Abstract
The gene encoding a novel α-l-arabinofuranosidase from Bifidobacterium longum B667, abfB, was cloned and sequenced. The deduced protein had a molecular mass of about 61 kDa, and analysis of its amino acid sequence revealed significant homology and conservation of different catalytic residues with α-l-arabinofuranosidases belonging to family 51 of the glycoside hydrolases. Regions flanking the gene comprised two divergently transcribed open reading frames coding for hypothetical proteins involved in sugar metabolism. A histidine tag was introduced at the C terminus of AbfB, and the recombinant protein was overexpressed in Lactococcus lactis under control of the tightly regulated, nisin-inducible nisA promoter. The enzyme was purified by nickel affinity chromatography. The molecular mass of the native protein, as determined by gel filtration, was about 260 kDa, suggesting a homotetrameric structure. AbfB was active at a broad pH range (pH 4.5 to 7.5) and at a broad temperature range (20 to 70°C), and it had an optimum pH of 6.0 and an optimum temperature of 45°C. The enzyme seemed to be less thermostable than most previously described arabinofuranosidases and had a half-life of about 3 h at 55°C. Chelating and reducing agents did not have any effect on its activity, but the presence of Cu2+, Hg2+, and Zn2+ markedly reduced enzymatic activity. The protein exhibited a high level of activity with p-nitrophenyl α-l-arabinofuranoside, with apparent Km and Vmax values of 0.295 mM and 417 U/mg, respectively. AbfB released l-arabinose from arabinan, arabinoxylan, arabinobiose, arabinotriose, arabinotetraose, and arabinopentaose. No endoarabinanase activity was detected. These findings suggest that AbfB is an exo-acting enzyme and may play a role, together with other glycosidases, in the degradation of l-arabinose-containing polysaccharides.
Arabinofuranosidases are hemicellulose-degrading enzymes that cleave l-arabinofuranosyl residues from different oligosaccharides and polysaccharides. These enzymes are part of a set of glycosidases required for complete degradation of polymeric substrates such as arabinan, arabinoxylan, and other polysaccharides, which are major components of plant cell walls (32, 39). Such hydrolases are endo- and exoenzymes that cleave the glycosidic linkages of the polymeric backbone and the various side chains, providing many microorganisms, both prokaryotic and eukaryotic, with soluble saccharides that can be used as carbon or energy sources (29, 32). Arabinans consist of a backbone of α-1,5-linked l-arabinofuranosyl residues branched with α-1,2- and α-1,3-linked side chains of l-arabinose in the furanose conformation (3). In arabinoxylans, the β-1,4-xylose backbone is mainly branched with α-l-arabinofuranose residues attached to the O-3 position or, occasionally, to both the O-2 and O-3 positions (30). The α-l-arabinofuranosidases (α-l-arabinofuranoside arabinofurane hydrolase; EC 3.2.1.55) are exo-type enzymes that hydrolyze terminal nonreducing α-l-arabinofuranosyl groups from l-arabinose-containing polysaccharides. They cooperate with endo-1,5-α-l-arabinanases (EC 3.2.1.99), which act by carrying out endohydrolysis of 1,5-α-l-arabinofuranosidic linkages in α-1,5-arabinans, resulting in their complete degradation.
Arabinofuranosidases have received a great deal of attention in recent years because of their potential applications in agroindustrial processes, such as the processing of fruits, vegetables, and cereals (45) and the conversion of hemicellulose to fuels and chemicals (1). These enzymes are particularly important in the genus Bifidobacterium, where they play a crucial physiological role. Bifidobacteria are anaerobic microorganisms that are generally considered to be probiotics. Some Bifidobacterium strains can colonize the gastrointestinal tract and are important components of the human intestinal microbiota, in which they occur at concentrations of 109 to 1010 cells/g of feces (41). Their presence has been associated with beneficial health effects, such as diarrhea and colon cancer prevention, immunomodulation, normalization of gut mucosal dysfunction, down-regulation of hypersensitivity reactions, etc. (15). Some of the main substrates for bacterial growth in the colon are dietary carbohydrates that are not digested in the upper gastrointestinal tract. These carbohydrates are assimilated preferentially by bifidobacteria, which promotes proliferation of the organisms in the gut. The ability of these microorganisms to metabolize nondigestible carbohydrates is dependent on their glycosidic activities and gives them a selective advantage over other bacteria in the intestinal ecosystem. The recent description of the genome sequence of Bifidobacterium longum (35) revealed an unexpectedly large number of predicted proteins (more than 8%) that seem to be related to the catabolism of polysaccharides and oligosaccharides released from nondigestible plant polymers. Among these, many different proteins displayed high levels of sequence homology with proteins involved in the metabolism of arabinose-containing sugars, such as intra- and extracellular α-l-arabinofuranosidases, endo-α-1-5 arabinanases, oligosaccharide transporters, transcriptional regulators, etc.
There has been considerable research dealing with the study of fermentation of nondigestible sugars by bifidobacteria. However, few reports have focused on the glycosidases involved in fermentation of arabinose-containing sugars, and no information concerning the functional characteristics of arabinofuranosidases with known sequences is available yet. In this paper we describe identification of a protein-encoding gene and overexpression, purification, and characterization of a novel α-l-arabinofuranosidase from B. longum B667.
MATERIALS AND METHODS
Substrates and chemicals.
DNase, l-arabinose, EDTA, dithiothreitol (DTT), β-mercaptoethanol, p-nitrophenyl (pNP) glycoside substrates, and standard proteins for gel filtration were purchased from Sigma Chemical Co. (St. Louis, Mo.). Sugar beet arabinan, wheat flour arabinoxylan, red debranched arabinan (RDA), arabinobiose, arabinotriose, arabinotetraose, and arabinopentaose were obtained from Megazyme (Bray, Wicklow, Ireland). NAD, β-galactose dehydrogenase, Taq DNA polymerase, and chloramphenicol were obtained from Roche Applied Science (Mannheim, Germany). Amersham Biosciences (Uppsala, Sweden) supplied all of the restriction enzymes. The Elongase amplification system and T4 DNA ligase were obtained from Invitrogen Co. (Carlsbad, Calif.), and Ni2+ nitriloacetic acid (Ni-NTA) agarose was supplied by Qiagen Inc. (Valencia, Calif.). Metal salts were purchased from Panreac Química, S.A. (Barcelona, Spain). All chemicals were reagent grade, and all solutions were made with distilled and deionized water.
Bacterial strains and culture conditions.
Escherichia coli CS1562 (2) was grown at 30°C in Luria-Bertani medium containing 0.5% glucose and supplemented with ampicillin when required. Vector pUC19 (48) was used to construct a genomic library of B. longum B667. Lactococcus lactis subsp. lactis NZ9000 and NZ9700 and B. longum B667 were obtained from NIZO Food Research, Ede, The Netherlands. B. longum was grown at 37°C under anaerobic conditions in MRS medium (Merck KGaA, Darmstadt, Germany) supplemented with 0.1% (wt/vol) l-cysteine (Merck). Lactococcus cells were cultivated at 30°C in M-17 medium (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) with 0.5% (wt/vol) glucose and with 5 μg of chloramphenicol per ml when they contained the pNZ8048 vector (19) or a derivative of this plasmid.
DNA manipulation, cloning of the abfB gene, and sequence analysis.
DNA manipulations were carried out as described by Sambrook et al. (33). Total DNA was obtained from B. longum B667 as described by Tanaka and coworkers (40). A genomic library was constructed in E. coli CS1562 by partial digestion of the total DNA of B. longum with Sau3AI. DNA fragments that were between 3 and 8 kbp long were cloned into the pUC19 vector digested with BamHI, as previously reported (4). Sequencing of a fragment that was about 3.1 kbp long revealed the presence of a region coding for a putative α-l-arabinofuranosidase designated AbfB. The structural gene was subsequently amplified by PCR by using two primers in a way that enabled in-frame cloning of the structural gene into the pNZ8048 expression vector. Given that the abfB gene contains an internal NcoI site, the enzyme RcaI, which yields compatible ends with NcoI, was used for cloning. Therefore, the gene was amplified by using an N-terminal primer (5′-TCTAGACCATCATGACCACTCACAACAGC-3′) that included an ATG translational start codon inside the RcaI restriction site (TCATGA) and a C-terminal primer (5′-GCCGACTCTAGATTATTAGCCGTGGAATTC-3′) containing an XbaI restriction site (TCTAGA) after the end of the gene. The PCR product was purified on an agarose gel, digested with RcaI and XbaI, and ligated with the NcoI/XbaI-digested pNZ8048 vector, resulting in pNAbfB. This plasmid was transformed into L. lactis NZ9000 electrocompetent cells by using the methods described previously (10), and transformants were screened by restriction analysis of the plasmid. Correct cloning was verified by DNA sequencing.
Two complementary primers were constructed for introduction of the six-histidine tag at the C terminus of the protein. Annealing of the forward (5′-AATTCCACGGCCATCACCATCACCATCACTAATAAA-3′) and re-verse (5′-AGCTTTTATTAGTGATGGTGATGGTGATGGCCGTGG-3′)primers generated a double-stranded DNA linker with an EcoRI end at the N terminus and a HindIII end at the C terminus, which included the coding sequence for the histidine tag. This linker was ligated with plasmid pNAbfB, previously linearized by treatment with EcoRI and HindIII. The resulting plasmid, pNHAbfB, was transformed into L. lactis NZ9000, and the mutated gene was sequenced to confirm that only the desired changes had been produced.
Inverse PCRs were carried out to amplify the regions flanking the abfb gene. Chromosomal DNA was digested by using a variety of restriction enzymes. The enzyme reaction mixtures were inactivated by incubation at 65°C for 20 min. Subsequent ligation mixtures (final volume, 20 μl) containing 0.5 μg of restricted DNA and 1 U of T4 DNA ligase were incubated for 3 h at 23°C and heat inactivated at 65°C for 20 min. The self-ligated DNA was precipitated with ammonium acetate and ethanol and resuspended in sterile distilled water to a final concentration of approximately 10 ng/μl. Inverse PCRs were performed with a Hybaid PCR Sprint thermocycler (Hybaid Ltd., Middlesex, United Kingdom) in 50-μl (final volume) mixtures by using the Elongase amplification system (Invitrogen) according to the manufacturer's instructions. The reaction mixtures contained 1 μl of self-ligated DNA, each deoxynucleoside triphosphate at a concentration of 200 μM, each primer at a concentration of 500 μM, 1 U of elongase, and 1.5 mM MgCl2 in 1× PCR buffer. The program used was as follows: 4-min hold at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 55 to 60°C (depending on the primer melting temperature), and 8 min at 68°C. The following four pairs of primers were subsequently used: (i) forward primer 5′-CAACAAGGGCGTCTTCGACAAG-3′ and reverse primer 5′-GGTTTCTGGCCGACGAGTTGAC-3′; (ii) forward primer 5′-CGACAACGACACCCTCAAGTCCAACG-3′ and reverse primer 5′-CCTTGAAGGAGGCGACCACGTGAGC-3′; (iii) forward primer 5′-GACTTGACCAGCTTGGTGCTCAGC-3′ and reverse primer 5′-CTGAACCATAGAGTGATGGTTCACG-3′; and (iv) forward primer 5′-GTTGATCGGGCGCCAATTGACG-3′ and reverse primer 5′-GACGCCGAGCAGATCAAGACCTGG-3′. DNA visualization in agarose gels after electrophoresis was carried out by using a Gel Doc 2000 system (Bio-Rad, Hercules, Calif.). The PCR products were sequenced by the Servicio de Secuenciación de DNA (Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas) with an ABI Prism 377 sequencer (Applied Biosystems, Foster City, Calif.). DNA and protein sequences were analyzed by using the computer program Clone Manager 5 (Scientific and Educational Software, Durham, N.C.). Homology searches with sequences in the GenBank/EMBL databases were carried out by using the BLAST server of the National Center for Biotechnology Information at the National Institutes of Health (Bethesda, Md.) (http://www.ncbi.nlm.nih.gov). Multiple alignments were constructed with CLUSTAL X software (42).
Overexpression and purification of the enzyme.
Transformed cells of L. lactis NZ9000 (19) were grown to an optical density at 600 nm of about 0.6. In order to trigger transcription of the abfB gene from the nisA promoter, 0.2% (vol/vol) supernatant from a culture of the nisin-producing strain L. lactis NZ9700 (18), containing approximately 10 ng of nisin A/ml, was added to this culture. Then the cells were incubated for 90 min at 30°C (which resulted in an optical density at 600 nm of about 1.2), harvested by centrifugation, washed with 100 mM potassium phosphate (pH 7.0), and resuspended in the same buffer. The cells were treated with 10 mg of lysozyme per ml for 30 min at 30°C and disrupted by passage through a French pressure cell (20,000 lb/in2). The cell suspension was then incubated for 20 min at 30°C with DNase (100 μg/ml) and 10 mM MgSO4. Unbroken cells and cell debris were removed by centrifugation at 40,000 × g for 30 min at 4°C.
Purification of the histidine-tagged AbfB was carried out at 4°C. Cell extract was mixed for 1 h at 4°C with Ni-NTA agarose (approximately 10 mg of AbfB/ml of resin) that had been preequilibrated in buffer A (50 mM potassium phosphate buffer [pH 8.0], 100 mM NaCl, 10% [vol/vol] glycerol) containing 10 mM imidazole. After incubation, the resin was transferred to a Bio-Spin column (Bio-Rad) and washed first with 20 column volumes of buffer A containing 10 mM imidazole and then with 40 column volumes of buffer A (pH 7.0) containing 30 mM imidazole. The protein was eluted with buffer A (pH 7.0) containing 160 mM imidazole and dialyzed against 1,000 volumes of buffer A (pH 7.0) for 16 h. Neither loss of activity nor proteolysis was detected during 1 month of storage of the purified enzyme preparation at 4°C. Proteins from cell extracts and from the eluted fractions were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by using a Mini-Protean II system (Bio-Rad). SDS-PAGE gels were stained with Coomassie brilliant blue, and densitometric scanning was carried out by using the Gel Doc 2000 system with the Quantity One software (Bio-Rad). Protein concentrations were determined by using the bicinchoninic acid method (Pierce, Rockford, Ill.) and following the manufacturer's instructions.
Determination of the molecular mass.
The apparent molecular mass of the native enzyme was determined by gel filtration on a Superdex 75 HR10/30 column (Amersham) with carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), β-amylase (200 kDa), and apoferritin (443 kDa) as the standard proteins.
Determination of enzymatic activities.
Unless specified otherwise, the activity of AbfB was assayed at 45°C in 0.5 ml (total volume) of 50 mM potassium phosphate buffer (pH 6.0). α-l-Arabinofuranosidase activity was determined by using 0.25 mM pNP α-l-arabinofuranoside (α-l-Abf) as the substrate and the appropriate diluted enzyme solutions. After incubation of the reaction mixture for 10 min, 0.5 ml of cold 1 M Na2CO3 was added to stop the reaction. The number of micromoles of pNP released was calculated based on the relationship of the A420 to a standard curve. One unit of activity was defined as the amount of enzyme that released 1 μmol of pNP per min. The activity of the enzyme with other pNP glycosides was assayed as previously described.
The ability of AbfB to release arabinose from a solution generally containing either arabinan or arabinoxylan at a concentration of 0.5% was tested by using the appropriate diluted enzyme solutions. After incubation for 30 min at 45°C, reactions were stopped by cooling the reaction mixtures on ice. The amount of arabinose released was determined by quantifying the reducing sugar equivalents formed by the β-galactose dehydrogenase-NAD method (22); l-arabinose was used as a standard. One unit of activity was defined as the amount of enzyme that produced 1 μmol of arabinose-reducing sugar equivalents per min. The endoarabinanase activity of AbfB was tested by using RDA as a substrate and following the manufacturer's instructions. After incubation, unhydrolyzed substrate was precipitated by addition of 4 volumes of ethanol, and the absorbance of the supernatant was measured at 520 nm.
Effects of pH and temperature on AbfB activity and stability.
The optimum pH for activity α-l-arabinofuranosidase was determined by incubation at 37°C for 10 min in the pH range from 3.5 to 8.5. Intervals of 0.25 pH unit were used between pH 5.5 and 7.0 whereas 0.5-pH unit intervals were used for the remaining pH range. The following buffers were used: 50 mM sodium acetate, pH 4.0 to 5.5; 50 mM potassium phosphate, pH 5.5 to 7.5; and 25 mM Tris-HCl, pH 7.5 to 8.5. To rule out the possibility that the buffer composition had any influence on enzymatic activity, experiments were also carried out in McIlvaine buffer (100 mM citric acid, 200 mM disodium phosphate) in the pH range from 3.5 to 8.5. The optimum temperature for enzymatic activity was determined in 50 mM potassium phosphate buffer by using temperatures ranging from 10 and 90°C at the optimum pH. The results were expressed as percentages of the activity obtained at either the optimum pH or the optimum temperature.
The thermostability of the purified enzyme was determined in microcentrifuge tubes (0.5 ml) containing 0.1 U of AbfB in 50 mM potassium phosphate buffer (pH 6.0). Samples were incubated at 40, 50, 55, 60, 65, and 70°C, and the tubes were removed at different times, maintained at room temperature for 30 min, and then kept at 4°C for another 30 min. To determine the effect of pH on the stability of AbfB, the enzyme solution was kept for 5 h at the optimum temperature in McIlvaine buffer at pH 3.5, 4.5, 5.5, 6.5, 7.5, and 8.5. The residual α-l-arabinofuranosidase activity was determined after each treatment.
Effects of chemical agents and metal cations on the enzyme activity.
The effects of several metals and chemical agents on the α-l-arabinofuranosidase activity of the purified enzyme were determined. HgCl2, CuSO4, ZnSO4, MgCl2, MnCl2, CaCl2, CoCl2, and NaCl were assayed at concentrations of 0.1 and 1 mM in the reaction mixture. The chelating agent EDTA and the reducing agents DTT and β-mercaptoethanol were assayed at concentrations of 1 and 10 mM. Activity was determined as described above and was expressed as a percentage of the activity obtained in the absence of the compound.
Substrate specificity and kinetic studies.
The substrate specificity of the enzyme was tested by using the following pNP glycosides: pNP N-acetyl-β-d-glucosaminide, pNP β-d-fucopyranoside, pNP α-d-galactopyranoside, pNP β-d-galactopyranoside, pNP α-d-glucopyranoside, pNP β-d-glucopyranoside, pNP α-l-rhamnopyranoside, pNP β-d-xylopyranoside, pNP α-d-xylopyranoside, pNP α-l-arabinopyranoside, pNP β-l-arabinopyranoside, and pNP α-l-Abf. Kinetic parameters of the purified enzyme were estimated in triplicate for pNP α-l-Abf, arabinan, and arabinoxylan at early stages of hydrolysis. The concentrations in the reaction mixtures ranged from 0.01 to 4 mM for pNP α-l-Abf, from 0.1 to 40 mg/ml for arabinan, and from 0.1 to 20 mg/ml for arabinoxylan. Michaelis-Menten kinetic parameters were determined by using the Sigma Plot enzyme kinetics module software (SPSS Science, Chicago, Ill.).
Enzymatic hydrolysis of arabinan, arabinoxylan, and arabinooligosaccharides.
Thin-layer chromatography was used to identify the products generated by AbfB from arabinan, arabinoxylan, and arabinooligosaccharides having from two to five arabinose residues. Each reaction mixture (1 ml) containing 0.2 U of the enzyme and a substrate at a concentration of 0.1% (wt/vol) in 50 mM potassium phosphate buffer (pH 6.0) was incubated at 40°C. Samples were withdrawn at regular intervals and heated for 5 min in a boiling water bath to stop the reaction. The reaction mixtures were centrifuged at 11,000 × g for 10 min and filtered through Centricon columns (Millipore, Bedford, Mass.) with a molecular mass cutoff of 10 kDa to eliminate unhydrolyzed substrate. Chromatography was performed by the ascending method on silica gel 60 F254 thin-layer chromatography plates (Merck) with a solvent system consisting of ethyl acetate, acetic acid, and water (2:1:1). Sugars on the plates were detected by heating the plates at 120°C for about 10 min after they were sprayed with 5% (vol/vol) sulfuric acid in ethanol.
Nucleotide sequence accession number.
The nucleotide sequence of the B. longum 667 gene cluster containing the α-l-arabinofuranosidase gene has been deposited in the GenBank database under accession number AY259087.
RESULTS
Identification and sequence analysis of abfB and its flanking regions.
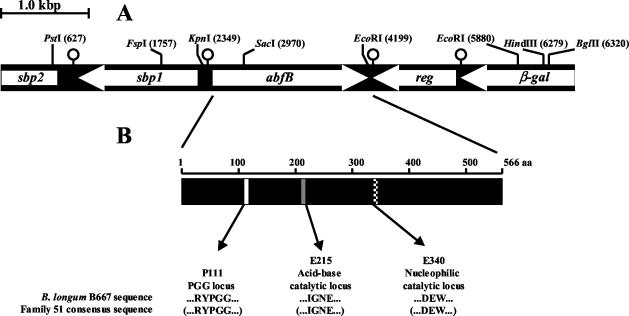
A 3,090-bp DNA fragment was identified during screening for possible glycosidase activities in B. longum (data not shown). Genetic analysis of the cloned fragment (positions 1,981 to 5,070) (Fig. 1) revealed the presence of a 1,698-bp open reading frame (ORF) encoding a hypothetical 566-amino-acid protein with a molecular mass of 61.548 kDa, identified by a database enquiry (BLASTP) as a putative family 51 α-l-arabinofuranosidase. A putative ribosome binding sequence, 5′-AGGAGA, was found 12 bp upstream of the potential ATG initiation codon. No signal sequence was detected with SIGNALP (27), indicating that the protein is not secreted. Downstream of abfB, an incomplete ORF with a high degree of similarity to LacI-type sugar-responsive repressors was found (26). Therefore, several inverse PCRs were performed in order to ascertain whether the abfB gene was included in a cluster coding for oligosaccharide degradation, a common feature in B. longum (35). Amplicons were obtained from ligation of restriction products that originated from PstI and BglII digestion of chromosomal DNA, which allowed sequencing of a 6,595-bp fragment. Two ORFs upstream of the abfB gene (encoding two sugar-binding proteins, one of which was incomplete) and two ORFs downstream of the abfB gene (encoding a Lac-I regulator and an incomplete β-galactosidase) were found. Possible transcriptional terminators corresponding to mRNA hairpin loops were found downstream of the termination codons of all predicted ORFs (Fig. 1A). The complete sequenced fragment was 99.41% identical to a DNA fragment of similar size from B. longum NCC2705 (GenBank accession number AE014741), which coded for the same hypothetical ORFs, including a putative α-l-arabinofuranosidase designated Abf2 (GenBank accession number AAN24971). The abfB gene and the protein translated from this gene exhibited 99.53% nucleotide identity and 99.29% amino acid sequence identity with the gene encoding Abf2 and with Abf2, respectively. A similar DNA cluster is also present in B. longum DJO10A (GenBank accession number NZ_AABF01000006).
FIG. 1.
(A) Organization of the B. longum B667 genomic region containing the abfB gene. reg is similar to the genes encoding several LacI repressors, β-gal resembles β-galactosidase genes, and sbp1 and sbp2 show significant homologies with the genes encoding sugar-binding proteins. The white arrows indicate the positions and directions of transcription of the ORFs. The pin-like symbols indicate predicted palindromes. Relevant restriction sites and their locations in the sequence are also indicated. (B) Modular structure of AbfB. Position 111 corresponds to the proline of the PGG conserved locus, and positions 215 and 340 correspond to the glutamic acid residues, which are predicted to be the acid-base and nucleophilic catalytic residues, respectively. The amino acid sequences in parentheses are the consensus sequences of family 51 glycoside hydrolases.
To date, the α-l-arabinofuranosidases have been assigned to two glycoside hydrolase families on the basis of substrate specificity (EC 3.2.1.55 and EC 3.2.1.99) and to five glycoside hydrolase families, families 3, 43, 51, 54 and 62, on the basis of sequence homology (CAZy classification; http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html). A comparison of the amino acid sequence of AbfB with the sequences of all members of family 51 enabled identification of three conserved loci. The conserved PGG locus (P111) (Fig. 1B) is probably related to structural features, since this motif has been suggested to confer additional flexibility to proteins (28). The α-l-arabinofuranosidase AbfA from Geobacillus stearothermophilus is the only member of this family for which structural and detailed mechanistic data are available (14). Based on sequence similarities with this enzyme, residues E215 and E340 are predicted to be the acid-base and nucleophilic catalytic residues necessary to carry out hydrolysis of the glycosidic bond (36, 37), respectively. These residues are conserved in most members of family 51 (Fig. 1B).
Overexpression of AbfB in L. lactis and purification of the enzyme.
The abfB gene was amplified by PCR, and the RcaI and XbaI sites were introduced at the 5′ and 3′ ends of the DNA fragment to allow cloning downstream of the nisin-inducible nisA promoter in pNZ8048, yielding pNAbfB. In order to facilitate purification of AbfB, a histidine tag was attached to the C terminus of the protein, yielding pNHAbfB. Addition of nisin A to L. lactis NZ9000 harboring pNAbfB or pNHAbfB resulted in synthesis of either the wild-type or recombinant protein. Cell extracts from the two types of L. lactis cells collected after nisin induction showed similar levels of activity against pNP α-l-Abf. This indicated that the histidine-tagged AbfB was functional and that addition of the affinity tag did not modify the activity of the protein. Control cells, harboring the empty vector pNZ8048, did not show any activity. To estimate the expression levels of AbfB, a densitometric scan of SDS-PAGE gels of total protein from L. lactis NZ9000/pNHAbfB was analyzed. The amount of AbfB was estimated to be about 10% of the total protein.
AbfB was purified from cell extracts of L. lactis NZ9000 harboring pNHAbfB by a single affinity chromatography step with a nickel chelate affinity resin (see Materials and Methods). Approximately 3.0 mg of AbfB was obtained from 1 liter of culture grown to an A600 of 1.0 to 1.2. The protein was more than 97% pure as determined by densitometric analysis (Fig. 2), and its absorption spectrum had very low absorbance in the range from 320 to 340 nm, indicating that aggregates of the protein were not formed during purification (data not shown).
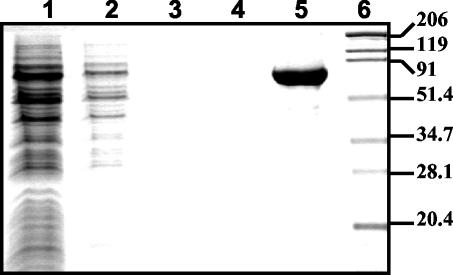
FIG. 2.
Purification of the histidine-tagged AbfB: Coomassie brilliant blue-stained SDS-PAGE gel showing intermediate steps in the purification procedure. Lane 1, total cell extract of L. lactis NZ9000/pNHAbfB; lane 2, flowthrough; lanes 3 and 4, protein eluted from the Ni-NTA resin during washing with 10 mM imidazole (lane 3) and 30 mM imidazole (lane 4); lane 5, purified protein eluted from the Ni-NTA column with 160 mM imidazole; lane 6, protein markers (molecular masses [in kilodaltons] are indicated on the right).
Characterization of the enzyme. (i) Molecular mass.
The molecular mass and the polymeric state of the histidine-tagged AbfB were estimated by permeation chromatography and SDS-PAGE. From the results of permeation chromatography, the molecular mass of the enzyme was estimated to be around 260 kDa, and from SDS-PAGE the molecular mass was estimated to be around 64 kDa. These results indicated that the native form of the enzyme is a polymeric structure, probably a homotetramer.
(ii) Effects of pH and temperature on activity and stability.
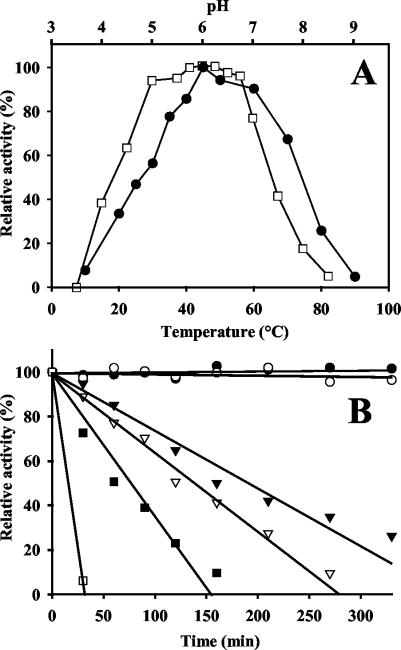
The purified enzyme was active at a broad pH range (pH 4.5 to 7.0), and the optimum pH was pH 6.0 in both McIlvaine buffer and potassium phosphate buffer (Fig. 3A). Considerable activity (about 40%) was still observed at pH 4.0 and 7.5. The optimal temperature for AbfB activity was 45°C. The enzymatic activity decreased significantly at temperatures below 35°C and above 60°C, although considerable levels of activity were still detected at 20 and 70°C.
FIG. 3.
(A) Effects of pH (□) and temperature (•) on AbfB activity as determined with pNP α-l-Abf as the substrate. The activities at the optimal pH and the optimal temperature were defined as 100%. (B) Stability of the protein at different temperatures. Residual AbfB activity was monitored after different times of incubation at 40°C (•), 50°C (○), 55°C (▾), 60°C (▿), 65°C (▪), and 70°C (□). The initial activity was defined as 100%.
The enzyme retained full activity at pH values from 5.5 to 7.5 (data not shown). After exposure to pH 4.5, the activity was reduced by 40%, and no residual activity was detected at pH values below 4.5 or above 7.5. The thermostability of AbfB fitted a first-order reaction. The enzyme exhibited full activity at 40 and 50°C and had a half-life of about 3 h at 55°C. The reduction in enzyme activity at high temperatures was not due to reduced stability during the incubation time (10 min) (Fig. 3B).
(iii) Effects of chemical agents and metal cations on the activity.
The effects of different metals on the activity of AbfB were determined by using pNP α-l-Abf as a substrate. No effect on activity was detected with Mg2+, Mn2+, Ca2+, Co2+, or Na+. Hg2+ and Cu2+ caused complete inhibition of activity, whereas 1 mM Zn2+ inhibited activity by 51%. Enzymatic activity was not affected by the chelating agent EDTA (10 mM) or by the reducing agents DTT and β-mercaptoethanol (10 mM).
(iv) Substrate specificity and kinetic analysis.
In a first experiment, the substrate specificity of the enzyme was tested at pH 6.0 and 45°C by using several pNP glycosides. AbfB was able to hydrolyze only pNP α-l-Abf, whereas no activity was detected with a variety of other pNP glycosides tested, including those with β-linkages and pyranoside conformations of arabinose (see Materials and Methods). The dependence of the rate of the enzymatic reaction on the pNP α-l-Abf concentration followed Michaelis-Menten kinetics, with apparent Km and Vmax values of 0.295 mM and 417 U/mg, respectively.
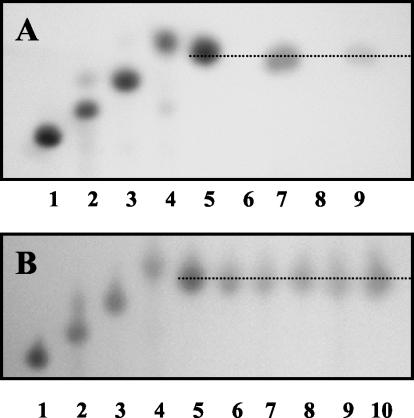
In order to ascertain whether AbfB could hydrolyze arabinose-containing oligosaccharides and polysaccharides and to define the mode of action of the enzyme (endo acting versus exo acting), hydrolysis of α-1,5-linked arabinooligosaccharides (containing from two to five arabinose residues) and some arabinose-containing polysaccharides was studied. The enzyme did not display activity against debranched linear sugar beet arabinan dyed with Procoin Red dye (RDA, containing 77% arabinose, 11% galactose, 5% rhamnose, and 7% galacturonic acid), indicating that there was no endoarabinanase activity. AbfB was active with sugar beet arabinan (containing 88% arabinose, 3% galactose, 2% rhamnose, and 7% galacturonic acid) and wheat arabinoxylan (containing 37% arabinose, 61% xylose, and 2% other sugars). It also exhibited activity against α-1,5-linked arabinobiose, arabinotriose, arabinotetraose, and arabinopentaose. The apparent Km values for degradation of arabinan and arabinoxylan were 51.19 and 21.42 mg/ml, respectively, and the Vmax values were 0.83 and 1.22 μmol of arabinose-reducing sugar equivalents min−1 mg−1, respectively. In all cases, only arabinose was detected as the final product, and no other residues were found by thin-layer chromatography (Fig. 4). These results indicated that AbfB is an exo-acting enzyme that hydrolyzes the nonreducing terminal l-arabinofuranose residues.
FIG. 4.
(A) Thin-layer chromatography of hydrolysis products obtained from sugar beet arabinan and wheat arabinoxylan. Lane 1, arabinopentaose; lane 2, arabinotetraose; lane 3, arabinotriose; lane 4; arabinobiose; lane 5, arabinose; lane 6, nontreated arabinan; lane 7, arabinan (1%, wt/vol) incubated with purified enzyme (0.1 U) for 16 h; lane 8, nontreated arabinoxylan; lane 9, arabinoxylan (1%, wt/vol) incubated with purified enzyme (0.1 U) for 24 h. (B) Thin-layer chromatography of hydrolysis products obtained from arabinooligosaccharides. Lane 1, arabinopentaose; lane 2, arabinotetraose; lane 3, arabinotriose; lane 4, arabinobiose; lane 5, arabinose; lanes 6, 7, 8, 9, and 10, arabinopentaose, arabinotetraose, arabinotriose, arabinobiose, and arabinose (50 μg each), respectively, after incubation with 0.1 U of enzyme for 16 h. All reactions were carried out in 1-ml tubes.
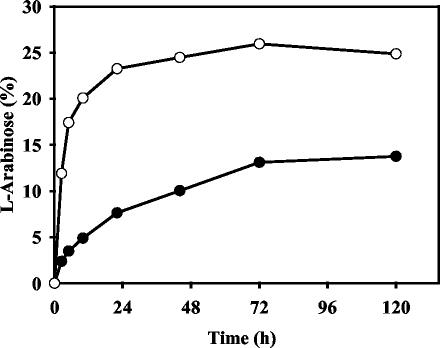
The time course of hydrolysis of arabinan and arabinoxylan is shown in Fig. 5. Under the conditions tested, AbfB poorly hydrolyzed these two water-soluble polysaccharides. Less than 25% of the total l-arabinose present in the arabinoxylan was released in a 72-h period, and less than 14% of the arabinose present in the arabinan was released after 5 days of incubation. These limits of hydrolysis were not significantly affected when there was an excess of the enzyme and/or when the reaction mixture was incubated for a longer time.
FIG. 5.
Time course of hydrolysis of sugar beet arabinan (•) and wheat arabinoxylan (○) by purified AbfB. A 0.1% (wt/vol) solution of polysaccharide was incubated at 45°C with 0.2 U of enzyme per ml. The amount of arabinose released is expressed as a percentage of the total amount of arabinose present in the polysaccharide.
DISCUSSION
Arabinose-containing polysaccharides have emerged in recent years as valuable prebiotics (health-promoting nondigestible food ingredients) (12), and their beneficial effects as bifidobacterium growth-promoting factors are being investigated (5, 7). Recently available data from the B. longum sequencing project revealed a large number of arabinosidases that are probably involved in the degradation of such polysaccharides. The arabinofuranosidase AbfB from B. longum identified in this study is the first exoarabinofuranosidase produced by a member of the genus Bifidobacterium genetically and biochemically characterized that exclusively releases nonreducing arabinose end residues from arabinan and arabinoxylan. Sequence analysis of abfB revealed significant similarities with genes coding for members of family 51 of the glycoside hydrolases. Furthermore, preliminary data obtained by our research group also revealed that more than 90% of Bifidobacterium strains tested have genes similar to abfB (data not shown), indicating that abfB homologues are widespread in this genus. Based on homology analysis of the amino acid sequences, glutamic acid residues at locations 215 and 340 were identified as putative acid-base and nucleophilic amino acids, respectively (36, 37). These residues, which are responsible for cleavage of the glycosidic bond, are conserved in all members of the family except AbjA (GenBank accession number Y16849) (6), an α-l-arabinofuranosidase from Thermobacillus xylanolyticus, in which the putative nucleophilic acid was not clearly located. These data strongly support the conclusion that AbfB should be included in family 51 of the glycoside hydrolases.
In order to elucidate the function of AbfB, we developed an efficient method for overexpression and purification almost to homogeneity of the histidine-tagged protein. The nisin-controlled gene expression system, based on the L. lactis nisA promoter (10, 19), was successfully used in this study to obtain high levels of expression of AbfB. The nisin-controlled gene expression system has been used recently to overexpress several homologous and heterologous proteins in L. lactis (20, 25). However, to the best of our knowledge, this is the first report of a Bifidobacterium protein produced in L. lactis that highlights the capacity of the host to express proteins encoded by genes having rather high G+C contents.
The purified native enzyme had a multimeric conformation; it was probably a tetramer. The multimeric quaternary structures of several arabinofuranosidases have been reported previously (9, 32). The optimal temperature for AbfB activity (45°C) was higher than the optimal temperatures for the arabinosidases AXH-d3 (30°C) and AXH-m23 (37°C) from Bifidobacterium adolescentis (43, 44) but lower than the optimal temperatures for most arabinofuranosidases from other bacteria and fungi (8, 9, 11, 16, 31, 39). The optimal pH of AbfB (pH 6.0) fell in the same range as the optimal pHs found for AXH-d3 and AXH-m2 (43, 44). Slightly acid pHs, between pH 5.5 and 7.0, have also been reported to be optimal for most bacterial arabinosidases, although fungal enzymes had lower optimal pHs (around pH 4.0) (32). The enzyme showed great stability during prolonged incubation at pHs ranging from 5.5 to 7.5 and at temperatures lower than 50°C, which are conditions found in the large bowel. This suggests that B. longum is effectively involved in polysaccharide assimilation in this environment. The activity of AbfB was completely inhibited by Hg2+ and Cu2+ and was partially inhibited by Zn2+; these are known inhibitors of other enzymes of this family (8, 14, 17, 39, 47). Hg2+ is known to react with sulfhydryl groups of proteins, as well as with histidine and tryptophan residues. As addition of the chelating agent EDTA and the reducing agents DTT and β-mercaptoethanol did not affect enzymatic activity, the enzyme does not seem to need any metal ion or disulfide bonds to carry out the hydrolysis reaction.
With respect to substrate specificity, AbfB was not able to hydrolyze pNP glycosides other than pNP α-l-Abf. The purified enzyme exhibited activity against sugar beet arabinan, wheat arabinoxylan, and α-1,5-linked oligosaccharides, yielding arabinose as the sole hydrolysis end product. However, AbfB was not able to hydrolyze RDA, a substrate that is cleaved only by endo-1,5-arabinanase activity (23), and dye molecules attached to the arabinose residues prevent the release of arabinosyl residues from the nonreducing end. On the other hand, single arabinose residues are predominantly linked to the main chain in sugar beet arabinan and wheat arabinoxylan through α-1,3 linkages, although α-1,2 arabinose bonds can also be found in this type of arabinoxylan (16, 32, 44). Therefore, we concluded from these results that AbfB of B. longum is an exo-acting enzyme that exhibites hydrolytic activity against α-1,3- and α-1,5-linked nonreducing terminal l-arabinofuranose residues but not against internal α-l-arabinosyl linkages. Nevertheless, the possibility of hydrolytic activity against α-1,2 linkages cannot be eliminated on the basis of our data. The substrate specificity of our enzyme was similar to that of α-l-arabinofuranosidases from Streptomyces diastaticus (38) and Aerobasidium pullulans (31). However, the affinity of AbfB toward arabinan and arabinoxylan was rather low, and the enzyme acted slowly on these substrates, releasing arabinose only after prolonged incubation. The limits of hydrolysis of arabinan and arabynoxylan were also quite low (13.75 and 24.85%, respectively), and most of the polysaccharide remained intact. In this respect, it has been reported that extension of side chains and the frequency of substitution on the backbone can influence the affinity of arabinofuranosidases towards polysaccharides (8).
Several arabinosidases from different microorganisms, including yeast (47), fungi (11, 13, 16, 31, 46), and bacteria (8, 9, 14, 17, 21, 24, 34, 39, 43, 44), have been documented and characterized to date. Among the bifidobacteria, two different arabinosidases from B. adolescentis, AXH-d3 and AXH-m23, have been partially characterized (43, 44). The genes encoding these enzymes have not been identified yet, but biochemical data have indicated that both enzymes are active against arabinoxylan-derived oligosaccharides and that AXH-d3 is also active against arabinoxylan polymers. However, neither of these enzymes was active against pNP α-l-Abf or arabinans. Therefore, since AbfB hydrolyzed pNP α-l-Abf and arabinan, a mode of action and substrate specificity different than those previously described for arabinofuranosidases in Bifidobacterium strains could be attributed to this enzyme. The activity of AbfB against polysaccharides derived from natural plant cell walls suggests that this enzyme may play an important role in the degradation of hemicellulose in the diet by B. longum in the large intestine. Results obtained in this study could represent a step forward in unraveling the complex systems that allow B. longum to survive and colonize the human intestinal tract.
Acknowledgments
This work was financially supported by a Marie Curie fellowship (category R) granted to Abelardo Margolles by the European Community, by the Ministerio de Ciencia y Tecnología of Spain (grant AGL2001-2296), and by European Union FEDER funding.
We gratefully thank NIZO Food Research for providing L. lactis strains NZ9000 and NZ9700 and plasmid pNZ8048. We are also indebted to Ana Hernández Barranco for her assistance with gel filtration chromatography.
REFERENCES
- 1.Aristidou, A., and M. Penttila. 2000. Metabolic engineering applications to renewable resource utilization. Curr. Opin. Biotechnol. 11:187-198. [DOI] [PubMed] [Google Scholar]
- 2.Austin, E. A., J. F. Graves, L. A. Hite, C. T. Parker, and C. A. Schnaitman. 1990. Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli K-12: insertion mutagenesis of the rfa locus. J. Bacteriol. 172:5312-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basic, A., P. J. Harris, and B. A. Stone. 1988. Structure and function of plant cell walls, p. 297-371. In J. Preiss (ed.), The biochemistry of plants, vol. 14. Academic Press, San Diego, Calif.
- 4.Buist, G., J. Kok, K. J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haandrikman. 1995. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J. Bacteriol. 177:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charalampopoulos, D., R. Wang, S. S. Pandiella, and C. Webb. 2002. Application of cereals and cereal components in functional foods: a review. Int. J. Food Microbiol. 79:131-141. [DOI] [PubMed] [Google Scholar]
- 6.Connerton, I., N. Cummings, G. W. Harris, P. Debeire, and C. Breton. 1999. A single domain thermophilic xylanase can bind insoluble xylan: evidence for surface aromatic clusters. Biochim. Biophys. Acta 1433:110-121. [DOI] [PubMed] [Google Scholar]
- 7.Crittenden, R., S. Karppinen, S. Ojanen, M. Tenkanen, R. Fagerström, J. Mättö, M. Saarela, T. Mattila-Sandholm, and K. Poutanen. 2002. In vitro fermentation of cereal dietary fibre carbohydrates by probiotic and intestinal bacteria. J. Sci. Food Agric. 82:781-789. [Google Scholar]
- 8.Debeche, T., N. Cummings, I. Connerton, P. Debeire, and M. J. O'Donohue. 2000. Genetic and biochemical characterization of a highly thermostable α-l-arabinofuranosidase from Thermobacillus xylanilyticus. Appl. Environ. Microbiol. 66:1734-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degrassi, G., A. Vindigni, and V. Venturi. 2003. A thermostable α-arabinofuranosidase from xylanolytic Bacillus pumilus: purification and characterization. J. Biotechnol. 101:69-79. [DOI] [PubMed] [Google Scholar]
- 10.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filho, E. X., J. Puls, and M. P. Coughlan. 1996. Purification and characterization of two arabinofuranosidases from solid-state cultures of the fungus Penicillium capsulatum. Appl. Environ. Microbiol. 62:168-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 13.Gielkens, M., L. González-Candelas, P. Sánchez-Torres, P. van de Vondervoort, L. de Graaff, J. Visser, and D. Ramón. 1999. The abfB gene encoding the major α-l-arabinofuranosidase of Aspergillus nidulans: nucleotide sequence, regulation and construction of a disrupted strain. Microbiology 145:735-741. [DOI] [PubMed] [Google Scholar]
- 14.Gilead, S., and Y. Shoham. 1995. Purification and characterization of α-l-arabinofuranosidase from Bacillus stearothermophilus T-6. Appl. Environ. Microbiol. 61:170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isolauri, E. 2001. Probiotics in human disease. Am. J. Clin. Nutr. 73:1142S-1146S. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko, S., T. Shimasaki, and I. Kusakabe. 1993. Purification and some properties of intracellular α-l-arabinofuranosidase from Aspergillus niger 5-16. Biosci. Biotechnol. Biochem. 57:1161-1165. [DOI] [PubMed] [Google Scholar]
- 17.Kosugi, A., K. Murashima, and R. H. Doy. 2002. Characterization of two noncellulosomal subunits, ArfA and BgaA, from Clostridium cellulovorans that cooperate with the cellulosome in plant cell wall degradation. J. Bacteriol. 184:6859-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 19.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 20.Margolles, A., M. Putman, H. W. van Veen, and W. N. Konings. 1999. The purified and functionally reconstituted multidrug transporter LmrA of Lactococcus lactis mediates the transbilayer movement of specific fluorescent phospholipids. Biochemistry 38:16298-16306. [DOI] [PubMed] [Google Scholar]
- 21.Matsuo, N., S. Kaneko, A. Kuno, H. Kobayashi, and I. Kusakabe. 2000. Purification, characterization and gene cloning of two α-l-arabinofuranosidases from Streptomyces chartreusis GS901. Biochem. J. 346:9-15. [PMC free article] [PubMed] [Google Scholar]
- 22.McCleary, B. V. 1981. Galactomannan quantitation in guar varieties and seed fractions. Lebensm.-Wiss. Technol. 14:188-191. [Google Scholar]
- 23.McCleary, B. V. 1988. Novel and selective substrates for the assay of endo-arabinanase, p. 291-300. In G. O. Phillips, D. J. Wedlock, and P. A. Williams (ed.), Gums and stabilisers for the food industry, vol. 5. IRL Press, Oxford, United Kingdom.
- 24.McKie, V. A., G. W. Black, S. J. Millward-Sadler, G. P. Hazlewood, J. I. Laurie, and H. J. Gilbert. 1997. Arabinanase A from Pseudomonas fluorescens subsp. cellulosa exhibits both an endo- and an exo-mode of action. Biochem. J. 323:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyoshi, A., I. Poquet, V. Azevedo, J. Commissaire, L. Bermudez-Humaran, E. Domakova, Y. le Loir, S. C. Oliveira, A. Gruss, and P. Langella. 2002. Controlled production of stable heterologous proteins in Lactococcus lactis. Appl. Environ. Microbiol. 68:3141-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller-Hill, B. 1998. Some repressors of bacterial transcription. Curr. Opin. Microbiol. 1:145-151. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 28.Pazos, F., P. Heredia, A. Valencia, and J. de las Rivas. 2001. Threading structural model of the manganese-stabilizing protein PsbO reveals presence of two possible beta-sandwich domains. Proteins 45:372-381. [DOI] [PubMed] [Google Scholar]
- 29.Pitson, S. M., A. G. Voragen, and G. Beldman. 1996. Stereochemical course of hydrolysis catalyzed by arabinofuranosyl hydrolases. FEBS Lett. 398:7-11. [DOI] [PubMed] [Google Scholar]
- 30.Puls, J., and J. Schuseil. 1993. Chemistry of hemicelluloses: relationship between hemicellulose structure and enzymes required for hydrolysis, p. 1-27. In M. P. Coughlan and J. P. Hazlewood (ed.), Hemicellulose and hemicellulase. Portland, London, United Kingdom.
- 31.Saha, B. C., and R. J. Bothast. 1998. Purification and characterization of a novel thermostable α-l-arabinofuranosidase from a color-variant strain of Aureobasidium pullulans. Appl. Environ. Microbiol. 64:216-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha, B. C. 2000. α-l-Arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol. Adv. 18:403-423. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sa-Nogueira, I., T. V. Nogueira, S. Soares, and H. de Lencastre. 1997. The Bacillus subtilis l-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology 143:957-969. [DOI] [PubMed] [Google Scholar]
- 35.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shallom, D., V. Belakhov, D. Solomon, G. Shoham, T. Baasov, and Y. Shoham. 2002. Detailed kinetic analysis and identification of the nucleophile in α-l-arabinofuranosidase from Geobacillus stearothermophilus T-6, a family 51 glycoside hydrolase. J. Biol. Chem. 277:43667-43673. [DOI] [PubMed] [Google Scholar]
- 37.Shallom, D., V. Belakhov, D. Solomon, S. Gilead-Gropper, T. Baasov, G. Shoham, and Y. Shoham. 2002. The identification of the acid-base catalyst of α-arabinofuranosidase from Geobacillus stearothermophilus T-6, a family 51 glycoside hydrolase. FEBS Lett. 514:163-167. [DOI] [PubMed] [Google Scholar]
- 38.Tajana, E., A. Fiechter, and W. Zimmermann. 1992. Purification and characterization of two α-l-arabinofuranosidases from Streptomyces diastaticus. Appl. Environ. Microbiol. 58:1447-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takao, M., K. Akiyama, and T. Sakai. 2002. Purification and characterization of thermostable endo-1,5-α-l-arabinase from a strain of Bacillus thermodenitrificans. Appl. Environ. Microbiol. 68:1639-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka, H., H. Hashiba, J. Kok, and I. Mierau. 2000. Bile salt hydrolase of Bifidobacterium longum—biochemical and genetic characterization. Appl. Environ. Microbiol. 66:2502-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tannock, G. W. 1999. A fresh look at the intestinal microflora, p. 5-14. In G. W. Tannock (ed.), Probiotics: a critical review. Horizon Scientific Press, Wymondham, United Kingdom.
- 42.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Laere, K. M., G. Beldman, and A. G. Voragen. 1997. A new arabinofuranohydrolase from Bifidobacterium adolescentis able to remove arabinosyl residues from double-substituted xylose units in arabinoxylan. Appl. Microbiol. Biotechnol. 47:231-235. [DOI] [PubMed] [Google Scholar]
- 44.van Laere, K. M., C. H. L. Voragen, T. Kroef, L. A. M. van den Broek, G. Beldman, and A. G. Voragen. 1999. Purification and mode of action of two different arabinoxylan arabinofuranohydrolases from Bifidobacterium adolescentis DSM 20083. Appl. Microbiol. Biotechnol. 51:606-613. [Google Scholar]
- 45.van Laere, K. M., R. Hartemink, M. Bosveld, H. A. Schols, and A. G. Voragen. 2000. Fermentation of plant cell wall derived polysaccharides and their corresponding oligosaccharides by intestinal bacteria. J. Agric. Food. Chem. 48:1644-1652. [DOI] [PubMed] [Google Scholar]
- 46.Wood, T. M., and S. I. McCrae. 1996. Arabinoxylan degrading enzyme system of the fungus Aspergillus awamori: purification and properties of an α-l-arabinofuranosidase. Appl. Microbiol. Biotechnol. 45:538-545. [DOI] [PubMed] [Google Scholar]
- 47.Yanai, T., and M. Sato. 2000. Purification and characterization of a novel α-l-arabinofuranosidase from Pichia capsulata X91. Biosci. Biotechnol. Biochem. 64:1181-1188. [DOI] [PubMed] [Google Scholar]
- 48.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]