakei 160K | 0d | 1.8 × 104 | 2.4 × 104 | 75 | 60 | 35 | 20 | 10 |
| L. sakei RV332 | 1c | 4.2 × 105 | 1.2 × 104 | 85 | 70 | 75 | 60 | 65 |
| L. sakei 23K | 0d | 3.1 × 105 | 1.4 × 106 | 95 | 85 | 60 | 40 | 20 |
| L. sakei 64F | 1d,e | 1 × 101 (1.4 × 105)g | 2 × 101 | 90 | 95 | 85 | 70 | 30 |
| L. plantarum ATCC 8014 | >3 | 4 × 101 (3.6 × 104)g | 90 | 90 | 65 | 55 | 30 | |
| L. plantarum ATCC 14917 | 2 | 0 (5.7 × 103)h | ||||||
| L. curvatus ATCC 20019 | 0 | 0 (1.1 × 104)h | ||||||
| L. curvatus 116 | ≥3c | 0 (0)g,h | ||||||
| L. curvatus T402 | >4c | 0 (1.1 × 104)h | ||||||
| L. curvatus T411 | 0 | 1 × 102 | 75 | 90 | 75 | 70 | 20 | |
| L. casei BL23 | 0f | >105 | 90 | 40 | 20 | 15 | 10 | |
Expressed as the number of erythromycin-resistant CFU per μg of DNA after electroporation.
Expressed as the ratio (%) of erythromycin-resistant CFU scored after growth for the indicated number of generations and corrected to the nearest 5%.
At least one of the plasmids strongly hybridized with pRV566.
Berthier et al. (6).
Langella et al. (33).
Gosalbes et al. (23).
For comparison, the transformation efficiency obtained with replicative plasmid pGhost4 is indicated in parentheses.
For comparison, the transformation efficiency obtained with replicative plasmid pIL253 is indicated in parentheses.
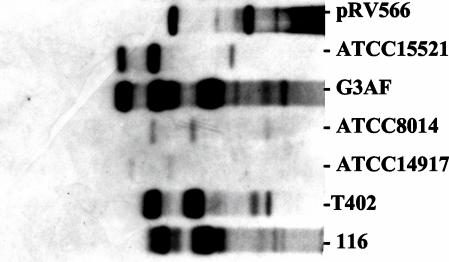
To understand why pRV566 could not be established or could be only poorly established in some strains, we examined the transformability of these strains with other plasmids for erythromycin resistance. All of these strains but one (Table 2) could be readily transformed by another plasmid. This indicated a specific low establishment or lack of establishment for pRV566 itself and not a low efficiency of the electroporation and selection procedure for these particular strains. We also examined the possible interference of resident plasmids with transformation efficiency of pRV566. The initial plasmid content of all the 13 strains was checked by agarose gel electrophoresis and hybridization to detect sequence similarity between resident plasmids and the incoming pRV566. Native plasmids giving strong positive signal with pRV566 were detected in four strains (L. sakei and closely related L. curvatus strains) (Fig. 6) in addition to L. sakei RV332, from which pRV500 was isolated. Three of these strains did not give transformants with pRV566 (Table 2). After the introduction of pRV566, no change in the resident plasmid profile was observed in transformable strains. However, the native pRV500 was lost in L. sakei RV332 after transformation by pRV566 (not shown). For all other transformed strains, no change in the initial resident plasmid profile was observed.
FIG. 6.
Hybridization pattern of plasmids extracted from lactobacillus strains poorly transformed or not transformed by pRV566. pRV566 was used as the probe. An equivalent amount (10 ng) of plasmid DNA extracted from various strains was run on the gel.
Copy number and segregational stability of pRV566.
Extraction of plasmid DNA of pRV566 or pRV500 from L. sakei yielded small quantities of DNA, suggesting that the copy number of the plasmids was low in that host. We determined the copy number of pRV500 in its native host RV332 and that of pRV566 after transformation in L. sakei strains 23K and RV332 (then containing only pRV566). By comparison of hybridization signals of plasmid target in known amounts of total DNA and of purified plasmid, we deduced that pRV566 and pRV500 are very-low-copy-number plasmids in L. sakei. They are at about the same copy number in the tested strains, which we estimate at around one copy per cell (not shown).
The erythromycin-resistant phenotype of transformed strains was used to study the stability of the plasmid in these hosts. Bacteria were cultivated without antibiotic selection pressure, and isolated colonies were scored for erythromycin resistance. Loss of erythromycin resistance, considered to indicate plasmid loss, varied with the strains (Table 2). pRV566 was maintained at a reasonable rate of 50% after 40 generations in all of the strains tested except for L. casei BL23.
DISCUSSION
pRV500 is, to our knowledge, the first plasmid of L. sakei to be entirely sequenced. Careful sequence analysis showed that pRV500 belongs to the pUCL287 family, itself related to class A theta-type plasmids of the pUCL22 and pSC101 families. Experimental evidence of the theta replication mode was obtained for pUCL287 itself (2). Although the minimum length of the pRV500 oriA region remains to be determined, it is likely to involve the same two kinds of DNA repeats as other pUCL287-type plasmids. The involvement of repetitive sequences of 22 bp (iterons) as a target for binding of the Rep protein was demonstrated for pSBO1 (40), and the upstream 11-bp repeated sequences were observed to be essential for pUCL287 replication (3).
Like other plasmids (14, 49), pRV500 appears to be a composite structure containing DNA segments from different sources. Some ORFs are shared by plasmids from both subfamilies pUCL22 and pUCL287; this applies to orf6 in pRV500 and ORFs of unknown function around the gene for a site-specific recombinase (orf10). Whether this recombinase is involved in mobility of adjacent regions is not known. The region also showed interspersed nucleotide conservation with pLH1, belonging to the pLS32 family of theta plasmids (49), and with pLP9000 (unpublished), whose family is unknown but in which we could not detect an ORF sharing similarity with Rep proteins of plasmids pUCL287, pUCL22, or pLH1. Short DNA sequences with high sequence conservation (>90% nucleotide identity) among the different plasmids were noticed. Notably, palindromic sequences, representing putative transcriptional terminators, were found at an edge of a region of putative DNA exchange. Those highly conserved regions, though small (40 bp), could serve as sites for facilitating DNA exchanges by recombination. A number of stem-loop structures identified in pLH1 were found to be conserved among different plasmids from LAB and/or at the boundary of regions of homology (49). Altogether, plasmids showing these sequence conservations can replicate in LAB. Paradoxically, although R-M systems are often carried by lactococcal plasmids of the pUCL22 family (8), the R-M gene cluster of pRV500 was found to be related to that of genomes of gram-negative bacteria by database similarity search. An entire R-M system of type I appears to be carried on pRV500 and, although we did not test it, is likely to be functional. Integrase/recombinase genes have already been observed to be associated with R-M systems. They might be involved in their mobility (38) or contribute to R-M polymorphism in conjunction with rearrangements in specificity genes (48). The structure of the pRV500 R-M cluster, with an additional truncated gene for a specificity subunit (hsdS′) and a nearby Int/rec gene (orf2), could be typical of a system able to generate polymorphism in the specificity subunits of restriction enzymes.
pRV500 and its pRV566 derivative were present at the same copy number in L. sakei, namely, about one copy per cell, indicating that elements involved in the apparently tight control of copy number are present on both plasmids. The DNA segment allowing replication encompasses orf9, orf8, oriA, repA, and orf6. It does not include orf11, whose product was found to be similar to Orf4 of plasmid pSRQ800, itself with similarity to regulatory proteins and plasmid copy number control proteins (8). Thus, orf11 does not appear to be necessary for the control of copy number of pRV500 in L. sakei. Another candidate could be Orf6, which has homologues in different plasmids of LAB (Fig. 4). A possible role in the control of plasmid copy number has been found for the related gene product (RepB) of pUCL287 (3). However, it was not confirmed for related proteins in other plasmids (18). Alternatively, copy number control could be simply exerted though handcuffing of the initiator protein at iterons (13). Low-copy-number plasmids often carry an active partition system which allows the plasmid to be correctly distributed in dividing cells through the generations and thus contribute to the segregational stability (for a review, see reference 22). We were not able to detect such a system in pRV500 by sequence analysis. Other components are known to participate in the maintenance of plasmids in their hosts, such as site-specific recombinases, R-M systems, and addiction modules. Addiction modules consist of two genes, with one coding for a stable toxin and the other coding for a labile antitoxin (for a review, see reference 19). They mediate programmed cell death in bacteria, and when borne by plasmids, they are responsible for postsegregational killing of plasmid-free segregant bacteria: an example of such a module is the kis/kid of plasmid R1. R-M systems can also behave as addiction modules: type II R-M complexes were shown to be responsible for postsegregational killing through chromosome cleavage and thereby to increase stability of plasmids encoding them (32, 39). XerC-like Int/rec recruited by ColE1 plasmids have been reported to increase segregation efficiency by multimer resolution (41). A resolvase (serine recombinase) is involved in multimer resolution in plasmids of the pAMβ1 family (43), a homologue of which was found on plasmid pLH1 of the pLS32 family (49) in addition to the Int/rec homologous to Orf10 of pRV500 mentioned in Table 1. In pRV500, we could identify by sequence analysis a type I R-M system and two recombinases. Although pRV566 is rather stable, it is progressively lost without selection pressure in some strains and appears less stable than its pRV500 parent, which could not be cured from its host strain by classical methods. None of the candidate genes for increasing segregational stability that are carried by pRV500 are present on pRV566. For practical applications, the stability of pRV566 is satisfactory, as growth in laboratory conditions for physiological studies is usually performed on a few generations. L. sakei strains used as starters for sausage are inoculated at high cell density and grow for only about 12 generations during the fermentation process.
Thus, pRV500 carries a replicon potentially useful for different applications in lactobacilli, and further developments may be considered: as a replicative plasmid, it could be useful for cloning large clusters of genes in various lactobacillus hosts and particularly in L. casei, in which the plasmid established efficiently. To our knowledge, no conditionally replicative vectors usable in L. sakei or other mesophilic lactobacilli are known. We hope to improve the procedure of gene replacement in L. sakei if such a derivative plasmid based on the pRV500 replicon could be obtained.
Acknowledgments
The contributions of Anne-Marie Dudez for running the sequencer and of Stéphane Chaillou for help with sequence analysis are gratefully acknowledged. Laurent Jannière is kindly acknowledged for helpful discussions and critical reading of the manuscript.
C.-A. A. received a grant as chercheur étranger from the Institut National de la Recherche Agronomique.
REFERENCES
- 1.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benachour, A., J. Frere, P. Boutibonnes, and Y. Auffray. 1995. Characterization and replication mode determination of the minimal replicon of Tetragenococcus halophila ATCC33315 plasmid pUCL287. Biochimie 77:868-874. [DOI] [PubMed] [Google Scholar]
- 3.Benachour, A., J. Frere, S. Flahaut, G. Novel, and Y. Auffray. 1997. Molecular analysis of the replication region of the theta-replicating plasmid pUCL287 from Tetragenococcus (Pediococcus) halophilus ATCC33315. Mol. Gen. Genet. 255:504-513. [DOI] [PubMed] [Google Scholar]
- 4.Benachour, A., J. Frere, and G. Novel. 1995. pUCL287 plasmid from Tetragenococcus halophila (Pediococcus halophilus) ATCC 33315 represents a new theta-type replicon family of lactic acid bacteria. FEMS Microbiol. Lett. 128:167-175. [DOI] [PubMed] [Google Scholar]
- 5.Berthier, F., and S. D. Ehrlich. 1999. Genetic diversity within Lactobacillus sakei and Lactobacillus curvatus and design of PCR primers for its detection using randomly amplified polymorphic DNA. Int. J. Syst. Bacteriol. 49:997-1007. [DOI] [PubMed] [Google Scholar]
- 6.Berthier, F., M. Zagorec, M. Champomier-Vergès, S. D. Ehrlich, and F. Morel-Deville. 1996. High-frequency transformation of Lactobacillus sake by electroporation. Microbiology 142:1273-1279. [DOI] [PubMed]
- 7.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boucher, I., E. Emond, M. Parrot, and S. Moineau. 2001. DNA sequence analysis of three Lactococcus lactis plasmids encoding phage resistance mechanisms. J. Dairy Sci. 84:1610-1620. [DOI] [PubMed] [Google Scholar]
- 9.Bringel, F., L. Frey, and J. C. Hubert. 1989. Characterization, cloning, curing, and distribution in lactic acid bacteria of pLP1, a plasmid from Lactobacillus plantarum CCM 1904 and its use in shuttle vector construction. Plasmid 22:193-202. [DOI] [PubMed] [Google Scholar]
- 10.Bruand, C., E. Le Chatelier, S. D. Ehrlich, and L. Janniere. 1993. A fourth class of theta-replicating plasmids: the pAMβ1 family from gram-positive bacteria. Proc. Natl. Acad. Sci. USA 90:11668-11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckenhüskes, H. J. 1993. Selection criteria for lactic acid bacteria to be used as starter cultures for various food commodities. FEMS Microbiol. Rev. 12:253-272. [Google Scholar]
- 12.Champomier, M. C., M. C. Montel, F. Grimont, and P. A. Grimont. 1987. Genomic identification of meat Lactobacilli as Lactobacillus sake. Ann. Inst. Pasteur Microbiol. 138:751-758. [DOI] [PubMed] [Google Scholar]
- 13.Chattoraj, D. K. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37:467-476. [DOI] [PubMed] [Google Scholar]
- 14.Danielsen, M. 2002. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98-103. [DOI] [PubMed] [Google Scholar]
- 15.Del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 17.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of Escherichia coli by high-voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Émond, E., R. Lavallée, G. Drolet, S. Moineau, and G. LaPointe. 2001. Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Appl. Environ. Microbiol. 67:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 20.Frere, J., M. Novel, and G. Novel. 1993. Molecular analysis of the Lactococcus lactis subspecies lactis CNRZ270 bidirectional theta replicating lactose plasmid pUCL22. Mol. Microbiol. 10:1113-1124. [DOI] [PubMed] [Google Scholar]
- 21.Gardner, M. N., S. M. Deane, and D. E. Rawlings. 2001. Isolation of a new broad-host-range IncQ-like plasmid, pTC-F14, from the acidophilic bacterium Acidithiobacillus caldus and analysis of the plasmid replicon. J. Bacteriol. 183:3303-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681-708. [DOI] [PubMed] [Google Scholar]
- 23.Gosalbes, M. J., V. Monedero, and G. Perez-Martinez. 1999. Elements involved in catabolite repression and substrate induction of the lactose operon in Lactobacillus casei. J. Bacteriol. 181:3928-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gravesen, A., A. vonWright, J. Josephsen, and F. K. Vogensen. 1997. Replication regions of two pairs of incompatible lactococcal theta-replicating plasmids. Plasmid 38:115-127. [DOI] [PubMed] [Google Scholar]
- 25.Gruss, A., and S. D. Ehrlich. 1989. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol. Rev. 53:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammes, W. P., and C. Hertel. 1998. New developments in meat starter cultures. Meat Sci. 49:125-138. [PubMed] [Google Scholar]
- 27.Kearney, K., G. F. Fitzgerald, and J. F. Seegers. 2000. Identification and characterization of an active plasmid partition mechanism for the novel Lactococcus lactis plasmid pCI2000. J. Bacteriol. 182:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiewiet, R., S. Bron, K. de Jonge, G. Venema, and J. F. Seegers. 1993. Theta replication of the lactococcal plasmid pWVO2. Mol. Microbiol. 10:319-327. [PubMed] [Google Scholar]
- 29.Kiewiet, R., J. Kok, J. F. Seegers, G. Venema, and S. Bron. 1993. The mode of replication is a major factor in segregational plasmid instability in Lactococcus lactis. Appl. Environ. Microbiol. 59:358-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klaenhammer, T., E. Altermann, F. Arigoni, A. Bolotin, F. Breidt, J. Broadbent, R. Cano, S. Chaillou, J. Deutscher, M. Gasson, M. van de Guchte, J. Guzzo, A. Hartke, T. Hawkins, P. Hols, R. Hutkins, M. Kleerebezem, J. Kok, O. Kuipers, M. Lubbers, E. Maguin, L. McKay, D. Mills, A. Nauta, R. Overbeek, H. Pel, D. Pridmore, M. Saier, D. van Sinderen, A. Sorokin, J. Steele, D. O'Sullivan, W. de Vos, B. Weimer, M. Zagorec, and R. Siezen. 2002. Discovering lactic acid bacteria by genomics. Antonie Van Leeuwenhoek 82:29-58. [DOI] [PubMed] [Google Scholar]
- 31.Klein, J. R., C. Ulrich, and R. Plapp. 1993. Characterization and sequence analysis of a small cryptic plasmid from Lactobacillus curvatus LTH683 and its use for construction of new Lactobacillus cloning vectors. Plasmid 30:14-29. [DOI] [PubMed] [Google Scholar]
- 32.Kulakauskas, S., A. Lubys, and S. D. Ehrlich. 1995. DNA restriction-modification systems mediate plasmid maintenance. J. Bacteriol. 177:3451-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langella, P., M. Zagorec, S. D. Ehrlich, and F. Morel-Deville. 1996. Intergenic and intragenic conjugal transfer of plasmids pAMβ1, pIL205 and pIP501 in Lactobacillus sake. FEMS Microbiol. Lett. 139:51-56. [Google Scholar]
- 34.Leloup, L., S. D. Ehrlich, M. Zagorec, and F. Morel-Deville. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malleret, C., R. Lauret, S. D. Ehrlich, F. Morel-Deville, and M. Zagorec. 1998. Disruption of the sole ldhL gene in Lactobacillus sakei prevents the production of both L- and D-lactate. Microbiology 144:3327-3333. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Bueno, M., E. Valdivia, A. Galvez, and M. Maqueda. 2000. pS86, a new theta-replicating plasmid from Enterococcus faecalis. Curr. Microbiol. 41:257-261. [DOI] [PubMed] [Google Scholar]
- 37.Meijer, W. J., A. J. de Boer, S. van Tongeren, G. Venema, and S. Bron. 1995. Characterization of the replication region of the Bacillus subtilis plasmid pLS20: a novel type of replicon. Nucleic Acids Res. 23:3214-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naderer, M., J. R. Brust, D. Knowle, and R. M. Blumenthal. 2002. Mobility of a restriction-modification system revealed by its genetic contexts in three hosts. J. Bacteriol. 184:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naito, T., K. Kusano, and I. Kobayashi. 1995. Selfish behavior of restriction-modification systems. Science 267:897-899. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura, M., K. Ogata, T. Nagamine, K. Tajima, H. Matsui, and Y. Benno. 2001. The replicon of the cryptic plasmid pSBO1 isolated from Streptococcus bovis JB1. Curr. Microbiol. 43:11-16. [DOI] [PubMed] [Google Scholar]
- 41.Nash, H. A. 1996. Site-specific recombination: integration, excision, resolution and inversion of defined DNA segments, p. 2363-2376. In Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 42.Nunes-Duby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pujol, C., S. D. Ehrlich, and L. Janniere. 1994. The promiscuous plasmids pIP501 and pAMβ1 from gram-positive bacteria encode complementary resolution functions. Plasmid 31:100-105. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Seegers, J. F., S. Bron, C. M. Franke, G. Venema, and R. Kiewiet. 1994. The majority of lactococcal plasmids carry a highly related replicon. Microbiology 140:1291-1300. [DOI] [PubMed] [Google Scholar]
- 46.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 47.Smith, M. C., and H. M. Thorpe. 2002. Diversity in the serine recombinases. Mol. Microbiol. 44:299-307. [DOI] [PubMed] [Google Scholar]
- 48.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, J. K., S. Foley, K. J. McConville, C. Nicholson, M. A. Collins, and R. D. Pridmore. 1999. Complete sequence of plasmid pLH1 from Lactobacillus helveticus ATCC15009: analysis reveals the presence of regions homologous to other native plasmids from the host strain. Plasmid 42:221-235. [DOI] [PubMed] [Google Scholar]
- 50.Wang, T. T., and B. H. Lee. 1997. Plasmids in Lactobacillus. Crit. Rev. Biotechnol. 17:227-272. [DOI] [PubMed] [Google Scholar]